Published online Mar 21, 2024. doi: 10.3748/wjg.v30.i11.1572
Peer-review started: January 7, 2024
First decision: February 1, 2024
Revised: February 7, 2024
Accepted: March 6, 2024
Article in press: March 6, 2024
Published online: March 21, 2024
Processing time: 74 Days and 0 Hours
Fecal microbiota transplantation (FMT) is a promising therapeutic approach for treating Crohn’s disease (CD). The new method of FMT, based on the automatic washing process, was named as washed microbiota transplantation (WMT). Most existing studies have focused on observing the clinical phenomena. However, the mechanism of action of FMT for the effective management of CD-particularly in-depth multi-omics analysis involving the metagenome, metatranscriptome, and metabolome-has not yet been reported.
To assess the efficacy of WMT for CD and explore alterations in the microbiome and metabolome in response to WMT.
We conducted a prospective, open-label, single-center clinical study. Eleven CD patients underwent WMT. Their clinical responses (defined as a decrease in their CD Activity Index score of > 100 points) and their microbiome (metagenome, metatranscriptome) and metabolome profiles were evaluated three months after the procedure.
Seven of the 11 patients (63.6%) showed an optimal clinical response three months post-WMT. Gut microbiome diversity significantly increased after WMT, consistent with improved clinical symptoms. Comparison of the metagenome and metatranscriptome analyses revealed consistent alterations in certain strains, such as Faecalibacterium prausnitzii, Roseburia intestinalis, and Escherichia coli. In addition, metabolomics analyses demonstrated that CD patients had elevated levels of various amino acids before treatment compared to the donors. However, levels of vital amino acids that may be associated with disease progression (e.g., L-glutamic acid, gamma-glutamyl-leucine, and prolyl-glutamine) were reduced after WMT.
WMT demonstrated therapeutic efficacy in CD treatment, likely due to the effective reconstruction of the patient’s microbiome. Multi-omics techniques can effectively help decipher the potential mechanisms of WMT in treating CD.
Core Tip: Fecal microbiota transplantation (FMT) is a promising therapeutic approach for treating Crohn’s disease (CD). The new method of FMT, based on the automatic washing process, was named as washed microbiota transplantation (WMT). However, most existing studies have focused on observing clinical phenomena. In the present study, we found that the efficacy of WMT in CD may be due to the effective remodeling of the intestinal micro-ecological balance of patients through the combined in-depth analysis of metagenomic, metatranscriptomic, and metabolomic technologies. Multi-omics techniques can effectively help decipher the underlying mechanism of WMT for CD.
- Citation: Chen SJ, Zhang DY, Wu X, Zhang FM, Cui BT, Huang YH, Zhang ZL, Wang R, Bai FH. Washed microbiota transplantation for Crohn’s disease: A metagenomic, metatranscriptomic, and metabolomic-based study. World J Gastroenterol 2024; 30(11): 1572-1587
- URL: https://www.wjgnet.com/1007-9327/full/v30/i11/1572.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i11.1572
The global incidence of Crohn’s disease (CD), a type of inflammatory bowel disease (IBD), has been steadily rising over recent years. The prevalence rate has increased from 79.5 cases per 100000 individuals in 1990 to 84.3 per 100000 in 2017[1]. This increase poses a significant risk to human health. The pathogenesis of CD is still not completely understood. However, it is widely believed that the onset of CD is influenced by genetic, environmental, and gut microbiota dysbiotic factors, especially the abnormal activation of the gastrointestinal immune system by disorders involving the gut microbiota, which have emerged as a research hotspot in recent years. CD patients have distinct differences in their gut microbiota compared to healthy individuals, including a general decrease in species diversity and abundance of certain commensal and beneficial bacteria (e.g., Firmicutes and Bacteroides spp.), accompanied by increased Proteobacteria. The pathotype adherent-invasive Escherichia coli (E. coli), in the phylum Aspergillus, has been identified as an important risk factor for CD[2]. It potentially contributes to inflammation by producing α-hemolysin, which can lead to the destruction of the intestinal mucosa[3]. On the other hand, Ruminococcus gnavus may be involved in CD development by producing a glucorhamnan polysaccharide that acts on toll-like receptor 4 to induce an inflammatory response[4].
The utilization of fecal microbiota transplantation (FMT) has shown potential as an effective treatment option for CD. The new method of FMT, based on the automatic washing process, was named washed microbiota transplantation (WMT)[5] in the consensus statement from the FMT-standardization Study Group in 2019[6]. Washed preparation of fecal microbiota improves transplantation-related safety, quantitative methods, and delivery of the microbiota suspension[5,7]. WMT has been used to effectively re-establish intestinal microecology and holds promise for treating CD. Interestingly, in comparison to manual FMT, WMT has significantly enhanced the safety of CD treatment, as evidenced by a decrease in adverse events from 21.7% (15/69) to 4.0% (35/882)[7]. Clinical responses to WMT in IBD treatment are primarily associated with successful Akkermansia fixation, and F. prausnitzii has been found to have a strong symbiotic relationship with it[8]. In addition, when the optimal timing of the second FMT was sought, it was determined that 125 d was the median time needed to maintain a clinical response to the first FMT[9].
A recent meta-analysis comprising 12 studies on 228 CD patients who underwent FMT revealed that 57% of patients achieved clinical remission between 2 and 4 wk after treatment[10]. However, most published studies are currently limited to observations of clinical phenomena, as the mechanisms of action by which FMT is effective in treating CD have not yet been fully identified, particularly regarding more extensive changes in the microbiome and metabolome. This identification is critical for expanding the high-quality evidence base for FMT in CD treatment.
In the first decade of microbiome research, microbial composition and genome structure were mainly elucidated using DNA sequencing techniques such as 16S rDNA and metagenomic sequencing[11]. Recently, researchers have employed transcriptomic sequencing to analyze bacterial communities to explore dynamic microbiome and gene expression. This approach can elucidate variations in microbiome function in specific environments and interactions with the host. It can also help to understand microbiome configurations across the transition from healthy to diseased states[11]. However, neither changes in the active gut microbiota (from metatranscriptomics) nor changes in fecal metabolites have been reported in CD patients treated with FMT. Therefore, this study aimed to reveal the multi-omics mechanism of WMT for the effective treatment of CD by evaluating and analyzing the metagenomic (DNA), metatranscriptomic (RNA), and metabolomic aspects of stool specimens before and after CD patients received WMT.
The study was a prospective, open-label, and single-center clinical trial conducted at the Second Affiliated Hospital of Nanjing Medical University. All the participants willingly agreed to participate in the research and provided written informed consent. The institution’s Institutional Review Board approved the study protocol (NCT01793831). The study protocol complied with the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the Second Affiliated Hospital of Nanjing Medical University (2022-KY-161-01).
Adult CD patients with a CD Activity Index (CDAI) score ≥ 150 were recruited from the Second Affiliated Hospital of Nanjing Medical University between March 2022 and March 2023. Exclusion criteria were applied as follows: (1) Patients who had used antibiotics, gastric acid suppressants (e.g., proton pump inhibitors), probiotics, and other medications that may interfere with the gut microbiota in the three months prior to the recruitment; (2) patients requiring antibiotics during hospitalization; (3) patients who had changed their medication in the four weeks prior to recruitment; (4) patients who had comorbidities, such as infectious diseases identified by examination that included (but were not limited to) bacterial infections [including Clostridioides difficile (C. difficile)], fungal infections, and viral infections; (5) pregnant women, or women of childbearing age planning to conceive; (6) presence of contraindications to gastrointestinal endoscopy or anesthesia; and (7) any other conditions deemed by the investigator to be unsuitable for inclusion in the study.
