Published online Jan 7, 2023. doi: 10.3748/wjg.v29.i1.190
Peer-review started: August 16, 2022
First decision: October 20, 2022
Revised: November 2, 2022
Accepted: November 21, 2022
Article in press: November 21, 2022
Published online: January 7, 2023
Processing time: 140 Days and 19.6 Hours
In recent years, associations between specific virulence markers of Helicobacter pylori (H. pylori) and gastrointestinal disorders have been suggested.
To investigate the presence of virulence factors including vacuolating cytotoxin A genotypes (s1m1, s1m2, s2m1, and s2m2), cytotoxin-associated gene A (CagA), and urease activity in H. pylori strains isolated from Arab and Jewish populations in northern Israel and to assess associations between these factors and patients’ demographics and clinical outcomes.
Patients (n = 108) who underwent gastroscopy at the Baruch Padeh Medical Center, Poriya due to symptomatic gastroduodenal pathologies as part of H. pylori diagnosis were enrolled in the study. Gastric biopsy specimens were collected from the antrum of the stomach. Clinical condition was assessed by clinical pathology tests. Bacteria were isolated on modified BD Helicobacter Agar (BD Diagnostics, Sparks, MD, United States). Bacterial DNA was extracted, and PCR was performed to detect CagA and vacuolating cytotoxin A genes. Urease activity was assessed using a rapid urease test.
A significant correlation was found between disease severity and patient ethnicity (P = 0.002). A significant correlation was found between CagA presence and the s1m1 genotype (P = 0.02), which is considered the most virulent genotype. Further, a higher level of urease activity was associated with isolates originating from the Jewish population. Moreover, higher urease activity levels were measured among CagA-/s1m1 and CagA-/s2m2 isolates.
Our study highlights the importance of incorporating molecular methods for detection of virulence markers of H. pylori in order to tailor optimal treatments for each patient. Further investigation should be performed regarding associations between H. pylori virulence factors and ethnicity.
Core Tip: In recent years, associations have been found between virulence markers of Helicobacter pylori and gastrointestinal disorders. In parallel, several physicians in northern Israel noted a higher treatment failure rate among Arab patients compared to Jewish patients. This work found a significant correlation between disease severity and patient ethnicity (P = 0.002). Further, a higher level of urease activity was associated with isolates originating from the Jewish population. Moreover, higher urease activity levels were measured among CagA-/s1m1 and CagA-/s2m2 isolates. These findings are expected to advance personalization of treatment to specific strains based on their virulence factors.
- Citation: Roshrosh H, Rohana H, Azrad M, Leshem T, Masaphy S, Peretz A. Impact of Helicobacter pylori virulence markers on clinical outcomes in adult populations. World J Gastroenterol 2023; 29(1): 190-199
- URL: https://www.wjgnet.com/1007-9327/full/v29/i1/190.htm
- DOI: https://dx.doi.org/10.3748/wjg.v29.i1.190
Helicobacter pylori (H. pylori) is a microaerophilic, Gram-negative bacterium that colonizes the gastric mucosa and infects the stomach epithelium, causing peptic ulcer disease[1]. Unrelenting H. pylori infections cause chronic inflammation, which can result in gastritis, intestinal metaplasia, and even gastric cancer[1].
The genetic variability of H. pylori and its host, combined with environmental factors, have been suggested to affect the clinical outcome[1-3]. H. pylori pathogenesis is mediated via distinct virulence factors, including the secreted vacuolating cytotoxin A (VacA), cytotoxin-associated gene A (CagA) protein, and urease[1,2]. VacA is one of the main toxins secreted by the bacterium; once bound to host cells and internalized, it causes “vacuolation,” characterized by the accumulation of large vesicles that disrupt protein trafficking pathways. Two polymorphic regions in the VacA gene sequence are the signal sequence region (s-region) and the mid region (m-region), with allelic variations classified as s1 or s2 and m1 or m2, respectively[3]. It was suggested that isolates with a s1/m1 genotype cause more severe chronic inflammation compared to the other VacA genotypes[4].
