Published online Oct 15, 1998. doi: 10.3748/wjg.v4.i5.385
Revised: September 16, 1998
Accepted: October 2, 1998
Published online: October 15, 1998
AIM: To study whether Helicobacter pylori is naturally transformable.
METHODS: Transformation was performed in BHI broth supplemented with horse serum and yeast extract. Genomic DNA extracted from a metronidazole resistant H.pylori strain was added to H. pylori broth culture. The mixture was incubated at microaerophilic atmosphere. The DNA-treated cells were plated on blood agar containing 8 mg/L metronidazole to select for transformants. Sterile distilled water was used as a negative DNA control. The DNA profiles of transformants were compared with that of their parent strains by randomly amplified polymorphic DNA (RAPD) fingerprinting.
RESULTS: Transformation of H. pylori with DNA from a metronidazole resistant strain as a marker was demonstrated. Out of the 12 strains of H. pylori tested, 9 (75%) strains were found to be transformable. The transformation frequencies ranged from 3.4 × 10-6 to 2.4 × 10-4. By RAPD, DNA fingerprints of the transformants and their parent strains showed no change in DNA profiles though transformants were all resistant to metronidazole as compared with their metronidazole-sensitive parent strains.
CONCLUSION: Helicobacter pylori is naturally transformable which might be one of the ways that H. pylori develops resistance to metronidazole.
- Citation: Hua JS, Zheng PY, Teo KF, Khin MM, Ho B. Helicobacter pylori acquistion of metronidazole resistance by natural transformation in vitro. World J Gastroenterol 1998; 4(5): 385-387
- URL: https://www.wjgnet.com/1007-9327/full/v4/i5/385.htm
- DOI: https://dx.doi.org/10.3748/wjg.v4.i5.385
Genetic transformation is a process by which a cell takes up naked DNA from the surrounding medium and incorporates it into its own genomic DNA to acquire an altered genotype. Natural transformation is widely distributed among bacteria. This process may enable bacteria to get advantageous mutations to escape and survive under unfavourable conditions. Helicobacter pylori is reconized as a major factor in the development of gastritis and peptic ulcer[1]. This bacterium can be eradicated from stomach by antibiotics. However, effective treatment of H. pylori has proved difficult with the development of resistance to some antimicrobials. An increase in prevalence of metronidazole resistant H. pylori has been reported[2]. In this study we test in vitro whether natural transformation could be one way for H. pylori to acquire metronidazole resistance.
H. pylori isolates obtained from patients with gastroduodenal diseases were used in this study. The strain was isolated on chocolate blood agar No. 2 medium supplemented with 5% horse blood and incubated at 37 °C under microaerophilic environment. The strain was identified by standard procedures as stated by Goodwin et al[3].
Plate culture of H. pylori was transferred into an Eppendorf tube and 1.5 mL volume of TE buffer (100 mM Tris-HCL and 1 mM EDTA) was added. The suspension was centrifuged at 8000 ×g and washed once with TE buffer. The pellet was suspended in 800 μL TE buffer. The bacterial suspension was incubated in 100 μL of 10 g/L lysozyme (Sigma) at 37 °C for 30 min, and then lysed with 100 μL of 10% sodium deodecyl sulfate for another 30 min at 37 °C. Following the addition of 5 μL of 10 g/L proteinase K (Boehringer), the mixture was incubated for 1 h at 56 °C. DNA was purified by extracting twice with equal volume of phenol and once with equal volume of chloroform, DNA was then precipitated overnight with two volumes of absolute ethanol and 20 μL 3M sodium acetate at -20 °C. The DNA precipitate was washed once with 70% ethanol. The pellet was vacumm-dried using speed-vac (Savant) ant resuspended in 200 µL sterile distilled water. This served as target DNA for PCR-based RAPD. DNA concentration was measured at λ 260 nm.