Montreal classification[12,13] was used to count the CD patients included in the study. The Montreal classification is based on the age at diagnosis (A1: < 16, A2: 17-40, A3: > 40), disease location (L1: Ileal, L2: Colonic, L3: Ileocolonic, L4: Upper gastrointestinal), and the disease behavior (B1: Non-stricturing/non-penetrating, B2: Stricturing, B3:penetrating, P: Perianal disease modifier). All the patients were assessed for CDAI and Harvey-Bradshaw Index (HBI) scores as well as clinically relevant indices (including general health, stool frequency, level of abdominal pain, presence of complications related to CD, abdominal masses, and Simple Endoscopy Score (SES) CD score by colonoscopy) before the WMT. Patients were also evaluated-three months post-WMT-for their CDAI and HBI scores and any modifications in their treatment medications and hospitalization. Patients were consistently monitored for any adverse events (AE) from when they underwent WMT until three months post-treatment. Stool samples were collected one week before and three months after WMT treatment for metagenomic, metatranscriptomic sequencing, and metabolomic testing.
WMT donors were selected from the universal stool bank (China Microbiota Transplantation Systemm, CMTS). The donor screening criteria have been described in detail in the Nanjing Consensus[6]. The donors in the present study were six individuals (four males and two females) between the ages of 18 and 25. No donors were related to the participating patients.
The steps for the preparation of washed microbiota suspensions were as follows: (1) An automatic purification system (GenFMTer, FMT Medical, Nanjing, China) was used to enrich the microbiota; (2) the fecal microbiota suspension was transferred to tubes for centrifugation and the supernatant was discarded (this step was repeated three times using sterile saline to make the suspension); and (3) the volume ratio of final precipitation/vector solution was 1:2 for making fresh suspensions[6]. The washed preparation of fecal microbiota changes the transplantation-related safety, quantitative method, and delivery[7]. Moreover, the “one-hour FMT protocol” was used to ensure that the interval between defecation and infusion of the fresh bacteria into the intestinal tract of patients was less than one hour[14]. Washed microbiota suspensions (1U) were delivered through either colonic or mid-gut transendoscopic enteral tubing (TET) once daily for two days (methods for colonic[15] and mid-gut[16] TET have been reported in detail in previous studies). Colonic TET offers a new and convenient method of delivering WMT that is painless and easily reproducible, resulting in high patient satisfaction[14].
The primary outcome was the clinical response (a decrease in CDAI score > 100) three months after WMT. Secondary outcome included clinical remission (CDAI < 150), improvement in HBI, SES CD score, reduction in the various inflammatory markers [erythrocyte sedimentation rate (ESR), high-sensitivity C-reactive protein (hs-CRP), fecal calprotectin], and improvement in different clinical symptoms three months after WMT. In particular, mucosal healing after WMT can be evaluated in patients willing to undergo colonoscopy three months after WMT.
The follow-up was conducted one week, one month, three months, and six months after WMT to assess the potential AE and concomitant medication changes. The correlation between AE and WMT was classified as either “unrelated,” “possible,” “probably,” or “definitive”[17].
The stool samples from patients and donors were collected and delivered to the laboratory within one hour and stored at -80 °C until processing. The samples were collected one week before WMT and three months after WMT. A total of 22 fecal samples were collected, of which 16 were from eight CD patients (including five responders and three non-responders), and six were from the six donors. All the samples were subjected to metagenomic, metatranscriptomic, and untargeted metabolomic analysis.
Total gut microbiota genomic DNA was extracted from the 22 thawed stool samples using the QIAGEN DNeasy PowerSoil Pro Kit (Qiagen, Germany) according to the manufacturer’s instructions. Total RNA was extracted from the samples using the QIAGEN RNeasy PowerMicrobiome Kit (Qiagen, Germany), employs the RiboMinus™ Transcriptome Isolation Kit (Invitrogen, United States), that utilizes rRNA-complementary probes to bind and remove rRNA. Subsequently, the mRNA was randomly divided into various short fragments of 250-300 bp with divalent cations in the NEB Fragmentation Buffer, and the first strand of cDNA was then synthesized using the fragmented RNA as a template and random oligonucleotides as potential primers. The DNA obtained by both methods was sequenced using the Illumina NovaSeq PE150 platform. The above experiments were performed at Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
The rRNA reads were removed using Bowtie2, based on the sortmerna-4.3 version of the rRNA database. The sequences were assembled using the MEGAHIT software, and gene prediction was conducted with Prodigal. The various redundant genes were removed from multiple samples using cd-hit. The obtained reference sequences and the reads were then compared to the reference sequences using the software BBMap Suite to calculate abundance. Both metagenome and metatranscriptome were analyzed using the software kraken2[18] to obtain abundance tables of species across different taxonomic levels. Intergroup differences were examined using linear discriminant analysis effect size (LEfSe).
The chromatographic separation of the target compounds was performed on a Waters ACQUITY UPLC BEH Amide (2.1 mm × 50.0 mm, 1.7 μm) liquid chromatography column using a Vanquish (Thermo Fisher Scientific) ultra-performance liquid chromatograph[19]. The liquid chromatographic phase A consisted of an aqueous phase containing 25 mmol/L ammonium acetate and 25 mmol/L ammonia, whereas the phase B was acetonitrile. The temperature of the sample tray was maintained at 4 °C, and the injection volume was 2 μL. The Orbitrap Exploris 120 mass spectrometer, controlled by Thermo’s Xcalibur software (version 4.4), could acquire primary and secondary mass spectrometry data. The raw data was converted into mzXML format by ProteoWizard software and then processed by an in-house written R package (kernel XCMS) for peak identification, extraction, alignment, and integration[20]. The substance annotation was then performed by matching it with the self-built secondary mass spectrometry database of BiotreeDB (V2.1), and the cut-off value for algorithm scoring was set to 0.3.
The data were log-transformed and centered (CTR) formatted using SIMCA software (V16.0.2, Sartorius Stedim Data Analytics AB, Umea, Sweden). After that, it was subjected to automated modeling analysis[21], and a Score scatter plot of the PCA model was obtained. For each set of comparisons, we calculated the Euclidean distance matrix for determining the quantitative values of the differential metabolites and clustered differential metabolites in a complete chaining approach, and these metabolites were subsequently presented in a heat map.
Based on the non-parametric test method, paired Wilcoxon tests were employed to obtain significant differences in metabolite level (P < 0.05), RNA level species annotated with LEfSe LDA greater than two, and significant differences (P < 0.05) in species annotated with LEfSe LDA. The correlation coefficients, R, were calculated with a P value using the Spearman method to create the heatmap.
The descriptive statistics included reporting the means and standard deviations for continuous variables and the frequencies and percentages for categorical variables. The data were examined using IBM SPSS 26.0. Wilcoxon signed-rank tests were employed to analyze paired data. P values < 0.05 were considered as significantly different.