H. pylori strains can be divided into two main subpopulations according to their ability to produce CagA, a 120-145 kDa protein associated with gastric carcinoma. The protein interferes with cellular signal transduction, provoking cellular dysfunction that can eventually lead to cell transformation and cancer[5].
Along with VacA and CagA, urease activity in H. pylori is an indicator of bacterial virulence. Urease is a metalloenzyme that requires nickel for its activity and consists of two components, UreA and UreB. The enzyme breaks down urea into ammonia and carbon dioxide, promoting bacterial survival in the acidic environment of the stomach. In addition, urease is involved in H. pylori colonization in the gastric tissue, prevention of phagocytosis, and induction of proinflammatory cytokines[6,7].
In addition to the virulence factors that may affect the efficacy of H. pylori eradication regimens, antibiotic resistance poses a major challenge to treatment success. Several physicians in northern Israel noted that the Arab population has a higher treatment failure rate compared to the Jewish population, a finding which triggered a study that examined antibiotic resistance of H. pylori among these two groups in Israel[8]. Indeed, isolates from the Arab population were more resistant to both clarithromycin and levofloxacin and exhibited simultaneous resistance to more antibiotics as compared to isolates from Jewish patients[8]. The current study further examined the characteristics of H. pylori in adult Arab and Jewish populations in northern Israel, with an emphasis on the three virulence markers of H. pylori and their correlation with clinical outcomes.
The study group included 108 isolates of patients who underwent gastroscopy at the Baruch Padeh Medical Center, Poriya, due to symptomatic gastroduodenal pathologies as part of the H. pylori workup between November 2018 and December 2019. We included the first 108 biopsies from which H. pylori was successfully isolated at the microbiology laboratory of the medical center. The study was approved by the Helsinki Committee of the Baruch Padeh Medical Center, Poriya (Approval no. POR 0007-20). The Institutional Review Board committee waived the need for participant approval. Gastric biopsy specimens were collected from the antrum of the stomach. Clinical pathology tests were performed to assess the patients’ clinical conditions. Demographic and clinical data were retrospectively collected from the patients’ medical records.
Gastric biopsy specimens were stained using hematoxylin and eosin staining method in order to identify H. pylori and to evaluate the degree of inflammation. The bacterium has an actively dividing spiral shape that changes to coccoid morphology under stressful environments.
All histologic slides were reviewed by a single blinded gastrointestinal pathologist at the pathology laboratory of the Baruch Padeh Medical Center, Poriya and graded unremarkable (none), mild, moderate, or severe, based on the presence of acute (polymorphonuclear cells) or chronic (monocytes, lymphocytes, plasma cells) inflammation, lymphoid aggregates, and metaplasia.
H. pylori identification was carried out in accordance with the routine identification tests of the clinical microbiology laboratory including a Gram stain, oxidase, catalase, and urease tests.
Biopsy specimens were manually minced with a sterile scalpel, seeded on modified BD Helicobacter Agar plates (BD Diagnostics, Sparks, MD), and incubated for 7 d at 35 °C in a microaerobic atmosphere (5% O2 and 10% CO2) produced by a gas-generating system adapted for Campylobacter (CampyGen™; Gamidor Diagnostics, Petah Tikva, Israel).
Final identification of the bacteria was performed by matrix-assisted laser desorption ionization-time of flight mass spectrometry[9], using the Bruker Biotyper system (Bruker Daltonics, Bremen, Germany) with MALDI BIOTYPER 3.3 (Bruker Daltonics) software.
DNA extraction: Tissue collected from gastroscopic biopsy was finely chopped with a sterile scalpel and then lysed by tissue lysis buffer supplemented with proteinase K enzyme (Bioneer, Daejeon, Korea). Total DNA was extracted using the AccuPrep Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea), according to the manufacturer’s instructions.
