Published online Jun 15, 1998. doi: 10.3748/wjg.v4.i3.268
Revised: May 5, 1998
Accepted: May 20, 1998
Published online: June 15, 1998
- Citation: Liu Y, Zhang BH, Qian GX, Chen H, Wu MC. Detection of blood AFPmRNA in nude mice bearing human HCC using nested RT-PCR and its significance. World J Gastroenterol 1998; 4(3): 268-270
- URL: https://www.wjgnet.com/1007-9327/full/v4/i3/268.htm
- DOI: https://dx.doi.org/10.3748/wjg.v4.i3.268
Prognosis of patients with HCC is estimated by several factors, such as histological differentiation of tumor cells, tumor size, and extent of lymhatic or hemotogenous spread. In this study, we detected the AFPmRNA in the blood of nude mice bearing human hepatocellular cacinoma (HCC) using nested reverse transcriptase polymerase chain reaction (nested RT-PCR) and study its significance and role in blood spread and distant metastasis.
Animal SMMC-LINM cell lines which can secrete AFP (provided by the pathology department of our university[1]) were inoculated into the neck and back of nude mice (BALB/C/NU, 4 weeks after birth), (107 HCC cells to each mouse). After 6 weeks, 20 nude mice (15 g-20 g in weight) with tumors growing to 2 cm-4 cm were used in the experiment.
Collection of samples Both eyes of the nude mice bearing HCC were scooped out and blood (about 1 mL) was collected and placed into a 12 mL centrifuge tube. The subcutaneous tumors were resected, and their integrity and relationship with the surrounding tissues were observed. The liver, lung, kidneys and other oragans of the mice were cut and examined pathologically.
Methods One mL whole blood was collected from the peripheralvein of each subject into a centrifuge tube. AFPmRNA was detected with nested RT-PCR. The detailed procedures were as follows.
Detection of AFPmRNA Heparinized whole blood was centrifuged and the plasma fraction was removed. The cellular fraction was enriched for mononuclear cells or possible tumor cells according to the method by Komeda[2]. Total cellular RNA was extracted by a single-step method of RNA isolation[3]. The reverse transcription reaction was carried out in 20 μL reaction mixture using a first-strand cDNA synthesis kit (Promega USA) according to the manufacturer’s instructions. Nested PCR was conducted by addition of 5 μL solution of cDNA to 100 μL reaction mixture containing 10 mM Tris HCl (pH 9.0), 50 mM potassium chloride, 4.5 mM magnesium chloride, 250 nM dNTP 15 pmol of each outer primer (EX-sense and EX-antisense) and 2.5 units of Taq DNA polymerase (Promega, USA). The reaction mixtures were subjected to 35 cycles of amplification in a programmable thermal cycler (Perking-Elmer Cetus, USA) using the following sequence: 94 °C for 1.5 min, 57 °C for 1.5 min and 72 °C for 2.5 min, and a final extension step at 72 °C for 10 min. A sample of 10 μL of the first amplification product was further amplified using an inner pair of primers (IN-sense and IN-antisense). To verify the amplified AFP DNA fragment, the samples were digested with the restriction enzyme Pst I and analysed by electrophoresis on a 2% agrose gel and stained with ethidium bromide for the specific bands of 174 base pairs (first amplification product) and 101 base pairs (second amplification product). Nested PCR was performed two or three times for samples with conflicting results. The external and inner pair of primers were designed as follows:
EX-sense 5’-ACTGAATCCACAACACTGCATAG-3’
EX-antisense 5’-TGCAGTCAATGCATCTTCACCA-3’
IN-sense 5’-TGGAATAGCTTCCATATTGGATTC-3’
IN-antisense 5’-AAGTGGCTTCTTGAACAAACTGG-3’
According to this design, the PCR products of 176 and 101 base pairs were amplified from AFPcDNA by external (EX-sense and EX-antisense) and internal (IN-sense and IN-antisense) primer pairs, respectively. EX-sense was located in exon 1 (AFPmRNA nucleotides 90-112), EX-antisense in exon 2 (AFPmRNA nucleotides 241-263), IN-sense over exon 1 and exon 2 (AFPmRNA nucleotides 122-145) and IN-antisense in exon 3 (AFPmRNA nucleotides 200-222). cDNA sequences followed the method reported previously[4].