The baseline characteristics of patients are depicted in Table 1 (Supplementary Table 1 for specific parameters). We found that seven of the 11 patients met the primary endpoint of CDAI improvement-a clinical response rate of 63.6%. All the responders achieved clinical remission. The remaining four non-responders experienced symptoms of disease within three months of treatment and returned to the hospital. The seven patients who reached the primary observation endpoint were followed up 3-6 months after the treatment. Among these, one patient experienced increased abdominal pain and was readmitted to the hospital. Two patients in remission requested a repeat WMT to consolidate their treatment. The remaining four patients remained in clinical remission. The different parameters in responders and non-responders three months after WMT-including CDAI, HBI, stool frequency, abdominal pain, CRP, ESR, and fecal calprotectin-are shown in Table 2.
| Characteristic | n (%) |
| Age (SD) | 35.6 (9.4) |
| Male | 8 (81.8) |
| Female | 3 (18.2) |
| Age at diagnosis (Montreal classification) | |
| A1 (age < 16 yr) | 1 (9.1) |
| A2 (age 17-40 yr) | 9 (81.8) |
| A3 (age > 40 yr) | 1 (9.1) |
| Location (Montreal classification) | |
| L1 (terminal ileum disease) | 5 (45.5) |
| L2 (colonic disease) | 3 (27.3) |
| L3 (ileocolonic disease) | 3 (27.3) |
| L4 (upper GI) | 0 (0.0) |
| Behavior (Montreal classification) | |
| B1 (inflammatory) | 3 (27.3) |
| B2 (stricturing) | 4 (36.4) |
| B3 (penetrating) | 2 (18.2) |
| B2 + B3 | 2 (18.2) |
| Perianal disease | 4 (36.4) |
| Duration of disease, mean (SD), yr | 8.8 (6.9) |
| Medication at the time of WMT | |
| None | 2 (18.2) |
| Mesalamines | 7 (63.6) |
| Steroids | 1 (9.1) |
| Immunomodulators | 3 (27.3) |
| Biologics therapy | 1 (9.1) |
| Bowel surgery | 2 (18.2) |
| Anal surgery | 4 (36.4) |
| Smoking status | |
| Current | 0 (0.0) |
| Ex-smoker | 2 (18.2) |
| Never smoker | 9 (81.8) |
| Disease severity | |
| Mild | 11 (100.0) |
| Moderate | 0 (0.0) |
| Severe | 0 (0.0) |
| Route of WMT | |
| Mid-gut TET | 4 (36.4) |
| Colonic TET | 7 (63.6) |
| Variable | Responders1 | Non-responders1 | ||||||
| n | Baseline, mean ± SD | 3-month post-WMT, mean ± SD | P value | n | Baseline, mean ± SD | 3-month post-WMT, mean ± SD | P value | |
| HBI (three-month post-WMT) | 7 | 5.7 ± 0.8 | 2.4 ± 0.5 | 0.014 | 4 | 5.0 ± 0.0 | 7.0 ± 2.3 | 0.157 |
| CDAI (three-month post-WMT) | 7 | 196.5 ± 21.7 | 76.4 ± 29.5 | 0.018 | 4 | 190.0 ± 13.3 | 238.5 ± 50.1 | 0.068 |
| Stool frequency (number/d) | 7 | 1.7 ± 1.0 | 0.6 ± 0.5 | 0.023 | 4 | 1.5 ± 0.6 | 3.5 ± 2.4 | 0.180 |
| Pain (scale 0-3) | 7 | 0.7 ± 0.5 | 0.3 ± 0.5 | 0.083 | 4 | 0.5 ± 0.6 | 0.5 ± 0.6 | 1.000 |
| hs-CRP (mg/L) | 7 | 3.6 ± 6.1 | 2.1 ± 2.0 | 0.500 | 4 | 14.8 ± 15.8 | 29.6 ± 19.0 | 0.144 |
| ESR (mm/h) | 7 | 15.9 ± 17.0 | 10.9 ± 11.2 | 0.340 | 4 | 43.3 ± 31.6 | 54.0 ± 33.0 | 0.273 |
| Fecal calprotectin (ug/g) | 6 | 367.6 ± 159.9 | 374.2 ± 290.4 | 0.753 | 4 | 391.6 ± 120.9 | 561.2 ± 140.2 | 0.144 |
| SES CD score | 4 | 5.3 ± 2.1 | 3.3 ± 2.5 | 0.285 | 4 | 9.3 ± 7.4 | 17.5 ± 7.1 | 0.068 |
Based on a decrease of more than 100 points in their CDAI score, the patients were categorized into responders (n = 7) and non-responders (n = 4). After three months of WMT, responders exhibited significant improvements in HBI, CDAI, and bowel frequency (P < 0.05). In addition, abdominal pain, hs-CRP, and ESR decreased to varying degrees, although the reduction was not statistically significant. The fecal calprotectin levels in responders remained constant before and after the treatment (Table 2). However, non-responders did not experience any improvement in CD-related clinical symptoms and inflammatory indexes; in some cases, their condition even deteriorated compared to the baseline period (Supplementary Table 2). Consistent with the clinical symptoms and laboratory indices, four patients in the responder group displayed a trend of decreasing SES CD scores compared with the previous period. Conversely, the non-responder group showed increased SES CD scores during the same period.
The incidence of AEs was 2/22 (9.1%). One patient experienced constipation (possibly related, mild) within one week of transplantation, which improved rapidly with laxative medication. In the other patient, a mild increase in defecation (possibly related, mild) was observed on the second day after transplantation and was not treated. Both patients' symptoms resolved spontaneously within 24 hours. Two other patients experienced painful episodes of urticaria and gout after treatment. Because these patients had previously relapsed with similar conditions, the treating physicians did not consider them to be transplant-related. All of these AE were clarified as short-term AE according to the cut-pont of 1 month-post WMT[22]. No severe AEs and long-term AE occurred during treatment and follow-up.