Multiplex PCR assay: DNA was amplified with a multiplex PCR, designed to detect the VacA and CagA genes in a single run, using specific primers (Table 1). For this purpose, 10 μL Taq ReadyMix2 (Hy labs, Rehovot, Israel) was added to 0.2 μL forward primer, 0.2 μL reverse primer, 4 μL template DNA, and 5.6 μL nuclease-free water. Reaction conditions were 35 cycles of: Denaturation of the pre-amplified templates at 95 °C for 1 min, followed by an annealing step at 72 °C for 1 min, an extension step at 25 °C for 1 min and one additional extension step for 7 min. PCR products were visualized by 1% agarose gel electrophoresis.
| Primer | Gene | Primer sequence | Product size (bp) |
| CAGAF | CagA | 5’-GATAACAGGCAAGCTTTTGAGG-3’ | 349 |
| CAGAR | 5’-CTGCAAAAGATTGTTTGGCAGA-3’ | ||
| VA1-F | VacA signal region | 5’-ATGGAAATACAACAAACACAC-3’ | 259/286 (s1/s2) |
| VA1-R | 5’-CTGCTTGAATGCGCCAAAC-3’ | ||
| VAG-F | VacA middle region | 5’-CAATCTGTCCAATCAAGCGAG-3’ | 567/642 (m1/m2) |
| VAG-R | 5’-GCGTCTAAATAATTCCAAGG-3’ |
Urease activity was quantified using the rapid urease test in a 96-well plate. For this purpose, bacteria were placed in a sterile Eppendorf tube containing sterile physiological solution until 0.5 McFarland turbidity was reached. Rapid Urease Test Brute solution (100 μL; Novamed, Jerusalem, Israel), containing 2% urea, pH 6.8, was then added to each well, along with 100 μL of the bacterial stock. Then, absorbance at 570 nm (optical density, O.D570) was measured after 1 min, 5 min and 10 min using the Multiskan™ FC Microplate Photometer (Thermo Fisher Scientific, Waltham, MA, United States)[10]. A change in the solution color from orange to pink was considered a positive result.
Continuous variables (age, urease activity) were presented as means and ranges with standard deviations and categorical variables (sex, disease severity, ethnicity, living area, genotypes) were presented as absolute numbers and percentages. T test or one-way analysis of variance was used to compare continuous variables of two or more groups and Fisher’s exact test to assess the relationship between categorical variables. P value < 0.05 indicated statistical significance. Statistical analysis was performed using R Statistical Software (version 4.1; R Foundation for Statistical Computing, Vienna, Austria).
In total, 108 patients [average of 42.3 (18.0-88.0) years] were enrolled in this study, 24% of whom were males and 76% of whom were females (Table 2). Within the study group, 56 were Arabs and 52 were Jews; 61.1% lived in a city and 33.9% lived in a village. A larger percentage of the Arab vs Jewish cohort lived in villages (62.5% vs 13.5%, respectively).
| Characteristic | Arabs, n = 56 | Jews, n = 52 | Total, n = 108 |
| Sex | |||
| Male | 14 (25) | 39 (23) | 26 (24) |
| Female | 84 (75) | 64 (77) | 82 (76) |
| Age, yr | |||
| Average (range) | 40.93 (18-81) | 43.65 (18-88) | 42.30 (18-88) |
| Area of residence | |||
| City | 21 (37.5) | 45 (86.5) | 66 (61.1) |
| Village | 35 (62.5) | 7 (13.5) | 42 (33.9) |
To characterize the common CagA and VacA genotypes in the patient population, 108 isolates were randomly selected, of which 56 originated from the Arab cohort and 52 from the Jewish cohort (Table 3). CagA was identified in 24 (22.2%) samples and was distributed equally between the two ethnic groups. However, it had a higher frequency among isolates from patients living in villages (67.7%) compared to isolates from those living in cities (33.3%) (P = 0.149). The most prevalent VacA genotype was s2m1. No statistically significant associations were noted between the different genotypes and demographic characteristics.