Universal primer for PCR-based RAPD was randomly chosen according to Akopyanz et al[4] (1992) to allow for the fingerprinting of the whole DNA content of cells. The primer used in this study was 5-AAGAGCCCGT-3. PCR reaction was carried out in 25µl volume. Fifty ng of H. pylori genomic DNA, 2 mM MgCl2, 20 pmol primer, 1 unit of Taq DNA polymerase and 250 mM each of dGTP, dCTP, dATP and dTTP were placed in standard PCR incubation buffer containing 10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin (Promega, USA). The reaction mixture was overlaid with a drop of mineral oil to prevent evaporation. PCR was performed with a thermal cycler (Amplitron, USA) consisting of an initial step of denaturation of target DNA at 94 °C for 5 min. This was followed by 39 cycles of denaturation at 94 °C for 1 min, annealing at 36 °C for 1 min and extension at 72 °C for 2 min. The microliters of the PCR products were electrophoresed in 1% horizontal agarose gels for 2 h at 80 V in TBE buffer. The gels were stained with ethidium bromide (1 mg/L) and photographed with filtered UV illumination on Polaroid type 667 film.
H. pylori transformation was performed in BHI broth supplemented with horse serum and yeast extract. Briefly, DNA was extracted from a metronidazole-resistant H. pylori strain, H38. Twelve metronidazole-sensitive strains of H. pylori grown respectively in Brain-heart infusion broth supplemented with 10% horse serum and 0.4% yeast extract were incubated under microaerobic conditions at 37 °C for 24 h. Aliquots of 50 µg DNA of H 38 were added into 1mL test H. pylori broth culture. The mixture was incubated at 37 °C for 6 h. The DNA-treated cells were plated on blood agar containing 8 mg/L metronidazole to select transformants. The transformation frequencies were calculated by dividing the number of transformants from the tatal number of viable cells on chocolate blood agar. Sterile distilled water was used as a negative DNA control. The DNA profiles of transformants were compared with that of their parent strains by RAPD fingerprinting.
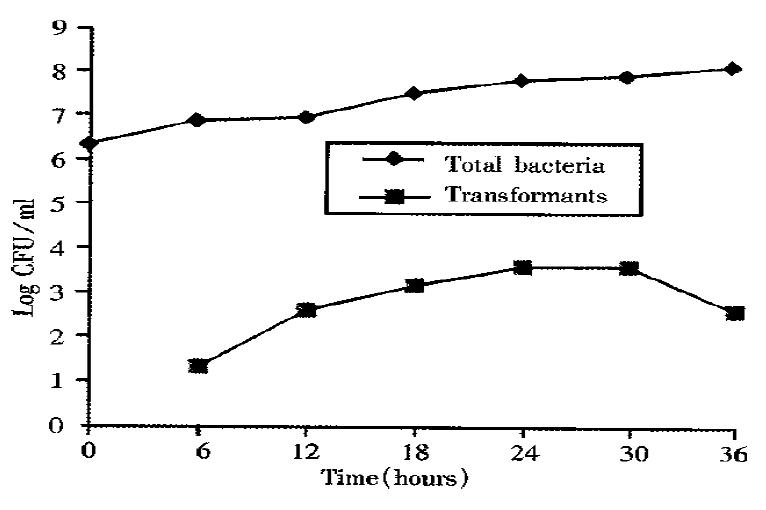
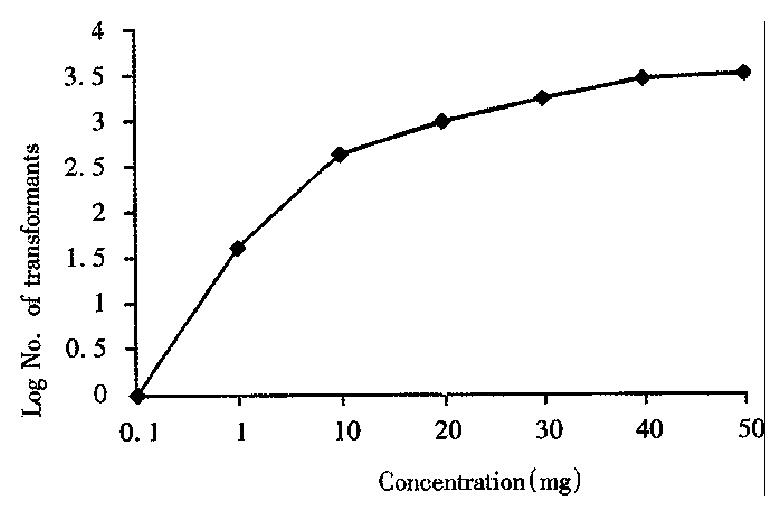
Transformation of H. pylori using metronidazole resistance as a marker was demonstrated. H. pylori H38 DNA (metronidazole resistance) was used as a donor to test for natural transformation competence in broth cultures. To optimize conditions for transformation of H. pylori, DNA of H38 was added to NCTC 11637 broth at 6-36 h after initial inoculation. Transformants were obtained at frequencies ranging from 2.8 × 10-6 to 5.9 × 10-5 (Figure 1 and Table 1). The highest number of transformants and frequency of transformation were found when DNA was added at 24 h. It was interesting to note that the transformation frequency of NCTC 11637 increased with increasing donor DNA concentration (Figure 2).