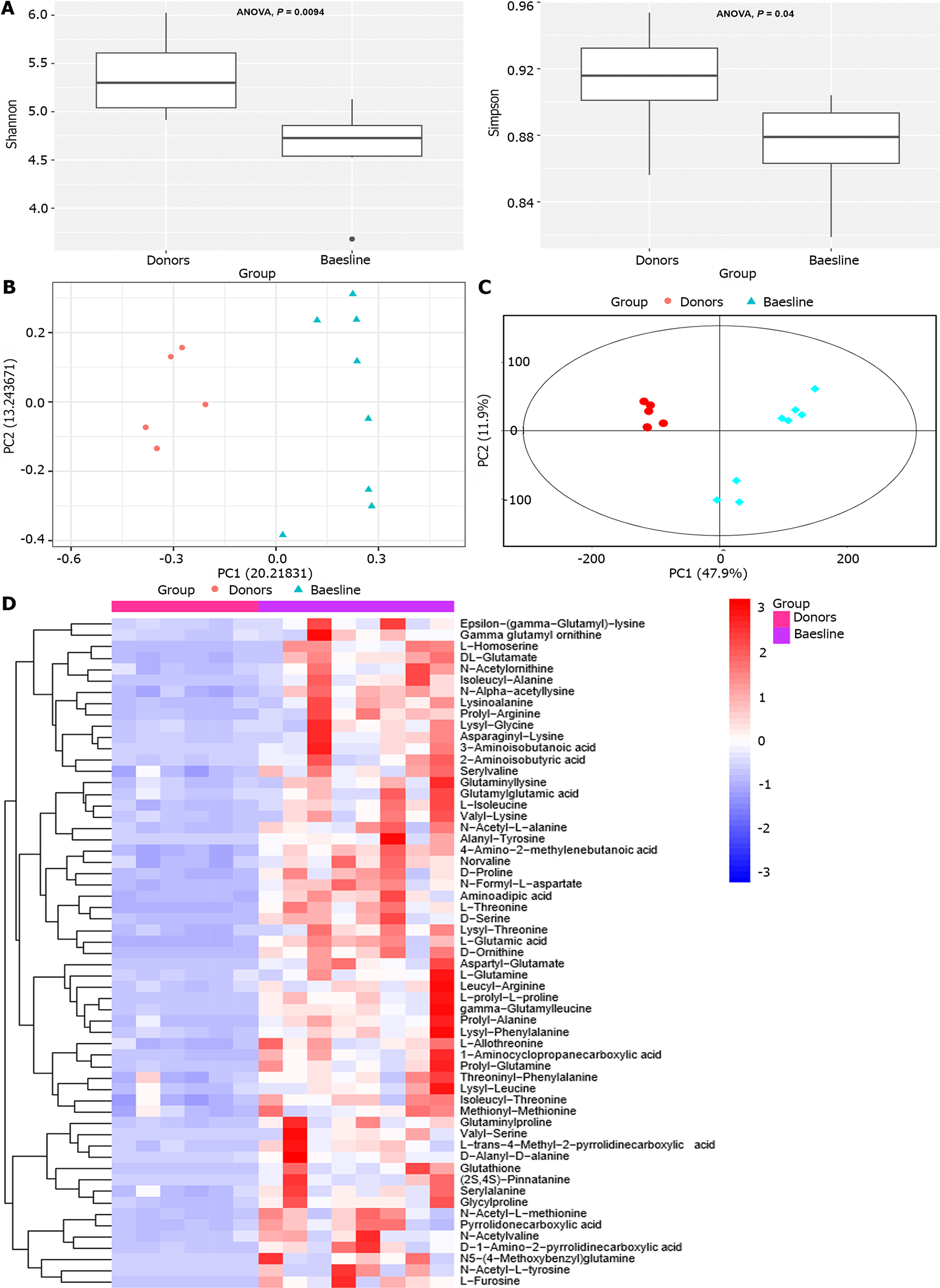
Metagenomic species-level annotation showed that CD patients had significantly decreased Shannon and Simpson indices compared to donors prior to WMT, indicating lower alpha diversity (ANOVA, P < 0.01; P < 0.05) (Figure 1A). Regarding β-diversity, we constructed a principal coordinate analysis plot of the categorical distances between the samples (Jaccard index in Figure 1B), revealing a significant separation between the two groups, with the donor group exhibiting a more compact clustering. At the metabolome level, a significant separation between the two groups was visible in the Score scatter plot of the PCA model. The donor group exhibited a significantly closer clustering than the CD group (Figure 1C). Differences in the amino acids composition of fecal samples were found between patients with baseline CD patients and donors (Figure 1D). Amino acids levels in the feces of CD patients were higher than those of donors.
LEfSe was employed in the metagenomic analysis to screen the two groups for the differential species based on LDA ≥ 2 and P < 0.05. The three species with the highest abundance in the patient group before the transplantation were Collinsella aerofaciens (LDA = 4.901 P < 0.05), Erysipelotrichaceae bacteriumI46 (LDA = 3.745, P < 0.05), and Clostridium scindens (LDA = 3.697, P < 0.01). In contrast, the three species with the highest abundance in the donor group were F. prausnitzii (LDA = 5.002, P < 0.01), Anaerostipes hadrus (A. hadrus, LDA = 4.259, P < 0.01), and Bacteroides vulgatus (LDA = 4.104, P < 0.01), respectively (Supplementary Figure 1).
The same LEFSe was employed in the metatranscriptomic analysis to screen for the differential species between baseline and donor in CD patients based on LDA ≥ 2 and P < 0.05. The top three species with the highest baseline abundance in CD patients were observed to be Phocaeicola coprophilus (LDA = 4.43, P < 0.01), Bacteroides fragilis (LDA = 4.178, P < 0.01) and Bacteroides sp. PHL 2737 (LDA = 4.035, P < 0.01). The three species with the highest abundance in the donor group were Bifidobacterium bifidum (LDA = 4.746, P < 0.05), A. hadrus (LDA = 4.638, P < 0.01), and Blautia producta (LDA = 4.518, P < 0.01) (Supplementary Figure 2). Interestingly, when analyzed in comparison with the metagenome assay results, elevated expression of F. prausnitzii, A. hadrus, and R. intestinalis in the donor group was found to be almost identical to the metatranscriptomic assay results (Supplementary Table 3).
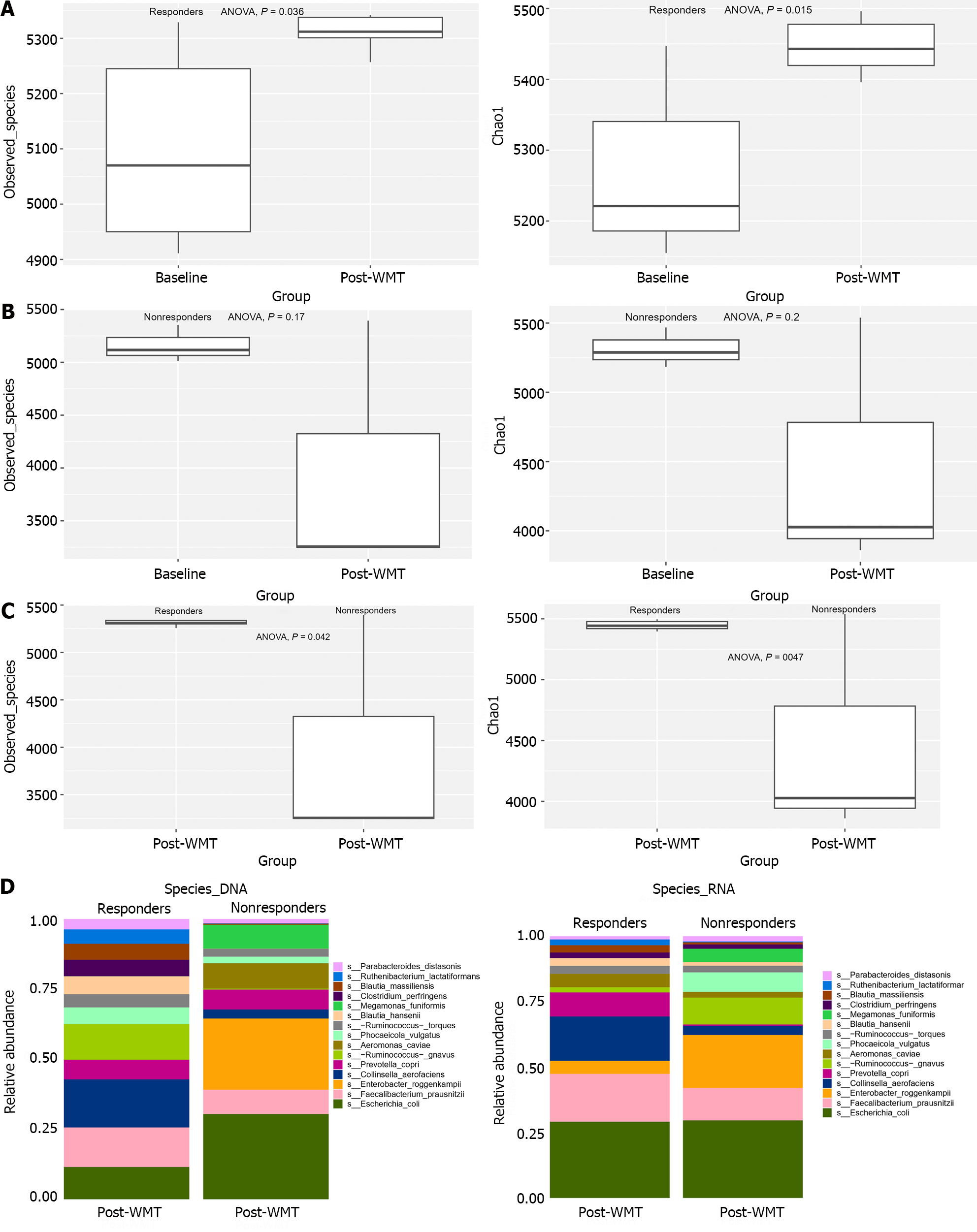
According to the metagenomic species level annotation, there was a significant increase in alpha diversity (as indicated by the observed species and chao1 index) in the responder group after WMT compared to the baseline (ANOVA, both P < 0.01) (Figure 2A). However, the non-responder group did not show any notable alterations in observed species and chao1 index following WMT compared to the baseline (ANOVA, both P < 0.05) (Figure 2B).