| Characteristic | CagA-positive, n = 24 | CagA-negative, n = 84 | P | VacA genotype | P | |||
| s1m1 | s1m2 | s2m1 | s2m2 | |||||
| Ethnicity | 0.834 | 0.729 | ||||||
| Arabs | 12 (50.0) | 40 (47.6) | 11 (21.2) | 11 (21.2) | 27 (51.9) | 3 (5.8) | ||
| Jewish | 12 (50.0) | 44 (52.4) | 8 (14.3) | 15 (26.8) | 31 (55.4) | 2 (3.6) | ||
| Sex | 0.623 | 0.214 | ||||||
| Male | 7 (29.2) | 29 (34.5) | 6 (16.7) | 5 (13.9) | 22 (61.1) | 3 (8.3) | ||
| Female | 17 (70.8) | 55 (65.5) | 13 (18.1) | 21 (29.2) | 36 (50.0) | 2 (2.8) | ||
| Residence | 0.149 | 0.055 | ||||||
| Village | 16 (67.7) | 42 (50.0) | 8 (13.8) | 17 (29.3) | 28 (48.3) | 5 (8.6) | ||
| City | 8 (33.3) | 42 (50.0) | 11 (22.0) | 9 (18.0) | 30 (60.0) | 0 (0) | ||
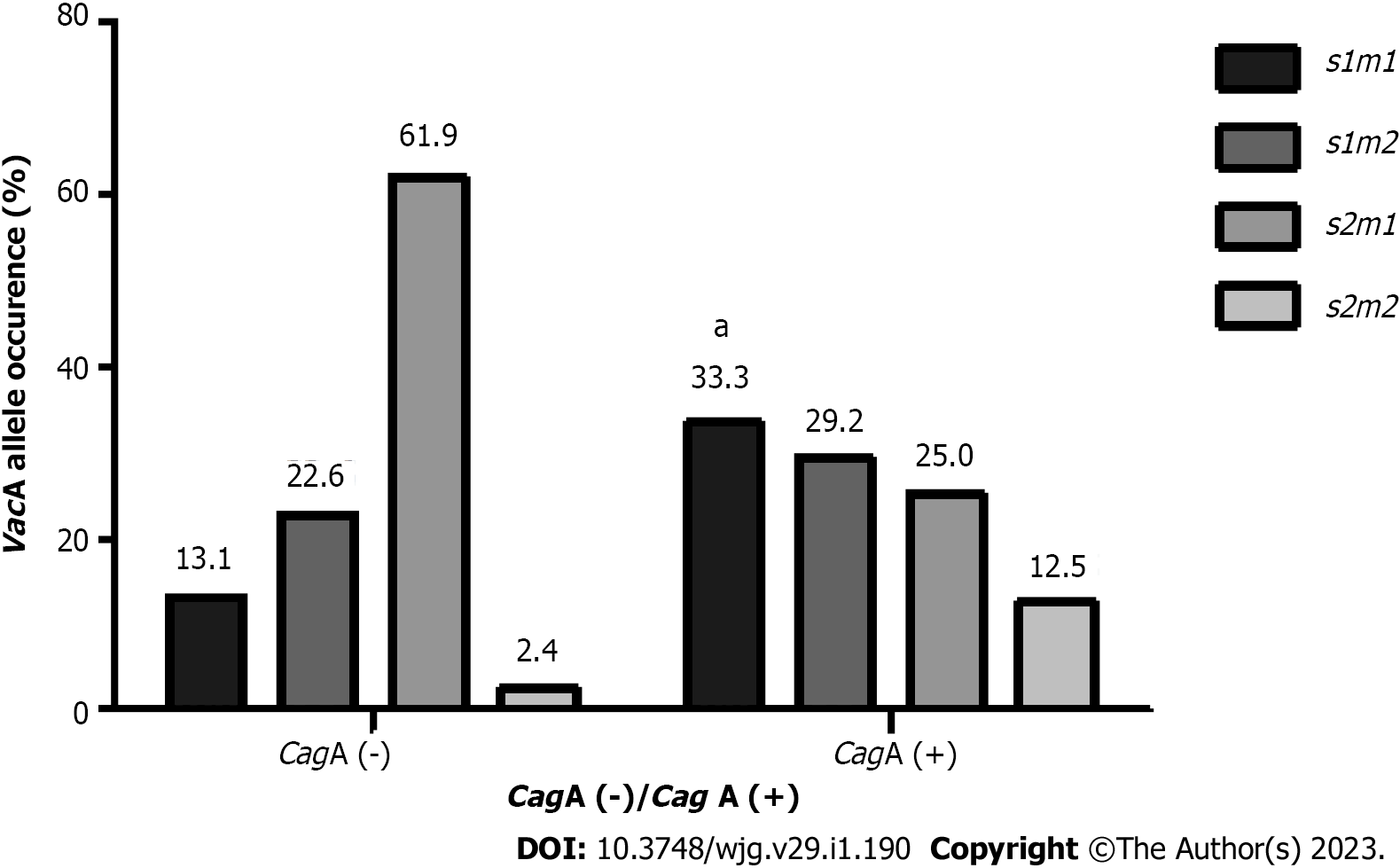
A significant association was found between the presence of CagA and specific VacA genotypes (P = 0.002) (Figure 1); 33.3% of the CagA-positive strains had the s1m1 genotype, which is considered the most virulent genotype[7]. Additionally, 61.9% of the CagA-negative strains had the s2m1 genotype.
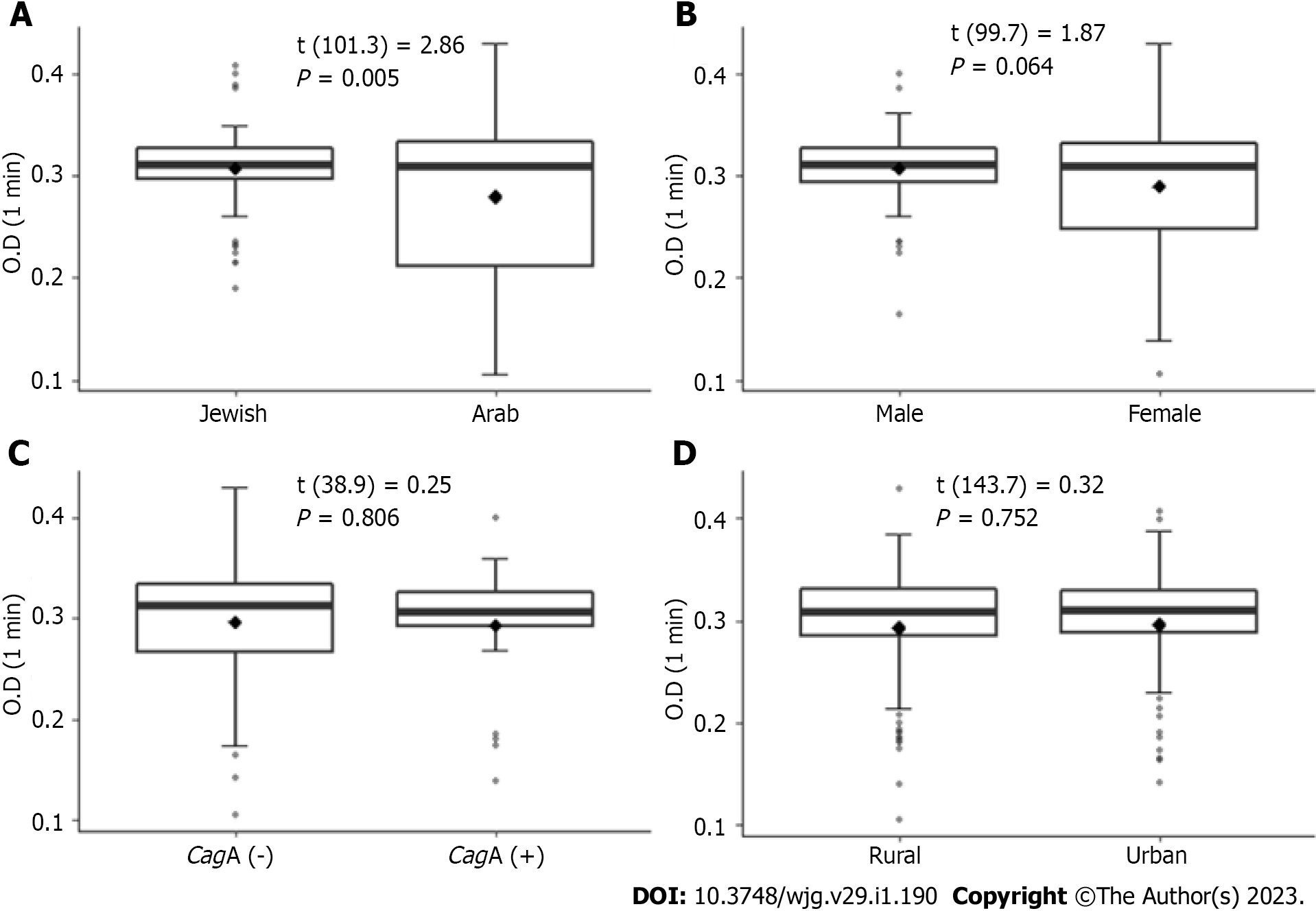