Statistical analysis The relationship between the AFPmRNA in peripheral blood and various clinical parameters was examined by Chi-square test.
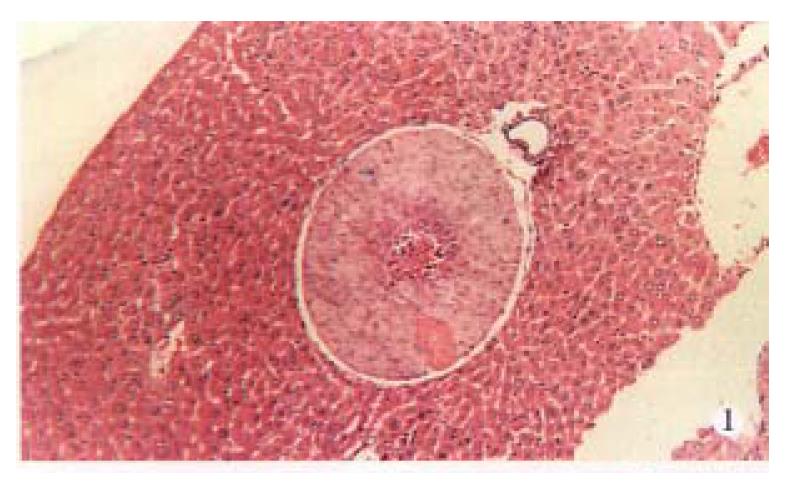
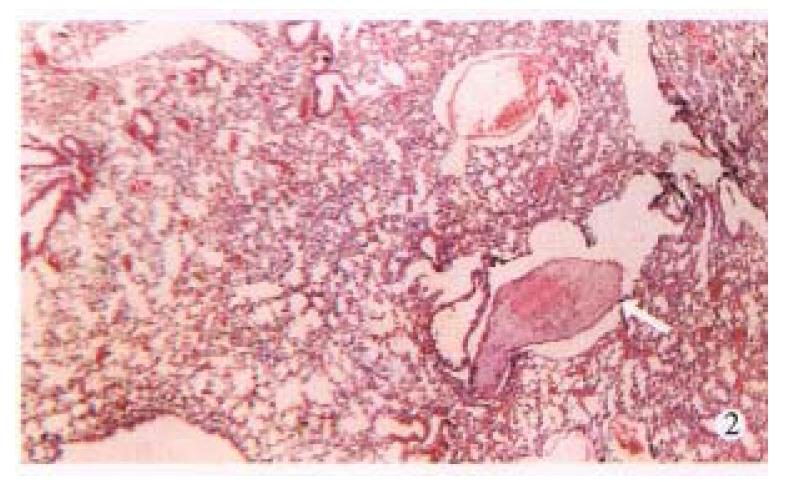
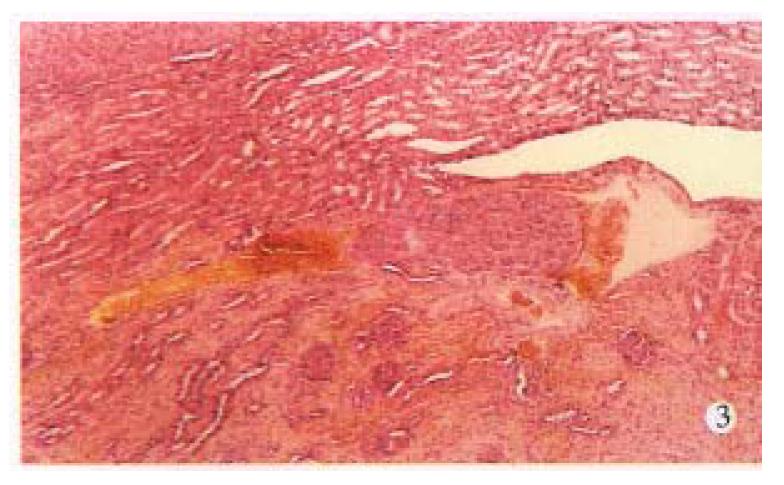
AFP mRNA was detected in the blood of 6 (30.0%) mice bearing HCC, 4 (66.67%) of 6 nude mice had distant metastasis in lungs, liver or kidneys (Figure 1, Figure 2, Figure 3, Table 1).