When analyzing the disparities in post-transplant microbiome diversity, the responder group presented a significant increase in observed species and chao1 index compared to the non-responder group. (ANOVA, both P < 0.05) (Figure 2C). Examining the disparity in species abundance at the DNA and RNA levels in both groups showed a marked resemblance between the two differential microbiomes (Figure 2D). There was a decrease in F. prausnitzii and Phocaeicola vulgatus and an increase in E. coli.
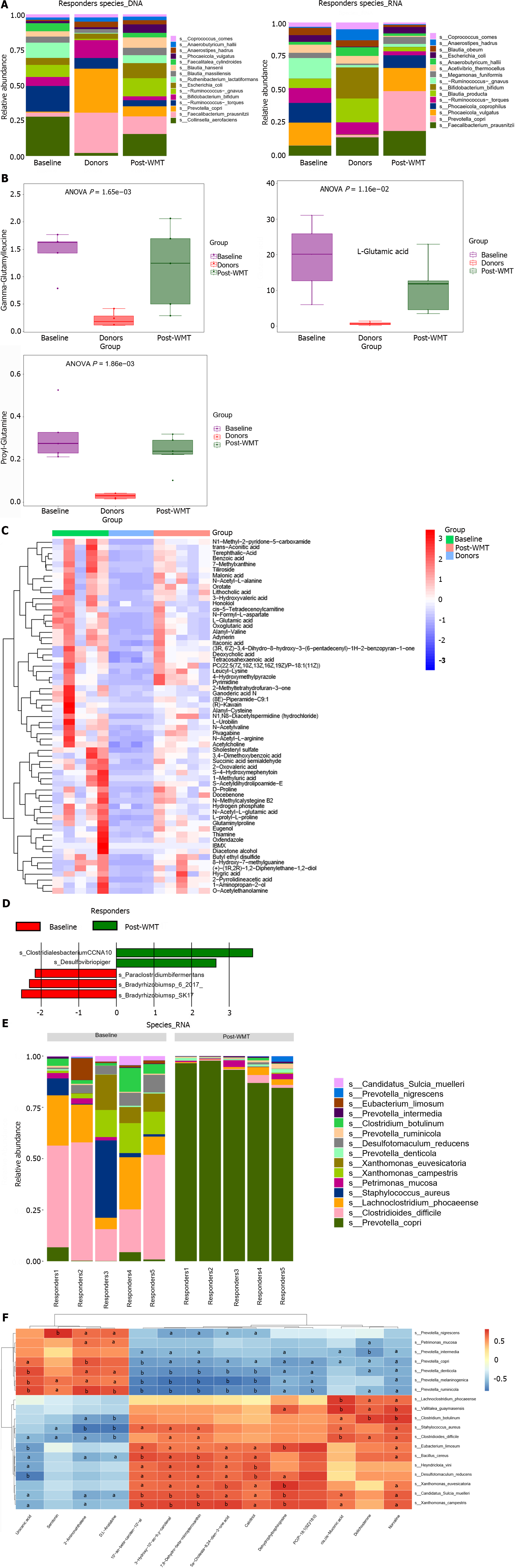
A thorough comparison of DNA levels between the baseline responder group, post-transplantation responders, and donors exhibited that the transfer of F. prausnitzii can be identified in both donor and responder samples, with corresponding variations observed in RNA levels (Figure 3A). The fecal metabolites L-glutamic acid, gamma-glutamyl-leucine, and prolyl-glutamine were significantly higher than the donors before WMT but decreased after WMT (Figure 3B). Furthermore, a number of metabolites exhibit a similar pattern of alteration (Figure 3C).
At the DNA level, the comparison between the community species pre-and post-WMT revealed an increase in Desulfovibrio piger (D. piger) and Clostridiales bacteriumCCNA10, and a decrease in Paraclostridium bifermentans (P. bifermentans), Bradyrhizobium sp 6 2017, and Bradyrhizobium sp SK17 (Figure 3D), according to the results from the comparative LEfSe analyses. Interestingly, the most significant changes at the RNA level included a decrease in C. difficile and a significant increase in Prevotella copri (P. copri) (Figure 3E). Analysis of fecal metabolites showed a significant decrease in several metabolites, such as isonicotinic acid, lutein, and norvaline (Supplementary Figure 3). Heatmaps were plotted to show differential species (RNA levels) and differential metabolite correlations before and after WMT in the responding group, as shown in Figure 3F.
In this prospective study on the effectiveness of WMT for CD, 63.6% of patients experienced notable improvements in CDAI (of more than 100 points) three months after WMT. This treatment efficacy was higher than in a previously reported study[10]. Although inflammatory indicators such as SES CD, fecal calprotectin, hs-CRP, and ESR decreased in the responding group, the small sample size prevented the identification of statistically differences (P > 0.05). Therefore, randomized controlled trials with larger sample sizes are needed for further validation of these findings.
We thoroughly analyzed fecal samples obtained from patients before and after WMT-and from donors-for metagenomic, metatranscriptomic, and non-targeted metabolomic assays. To our knowledge, this is the first time that metatranscriptomic and metabolomic techniques have been used to analyze pre- and post-transplant fecal samples in a study of FMT for CD. Metagenomic-based species annotation studies revealed that patient clinical phenotypes were significantly associated with alpha diversity (Figure 1A; Figure 2A-C). For example, the alpha diversity of the donor gut microbiome was significantly higher than that of baseline patients. The alpha diversity of the post-WMT responders was significantly higher than that of the baseline. However, the alpha diversity of the non-responders was not significantly altered pre- and post-WMT. These findings were consistent with those reported in previous studies[23]. In terms of beta diversity, it was observed that there was a distinct separation between the donor group and the baseline patients, and the samples were clustered more tightly between the donor groups, which also aligns with previous reports[24]. A similar phenomenon was observed in metabolomic analyses (Figure 1C).