Urease activity was found to be faster in isolates from the Jewish population compared to isolates from the Arab population. This was observed at all tested time points (1 min, 5 min, and 10 min; P < 0.05 for all). The results of the first-minute measurements are presented in Figure 2. No significant associations were found between urease activity and patient’s sex, place of residence and the presence of the CagA gene.
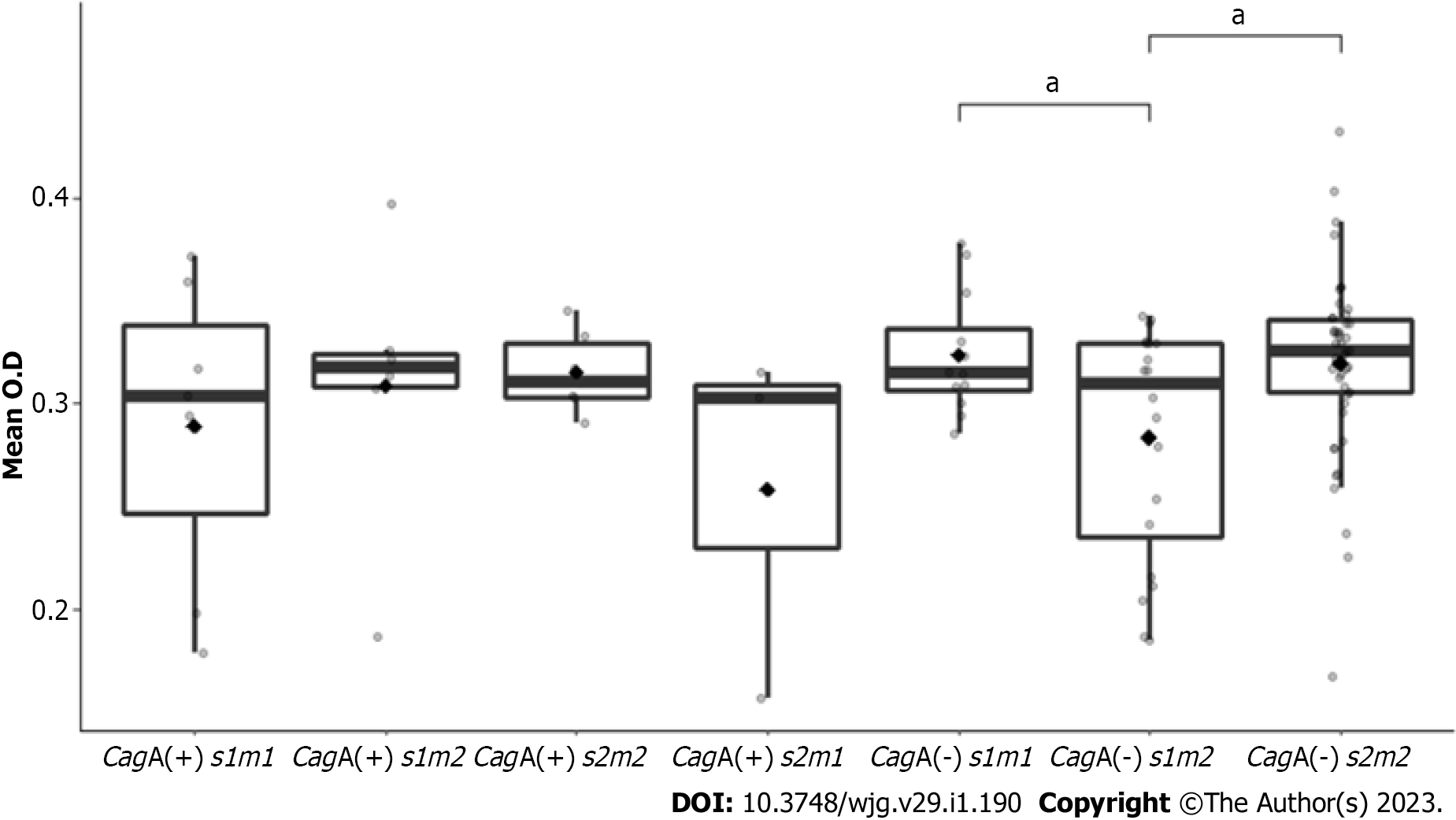
In addition, urease activity in CagA (-) s2m2 strains (mean O.D 0.33) and in CagA (-) s1m1 isolates (mean O.D 0.32) were significantly higher than urease activity in CagA (-) s1m2 (mean O.D 0.28), (P = 0.013 and P = 0.016, respectively) (Figure 3).
A significant association was found between disease severity and ethnicity (P =0.002) (Table 4); in patients with mild disease, 58% were Jews while 42% were Arabs. In patients with moderate disease, 75.7% were Arabs compared to 24.3% Jews. Finally, the severe group included 66.7% Arabs compared to 33.3% Jews. No significant links were found between disease severity and urease activity, CagA gene occurrence, VacA alleles, and genotype combinations.