| 6 h | 12 h | 18 h | 24 h | 30 h | 36 h | |
| Total bacteria | 7.1 × 106 | 8.1 × 106 | 2.8 × 107 | 5.4 × 107 | 6.3 × 107 | 9.8 × 107 |
| Transformants | 20 | 370 | 1300 | 3200 | 3100 | 290 |
| Frequencies | 2.8 × 10-6 | 4.6 × 10-5 | 4.6 × 10-5 | 5.9 × 10-5 | 4.9 × 10-5 | 3.0 × 10-6 |
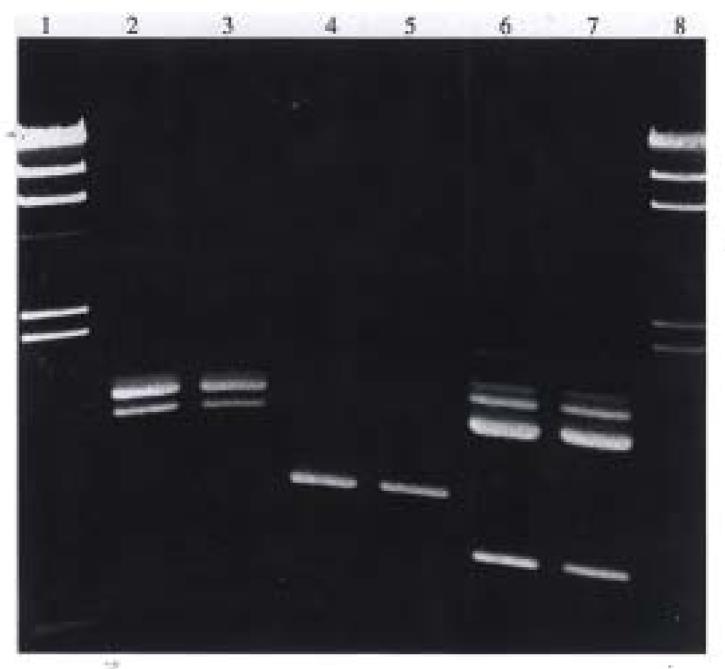
Using DNA of metronidazole resistant strain, H38, as a marker for transformation, 9 (75%) of 12 H. pylori strains tested were found to be transformable. The transformants were all resistant to metronidazole as compared with their metronidazole-sensitive parent cells. The transformation frequencies ranged from 3.4 × 10-6 to 2.4 × 10-4 (Table 2). By RAPD, the DNA fingerprints of the transformants and their parent strains showed nochange in DNA profiles (Figure 3).