The microbiome characteristics of the baseline CD patient samples were also analyzed. The significant reductions in R. intestinalis, F. prausnitzii, and Akkermansia muciniphila in the DNA-level species-richness-based differential analyses (Supplementary Figure 1) were very similar to the results of other recently reported studies of CD patients and healthy controls[25]. However, despite the presence of a gene in the DNA, its functionality and transcriptional activity are not guaranteed as they rely on the type and quantity of RNA transcribed from it[26]. The metatranscriptome can offer improved “resolution” for species identification compared to the metagenome[11]. In this study, we compared the metagenomic and metatranscriptomic analyses and observed that the changes in certain strains were consistent. When studying the species differences between the donors and baseline CD patients, it was discovered that F. prausnitzi, A. hadrus, and R. intestinalis were significantly more abundant in the donors than in baseline CD patients based on both DNA and RNA species annotation (Supplementary Table 3). All three bacteria are producers of short-chain fatty acid (SCFA)[25]. SCFA is an important metabolite for maintaining intestinal homeostasis and is thought to have some therapeutic potential for IBD[27]. F. prausnitzi and R. intestinalis are recognized as beneficial bacteria in IBD and can exert anti-inflammatory effects through the regulation of multiple pathways[25,28]. When comparing the differences in bacterial microbiomes between the responder and non-responder groups post-transplantation, an increase in SCFA-producing bacteria[25], such as F. prausnitzii and Phocaeicola vulgatus, as well as a decrease in the potentially pathogenic bacterium E. coli, were also found in the responder group at both the DNA and RNA levels (Figure 2D).
The effectiveness of WMT lies in its ability to restore the patient’s microbial balance effectively. We observed that, whether based on investigation of DNA or RNA levels, the abundance of F. prausnitzii showed a remarkable increase after transplantation in the responders compared to baseline and was higher than in the non-responders group (Figures 2D and 3A). It was observed that Ruminococcus gnavus, a well-established CD causative agent[4], significantly decreased in the responders after transplantation at the RNA level. However, contrasting results were observed at the DNA level. (Figure 3A). Thus, RNA-based transcriptional activity of the bacterial population could be more relevant to the clinical outcomes.
Furthermore, a significant increase in the abundance of D. piger in the responding group (based on DNA levels) was noted in this study. Around 50% of individuals carry sulfate-reducing bacteria (SRB) in their gut. Specifically, D. piger was reported to be the most common SRB in a cohort of healthy adults in the United States[29]. In addition, prior studies have shown that a higher abundance of D. piger is resistant to invasion by Salmonella enteritidis in a chick preclinical model[30]. Moreover, the abundance of potentially pathogenic bacteria, such as P. bifermentans and Bradyrhizobium sp 6 2017, was significantly reduced in the responder group after the transplantation. P. bifermentans was the first strain reported to cause worsening of the pathological scores in a mouse model of ulcerative colitis[31]. In another elegant study, Bradyrhizobium enterica (B. enterica) nucleotide sequences were identified by PCR from the biopsy specimens of patients with umbilical cord colitis, thereby suggesting that B. enterica may be an opportunistic human pathogen[32].
Interestingly, the decreased abundance of C. difficile and increased abundance of P. copri was predominantly found at the RNA level (Figure 3E). C. difficile is an opportunistic pathogen with increased abundance in the intestines of dysbiotic intestinal hosts and is a leading pathogen in pseudomembranous colitis. Several previous studies have shown a significant decrease in P. copri in pediatric CD patients and in a canine model of CD[33], whereas P. copri abundance was significantly increased after WMT intervention. Based on these findings, it can be suggested that WMT can improve patients’ clinical outcomes by optimizing microbiome structure, boosting the presence of helpful bacteria, and reducing the colonization of pathogenic bacteria.
Increased levels of many amino acids were observed in the baseline samples collected from CD patients (Figure 1D). Previous studies have shown that there is a noticeable increase in specific metabolites related to glutathione metabolism (including glutamate, glycine, cysteine, and pyroglutamate) in the urine of pediatric IBD patients[34], and it has also been suggested that CD patients tend to have enhanced fecal amino acids[35]. Furthermore, various studies in preclinical models have indicated that bacterial urease expression can effectively transfer host-sourced nitrogen to the gut microbiota to facilitate amino acid synthesis during gut microbiota dysregulation and inflammation[35].
Interestingly, several amino acids (including L-glutamic acid, gamma-glutamyl-leucine, and prolyl-glutamine) were elevated in the baseline samples compared to the donors but decreased significantly after WMT (Figure 3B). Among these, glutamic acid has been recognized as an inflammatory marker in an immunocompromised mouse model of a specific type of ulcerative colitis[36]. Moreover, another study indicates that increased levels of gamma-glutamyl-leucine-a biologically active peptide implicated in inflammation, oxidative stress, and glucose management-are causally associated with cardiometabolic risk[37]. In addition, the down-regulation of prolyl-glutamine has been linked with immune enhancement in mice[38]. We have illustrated the presence of several metabolites showing similar patterns (Figure 3C). One of these metabolites, N1-methyl-2-pyridone-5-carboxamide (M2Py), is known to be a uremic toxin[39]. The reduction in M2Py levels after transplantation through WMT suggests that WMT could be a potentially effective approach in protecting the renal function of patients suffering from renal failure[40]. In addition, the levels of many anti-inflammatory substances (including isonicotinic acid, lupulone, and norvaline) were elevated in the responders after WMT. The combination of isoniazid and salazosulfapyridine has demonstrated anti-colitis effects in preclinical experiments by suppressing the pathology of inflammation and fibrosis in a model of colitis[41]. Lupulone, a component found in hops extracts, is recognized for its significant role in exhibiting antimicrobial effects and its potential as an antioxidant, anti-inflammatory, and antibacterial agent[42]. Norvaline demonstrates anti-inflammatory properties by inhibiting arginase, and its effectiveness in reducing inflammation is partially due to its ability to suppress inhibited p70s6k[43].
Correlation heatmap display analyses were performed for both differential metabolites and differential species at the RNA level pre-and post-WMT in the responders (Figure 3F). It was observed that species such as Eubacterium limosum and Bacillus cereus were positively correlated with the metabolite norvaline, which is associated with anti-inflammatory properties. In addition, previous studies have indicated that Eubacterium spp. and Bacillus cereus can exhibit significant anti-inflammatory effects related to intestinal inflammation[25,44]. Several species within the genus Prevotella, including P. copri, are negatively correlated with the immune-related metabolite dehydrophytosphingosine[45] but positively correlated with the anti-inflammatory metabolite norvaline[43] and urocanic acid[46].
A limitation of this study is its small sample size, which is related to our strict inclusion criteria. To minimize the interference of drugs on the results of microbiome testing, we excluded patients who had used drugs (such as antibiotics, proton pump inhibitors, and probiotics) within three months before recruitment and three months after WMT. We also excluded some patients with moderately to severely active CD. Nonetheless, although not designed as a randomized controlled trial, this study closely integrates microbiomics and metabolomic analyses, providing new evidence for understanding FMT in treating CD and thus advancing the field of microbiota medicine[47].
This study demonstrates that WMT is effective in CD treatment. The clinical phenotype of the patients was closely related to the altered diversity of the gut microbiota, and metagenomic and metatranscriptomic techniques were effective and complementary in improving the “resolution” of the active microbiome, which could potentially have significant implications. To better understand the role of the microbiome in WMT treatment and uncover the underlying mechanisms, it remains crucial to utilize additional genomic techniques such as macro-proteomics and culture genomics in future studies.
Incidence of Crohn’s disease (CD) is increasing every year, posing a serious threat to human health. Fecal microbial transplantation (FMT) is a promising therapeutic approach for the treatment of CD. The new methodology of FMT, based on the automatic washing process, was named as washed microbiota transplantation (WMT).
Most existing studies have focused on observing clinical phenomena. However, a combined multi-omics (metagenomic, metatranscriptomic, and metabolomic) analysis of FMT for the effective treatment of CD has not been reported.
To examine the effects of two consecutive fixed WMT doses on clinical and endoscopic outcomes in CD patients. A secondary aim was to explore alterations in the microbiome and metabolome in response to WMT.