| Types | Cases | Cases of metastasis | Metastasis rate |
| AFPmRNA (+) | 6 | 4 | 66.67b |
| AFPmRNA (-) | 14 | 0 | 0.00 |
The diameters of HCC in 6 mice with positive AFP mRNA in blood were more than 3 cm, no distant metastasis occurred in tumors below 3 cm in diameter (Table 2).
| Diameter | Cases | Cases of AFPmRNA (+) | Positive rate |
| > 3 cm | 9 | 6 | 66.67b |
| < 3 cm | 11 | 0 | 0.00 |
AFPmRNA was detected among 12 (50%) nude mice bearing HCC with serum AFP levels beyond 4000 μg/L while no AFPmRNA was found in 8 nude mice with serum AFP levels below 4000 μg/L (Table 3).
| AFP levels (μg/L) | Cases | AFPmRNA (+) | Positive rate |
| > 4000 | 12 | 6 | 50.0b |
| < 4000 | 8 | 0 | 0.0 |
The first-cycle PCR products, 174 base pairs, can be cut into two pieces of 102 and 72 base pairs by restriction enzyme Pst I. The second-cycle PCR product, 101 base pairs, can be cut into two pieces of 60 and 41 base pairs (Figure 4).
Recently, there have been some reports about the employment of the reverse transcriptase-polymerse chain reaction (RT-PCR) technique to detect tumor cells spreading into the peripheral blood, bone marrow, and lymph nodes[5-7]. There trials have aimed to amplify tumor-specific gene transcripts which can not be detected in these tissues under normal conditions. Free mRNA is so fragile under conditions of abundant RNAse activity that the specific mRNA in blood can indicate the presence of intact cells producing such proteins just before the extraction of RNA.
This study showed that AFPmRNA was detected in 6 (30.0%) of 20 nude mice bearing human HCC 4 (66.67%) of them had distant metastasis. None of 14 nude mice with negative AFPmRNA in blood had distant metastasis (P < 0.01), suggesting that the distant metastasis occurred via blood circulation. The detectable rates of AFPmRNA was significantly related with AFP levels, tumor size and distant metastasis (P < 0.01). In other words, AFPmRNA in blood may be a prerequisite for distant metastasis of HCC.
In conclution, AFPmRNA may be an effective and sensitive marker for HCC matestasis in blood and distant metastasis as well.
Project supported by the Fund for Key Laboratories of PLA.
| 1. | Ji YY, Liu YF, Chen ZN. Radioimmunodetection and autoradiographic localization of monoclonal antibody against human hepatocellular carcinoma in xenografts. Cancer. 1992;69:2055-2059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Komeda T, Fukuda Y, Sando T, Kita R, Furukawa M, Nishida N, Amenomori M, Nakao K. Sensitive detection of circulating hepatocellular carcinoma cells in peripheral venous blood. Cancer. 1995;75:2214-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 3. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3020] [Cited by in RCA: 13179] [Article Influence: 346.8] [Reference Citation Analysis (0)] |
| 4. | Morinaga T, Sakai M, Wegmann TG, Tamaoki T. Primary structures of human alpha-fetoprotein and its mRNA. Proc Natl Acad Sci USA. 1983;80:4604-4608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 181] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 5. | Schoenfeld A, Luqmani Y, Smith D, O'Reilly S, Shousha S, Sinnett HD, Coombes RC. Detection of breast cancer micrometastases in axillary lymph nodes by using polymerase chain reaction. Cancer Res. 1994;54:2986-2990. [PubMed] |
| 6. | Ghossein RA, Scher HI, Gerald WL, Kelly WK, Curley T, Amsterdam A, Zhang ZF, Rosai J. Detection of circulating tumor cells in patients with localized and metastatic prostatic carcinoma: clinical implications. J Clin Oncol. 1995;13:1195-1200. [PubMed] |
| 7. | Pfleiderer C, Zoubek A, Gruber B, Kronberger M, Ambros PF, Lion T, Fink FM, Gadner H, Kovar H. Detection of tumour cells in peripheral blood and bone marrow from Ewing tumour patients by RT-PCR. Int J Cancer. 1995;64:135-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 2.2] [Reference Citation Analysis (0)] |