| Recipient | Donor | Total bacteria | Transformants | Frequencies |
| H 1 | H 38 | 1.8 × 108 | 43000 | 2.4 × 104 |
| H 9 | 5.2 × 107 | 8900 | 1.7 × 104 | |
| H 11 | 9.3 × 108 | No | ||
| H 13 | 8.4 × 107 | 4100 | 4.9 × 105 | |
| H 29 | 5.6 × 106 | No | ||
| H 41 | 8.3 × 105 | 40 | 4.8 × 105 | |
| H 43 | 5.1 × 106 | 30 | 5.9 × 106 | |
| H 46 | 5.8 × 106 | 20 | 3.4 × 106 | |
| H 50 | 3.2 × 108 | 4500 | 1.4 × 105 | |
| H 53 | 3.5 × 107 | 320 | 9.1 × 106 | |
| H 62 | 3.9 × 105 | No | ||
| H 68 | 8.8 × 107 | 740 | 8.4 × 106 |
Natural transformation competence was found among prokaryotes, such as Streptococcus pneumoniae[5], Haemophilus influenzae[6] and Bacillus subtilis[7]. Stewart[8] reported that competence was internally regulated and was in a stable state once developed. Natural transformation in H. pylori has been demonstrated by Nedenskow-S-rensen et al[9] and Wang et al[10]. This study has confirmed that H. pylori can acquire resistance to metronidazole by natural transformation. Nine (75%) of 12 strains were found to be transformable in this study. The transformation frequencies ranged from 3.4 × 10-6 to 2.4 × 10-4. Furthermore, we examined the DNA fingerprints of recipient cell and its progeny by RAPD. DNA fingerprinting showed that no significant DNA profile change occurred. This is not unexpected. The size of metronidazole resistance gene may be insignificant compared with the entire chromosomal length. RAPD only can detect a small fraction of target DNA. It is possible that the universal primer used for RAPD in study could not recognise this slight difference especially if a point mutation is involved.
Ling et al[11] found that H. pylori strains resistance to metronidazole increased from 29% in 1991 to 73% in 1995 in Hong Kong. We believe that the densely populated environment in Hong Kong and the increased use of metronidazole and other imidazoles in the population had contributed to this phenomenon. This study shows a 75% transformable frequency in vitro. The results indicate that natural transformation of metronidazole resistance may play an important role in the development of antibiotic resistance. Natural transformation might promote H. pylori in acquiring advantageous genes from other strains in order to adapt and survive in some particular environments.
In this study natural transformation of H. pylori was demonstrated in vitro. It might be one of the means by which H. pylori develops resistance to metronidazole.
| 1. | Dick JD. Helicobacter (Campylobacter) pylori: a new twist to an old disease. Annu Rev Microbiol. 1990;44:249-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Glupczynski Y, Burette A, De Koster E, Nyst JF, Deltenre M, Cadranel S, Bourdeaux L, De Vos D. Metronidazole resistance in Helicobacter pylori. Lancet. 1990;335:976-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 100] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 3. | Goodwin CS, Armstrong JA. Microbiological aspects of Helicobacter pylori (Campylobacter pylori). Eur J Clin Microbiol Infect Dis. 1990;9:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 92] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 4. | Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137-5142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 564] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 5. | Morrison DA, Mannarelli B, Vijayakumar MN. Competence for transforma-tion in Streptococcus pneumoniae: an inducible high-capacity system for genetic exchane. In: Schlessinger D.ed Microbiology. Am Soc Microbiol: Wash-ington DC. 1982;136-138. |
| 6. | Albritton WL, Setlow JK, Thomas M, Sottnek F, Steigerwalt AG. Heterospecific transformation in the genus Haemophilus. Mol Gen Genet. 1984;193:358-363. [PubMed] |
| 7. | Harford N, Mergeay M. Interspecific transformation of rifampicin resistance in the genus Bacillus. Mol Gen Genet. 1973;120:151-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 7] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 8. | Stewart GJ, Carlson CA. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 132] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Nedenskov-Sørensen P, Bukholm G, Bøvre K. Natural competence for genetic transformation in Campylobacter pylori. J Infect Dis. 1990;161:365-366. [PubMed] |
| 10. | Wang Y, Roos KP, Taylor DE. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485-2493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 140] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | Ling TK, Cheng AF, Sung JJ, Yiu PY, Chung SS. An increase in Helicobacter pylori strains resistant to metronidazole: a five-year study. Helicobacter. 1996;1:57-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 2.7] [Reference Citation Analysis (0)] |