WMT was administered to 11 patients with active CD. Their clinical response (defined as a decrease in CD activity index score > 100 points) was assessed three months after transplantation. Fecal samples collected 1 wk before and 3 months after WMT were subjected to combined metagenomic, metatranscriptomic, and metabolomic analyses.
Seven of 11 patients (63.6%) demonstrated response 3 months after WMT. There was a significant increase in the diversity of the gut microbiota after WMT, consistent with improved clinical symptoms. A comparison of metagenomic and metatranscriptomic analyses revealed constant changes in certain strains, such as Faecalibacterium prausnitzii, Roseburia intestinalis, and Escherichia coli. Metabolomic analysis of the responder group identified certain amino acids that may be associated with disease progression (e.g., L-glutamic acid, gamma-glutamyl-leucine, and prolyl-glutamine) that were higher in the pre-transplant than in the donor but lower in the post-transplant.
WMT has shown efficacy in CD treatment, possibly due to the effective reconstitution of the patient’s microbiome. Combined metagenomic, metatranscriptomic, and metabolomic analyses can effectively help decipher the underlying mechanisms of WMT for CD.
The exact mechanism by which FMT treats CD still needs to be better understood. Future studies need to clarify the underlying mechanisms by utilizing additional histological techniques (e.g., macro-proteomics and culture genomics etc.).
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Snyder AM, United States S-Editor: Chen YL L-Editor: A P-Editor: Cai YX
| 1. | GBD 2017 Inflammatory Bowel Disease Collaborators. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17-30. [PubMed] [DOI] [Full Text] |
| 2. | O’Brien CL, Bringer MA, Holt KE, Gordon DM, Dubois AL, Barnich N, Darfeuille-Michaud A, Pavli P. Comparative genomics of Crohn’s disease-associated adherent-invasive Escherichia coli. Gut. 2017;66:1382-1389. [PubMed] [DOI] [Full Text] |
| 3. | Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin Microbiol Rev. 2019;32. [PubMed] [DOI] [Full Text] |
| 4. | Pal D, Naskar M, Bera A, Mukhopadhyay B. Chemical synthesis of the pentasaccharide repeating unit of the O-specific polysaccharide from Ruminococcus gnavus. Carbohydr Res. 2021;507:108384. [PubMed] [DOI] [Full Text] |
| 5. | Zhang T, Lu G, Zhao Z, Liu Y, Shen Q, Li P, Chen Y, Yin H, Wang H, Marcella C, Cui B, Cheng L, Ji G, Zhang F. Washed microbiota transplantation vs. manual fecal microbiota transplantation: clinical findings, animal studies and in vitro screening. Protein Cell. 2020;11:251-266. [PubMed] [DOI] [Full Text] |
| 6. | Fecal Microbiota Transplantation-standardization Study Group. Nanjing consensus on methodology of washed microbiota transplantation. Chin Med J (Engl). 2020;133:2330-2332. [PubMed] [DOI] [Full Text] |
| 7. | Lu G, Wang W, Li P, Wen Q, Cui B, Zhang F. Washed preparation of faecal microbiota changes the transplantation related safety, quantitative method and delivery. Microb Biotechnol. 2022;15:2439-2449. [PubMed] [DOI] [Full Text] |
| 8. | Zhang T, Li P, Wu X, Lu G, Marcella C, Ji X, Ji G, Zhang F. Alterations of Akkermansia muciniphila in the inflammatory bowel disease patients with washed microbiota transplantation. Appl Microbiol Biotechnol. 2020;104:10203-10215. [PubMed] [DOI] [Full Text] |
| 9. | Li P, Zhang T, Xiao Y, Tian L, Cui B, Ji G, Liu YY, Zhang F. Timing for the second fecal microbiota transplantation to maintain the long-term benefit from the first treatment for Crohn’s disease. Appl Microbiol Biotechnol. 2019;103:349-360. [PubMed] [DOI] [Full Text] |
| 10. | Zhou S, Cui Y, Zhang Y, Zhao T, Cong J. Fecal microbiota transplantation for induction of remission in Crohn’s disease: a systematic review and meta-analysis. Int J Colorectal Dis. 2023;38:62. [PubMed] [DOI] [Full Text] |
| 11. | Bashiardes S, Zilberman-Schapira G, Elinav E. Use of Metatranscriptomics in Microbiome Research. Bioinform Biol Insights. 2016;10:19-25. [PubMed] [DOI] [Full Text] |
| 12. | Gajendran M, Loganathan P, Catinella AP, Hashash JG. A comprehensive review and update on Crohn’s disease. Dis Mon. 2018;64:20-57. [PubMed] [DOI] [Full Text] |
| 13. | Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749-753. [PubMed] [DOI] [Full Text] |
| 14. | Zhang F, Cui B, He X, Nie Y, Wu K, Fan D; FMT-standardization Study Group. Microbiota transplantation: concept, methodology and strategy for its modernization. Protein Cell. 2018;9:462-473. [PubMed] [DOI] [Full Text] |
| 15. | Zhang T, Long C, Cui B, Buch H, Wen Q, Li Q, Ding X, Ji G, Zhang F. Colonic transendoscopic tube-delivered enteral therapy (with video): a prospective study. BMC Gastroenterol. 2020;20:135. [PubMed] [DOI] [Full Text] |
| 16. | Long C, Yu Y, Cui B, Jagessar SAR, Zhang J, Ji G, Huang G, Zhang F. A novel quick transendoscopic enteral tubing in mid-gut: technique and training with video. BMC Gastroenterol. 2018;18:37. [PubMed] [DOI] [Full Text] |
| 17. | Kelly CR, Kunde SS, Khoruts A. Guidance on preparing an investigational new drug application for fecal microbiota transplantation studies. Clin Gastroenterol Hepatol. 2014;12:283-288. [PubMed] [DOI] [Full Text] |
| 18. | Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. [PubMed] [DOI] [Full Text] |
| 19. | Wang JL, Zhang T, Shen XT, Liu J, Zhao DL, Sun YW, Wang L, Liu YJ, Gong XY, Liu YX, Zhu ZJ, Xue FZ. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics. 2016;12:116. [DOI] [Full Text] |
| 20. | Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779-787. [PubMed] [DOI] [Full Text] |
| 21. | Wiklund S, Johansson E, Sjöström L, Mellerowicz EJ, Edlund U, Shockcor JP, Gottfries J, Moritz T, Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal Chem. 2008;80:115-122. [PubMed] [DOI] [Full Text] |
| 22. | Marcella C, Cui B, Kelly CR, Ianiro G, Cammarota G, Zhang F. Systematic review: the global incidence of faecal microbiota transplantation-related adverse events from 2000 to 2020. Aliment Pharmacol Ther. 2021;53:33-42. [PubMed] [DOI] [Full Text] |
| 23. | Cheng F, Huang Z, Wei W, Li Z. Fecal microbiota transplantation for Crohn’s disease: a systematic review and meta-analysis. Tech Coloproctol. 2021;25:495-504. [PubMed] [DOI] [Full Text] |
| 24. | Goyal A, Yeh A, Bush BR, Firek BA, Siebold LM, Rogers MB, Kufen AD, Morowitz MJ. Safety, Clinical Response, and Microbiome Findings Following Fecal Microbiota Transplant in Children With Inflammatory Bowel Disease. Inflamm Bowel Dis. 2018;24:410-421. [PubMed] [DOI] [Full Text] |
| 25. | Wu R, Xiong R, Li Y, Chen J, Yan R. Gut microbiome, metabolome, host immunity associated with inflammatory bowel disease and intervention of fecal microbiota transplantation. J Autoimmun. 2023;141:103062. [PubMed] [DOI] [Full Text] |
| 26. | Mallick H, Ma S, Franzosa EA, Vatanen T, Morgan XC, Huttenhower C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18:228. [PubMed] [DOI] [Full Text] |
| 27. | Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, Hermoso MA. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol. 2019;10:277. [PubMed] [DOI] [Full Text] |
| 28. | Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, Dekens J, Peters V, Voskuil MD, Visschedijk MC, van Dullemen HM, Keszthelyi D, Swertz MA, Franke L, Alberts R, Festen EAM, Dijkstra G, Masclee AAM, Hofker MH, Xavier RJ, Alm EJ, Fu J, Wijmenga C, Jonkers DMAE, Zhernakova A, Weersma RK. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10. [PubMed] [DOI] [Full Text] |
| 29. | Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci U S A. 2013;110:13582-13587. [PubMed] [DOI] [Full Text] |
| 30. | Wu S, Cong G, Zhang Q, Yao H, Wang Z, Kang K, He X, Shi S. Infection Heterogeneity and Microbiota Differences in Chicks Infected by Salmonella enteritidis. Microorganisms. 2021;9. [PubMed] [DOI] [Full Text] |
| 31. | Kutsuna R, Tomida J, Morita Y, Kawamura Y. Paraclostridium bifermentans exacerbates pathosis in a mouse model of ulcerative colitis. PLoS One. 2018;13:e0197668. [PubMed] [DOI] [Full Text] |
| 32. | Bhatt AS, Freeman SS, Herrera AF, Pedamallu CS, Gevers D, Duke F, Jung J, Michaud M, Walker BJ, Young S, Earl AM, Kostic AD, Ojesina AI, Hasserjian R, Ballen KK, Chen YB, Hobbs G, Antin JH, Soiffer RJ, Baden LR, Garrett WS, Hornick JL, Marty FM, Meyerson M. Sequence-based discovery of Bradyrhizobium enterica in cord colitis syndrome. N Engl J Med. 2013;369:517-528. [PubMed] [DOI] [Full Text] |
| 33. | Maldonado-Contreras A, Ferrer L, Cawley C, Crain S, Bhattarai S, Toscano J, Ward DV, Hoffman A. Dysbiosis in a canine model of human fistulizing Crohn’s disease. Gut Microbes. 2020;12:1785246. [PubMed] [DOI] [Full Text] |
| 34. | Martin FP, Su MM, Xie GX, Guiraud SP, Kussmann M, Godin JP, Jia W, Nydegger A. Urinary metabolic insights into host-gut microbial interactions in healthy and IBD children. World J Gastroenterol. 2017;23:3643-3654. [PubMed] [DOI] [Full Text] |
| 35. | Ni J, Shen TD, Chen EZ, Bittinger K, Bailey A, Roggiani M, Sirota-Madi A, Friedman ES, Chau L, Lin A, Nissim I, Scott J, Lauder A, Hoffmann C, Rivas G, Albenberg L, Baldassano RN, Braun J, Xavier RJ, Clish CB, Yudkoff M, Li H, Goulian M, Bushman FD, Lewis JD, Wu GD. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med. 2017;9. [PubMed] [DOI] [Full Text] |
| 36. | Jodeleit H, Winkelmann P, Caesar J, Sterz S, Holdt LM, Beigel F, Stallhofer J, Breiteneicher S, Bartnik E, Leeuw T, Siebeck M, Gropp R. Head-to-head study of oxelumab and adalimumab in a mouse model of ulcerative colitis based on NOD/Scid IL2Rγnull mice reconstituted with human peripheral blood mononuclear cells. Dis Model Mech. 2021;14. [PubMed] [DOI] [Full Text] |
| 37. | Wu Q, Li J, Zhu J, Sun X, He D, Cheng Z, Zhang X, Xu Y, Chen Q, Zhu Y, Lai M. Gamma-glutamyl-leucine levels are causally associated with elevated cardio-metabolic risks. Front Nutr. 2022;9:936220. [PubMed] [DOI] [Full Text] |
| 38. | Wu X, Cao J, Li M, Yao P, Li H, Xu W, Yuan C, Liu J, Wang S, Li P, Wang Y. An integrated microbiome and metabolomic analysis identifies immunoenhancing features of Ganoderma lucidum spores oil in mice. Pharmacol Res. 2020;158:104937. [PubMed] [DOI] [Full Text] |
| 39. | Carrey EA, Smolenski RT, Edbury SM, Laurence A, Marinaki AM, Duley JA, Zhu L, Goldsmith DJ, Simmonds HA. Origin and characteristics of an unusual pyridine nucleotide accumulating in erythrocytes: positive correlation with degree of renal failure. Clin Chim Acta. 2003;335:117-129. [PubMed] [DOI] [Full Text] |
| 40. | Zhong HJ, Xie X, Chen WJ, Zhuang YP, Hu X, Cai YL, Zeng HL, Xiao C, Li Y, Ding Y, Xue L, Chen M, Zhang J, Wu Q, He XX. Washed microbiota transplantation improves renal function in patients with renal dysfunction: a retrospective cohort study. J Transl Med. 2023;21:740. [PubMed] [DOI] [Full Text] |
| 41. | Yaghoubi A, Davoodi J, Asgharzadeh F, Rezaie S, Nazari E, Khazaei M, Soleimanpour S. Therapeutic effect of an anti-tuberculosis agent, isoniazid, and its nano-isoform in ulcerative colitis. Int Immunopharmacol. 2021;96:107577. [PubMed] [DOI] [Full Text] |
| 42. | Li Y, Dalabasmaz S, Gensberger-Reigl S, Heymich ML, Krofta K, Pischetsrieder M. Identification of colupulone and lupulone as the main contributors to the antibacterial activity of hop extracts using activity-guided fractionation and metabolome analysis. Food Res Int. 2023;169:112832. [PubMed] [DOI] [Full Text] |
| 43. | Ming XF, Rajapakse AG, Carvas JM, Ruffieux J, Yang Z. Inhibition of S6K1 accounts partially for the anti-inflammatory effects of the arginase inhibitor L-norvaline. BMC Cardiovasc Disord. 2009;9:12. [PubMed] [DOI] [Full Text] |
| 44. | Sheng K, Xu Y, Kong X, Wang J, Zha X, Wang Y. Probiotic Bacillus cereus Alleviates Dextran Sulfate Sodium-Induced Colitis in Mice through Improvement of the Intestinal Barrier Function, Anti-Inflammation, and Gut Microbiota Modulation. J Agric Food Chem. 2021;69:14810-14823. [PubMed] [DOI] [Full Text] |
| 45. | Kunisawa J, Kiyono H. Immunological function of sphingosine 1-phosphate in the intestine. Nutrients. 2012;4:154-166. [PubMed] [DOI] [Full Text] |
| 46. | Albert E, Walker J, Thiesen A, Churchill T, Madsen K. cis-Urocanic acid attenuates acute dextran sodium sulphate-induced intestinal inflammation. PLoS One. 2010;5:e13676. [PubMed] [DOI] [Full Text] |
| 47. | Zhang F, Wang W, Nie Y, Li J, He X. From microbial technology to microbiota medicine as a clinical discipline: Sustainable development goal. Microb Biotechnol. 2023;16:1705-1708. [PubMed] [DOI] [Full Text] |