| Disease severity | P value | |||||
| Unremarkable, n = 18 | Mild, n = 50 | Moderate, n = 37 | Severe, n = 3 | Total, n = 108 | ||
| Ethnicity | 0.002 | |||||
| Arab | 5 (27.8) | 21 (42.0) | 28 (75.7) | 2 (66.7) | 56 (51.9) | |
| Jewish | 13 (72.2) | 29 (58.0) | 9 (24.3) | 1 (33.3) | 52 (48.1) | |
| Urease activity | ||||||
| t1 | 0.32 (0.05) | 0.29 (0.06) | 0.29 (0.07) | 0.30 (0.06) | 0.30 (0.06) | 0.273 |
| t5 | 0.32 (0.05) | 0.30 (0.05) | 0.29 (0.07) | 0.34 (0.02) | 0.30 (0.06) | 0.174 |
| t10 | 0.32 (0.05) | 0.30 (0.06) | 0.30 (0.06) | 0.35 (0.04) | 0.30 (0.06) | 0.171 |
| t15 | 0.33 (0.04) | 0.31 (0.05) | 0.31 (0.06) | 0.36 (0.03) | 0.31 (0.05) | 0.245 |
| CagA gene | 0.534 | |||||
| CagA- | 13 (72.2) | 41 (82.0) | 27 (73.0) | 3 (100) | 84 (77.8) | |
| CagA+ | 5 (27.8) | 9 (18.0) | 10 (27.0) | 0 (0) | 24 (22.2) | |
| VacA s allele | 0.651 | |||||
| VacA s1 | 5 (27.8) | 22 (44.0) | 16 (43.2) | 1 (33.3) | 44 (40.7) | |
| VacA s2 | 13 (72.2) | 28 (56.0) | 21 (56.8) | 2 (66.7) | 64 (59.3) | |
| VacA m Allele | 0.652 | |||||
| VacA m1 | 3 (16.7) | 14 (28.0) | 7 (18.9) | 1 (33.3) | 25 (23.1) | |
| VacA m2 | 15 (83.3) | 36 (72.0) | 30 (81.1) | 2 (66.7) | 83 (76.9) | |
| Genotype | 0.926 | |||||
| CagA+/s1m1 | 0 (0) | 3 (6.0) | 4 (10.8) | 0 (0) | 7 (6.5) | |
| CagA+/s1m2 | 1 (5.6) | 2 (4.0) | 3 (8.1) | 0 (0) | 6 (5.6) | |
| CagA+/s2m2 | 2 (11.1) | 2 (4.0) | 2 (5.4) | 0 (0) | 6 (5.6) | |
| CagA+/s2m1 | 1 (5.6) | 1 (2.0) | 1 (2.7) | 0 (0) | 3 (2.8) | |
| CagA-/s1m1 | 2 (11.1) | 7 (14.0) | 2 (5.4) | 1 (33.3) | 12 (11.1) | |
| CagA-/s1m2 | 2 (11.1) | 11 (22.0) | 7 (18.9) | 0 (0) | 20 (18.5) | |
| CagA-/s2m2 | 10 (55.6) | 22 (44.0) | 18 (48.6) | 2 (66.7) | 52 (48.1) | |
| CagA-/s2m1 | 0 (0) | 2 (4.0) | 0 (0) | 0 (0) | 2 (1.9) | |
The main purpose of this study was to evaluate and compare the virulence markers of H. pylori among two adult populations in northern Israel and their correlation with clinical outcomes. It should be noted that, due to its special growth requirements and slow growth, cultivation of H. pylori is difficult. As a result, the diagnosis of H. pylori infection is usually performed, as opposed to diagnosis of other bacterial infections, on the basis of indirect tests that do not require bacterial isolation[11]. This is the reason why there is limited data on the distribution of virulence factors and their associations with clinical outcomes among H. pylori isolates in Israel.
CagA was found in 22.2% of the detected strains. This rate is a bit lower than expected according to previous studies; the prevalence of the cytotoxin-associated gene pathogenicity island, which includes CagA, varies between different geographic areas, ranging from 95% in Western and South Africa and East and Central Asia to 28% in Latin America. In Europe, approximately 58% of the H. pylori strains carry the CagA gene, while in the Middle East, CagA was detected in approximately 50% of the strains[12,13]. As only a sample of our isolates were tested for the presence of CagA, it is possible that a higher prevalence of this gene exists among H. pylori strains in Israel.
We did not find significant differences in the distribution of CagA between the two populations. Interestingly, Muhsen et al[14], who also investigated H. pylori isolates in Arab and Jewish populations, demonstrated higher CagA IgG antibodies in the Arab population. Another study, performed in Israeli children and adolescents, found higher H. pylori seroprevalence among Arab participants as compared to Jewish participants[15].
Regarding the virulence factor VacA, s2m1 genotype was the most common, present in 53.3% of the isolates, while only 17.6% carried the s1m1 genotype. In a study conducted in South Africa, s1m1 was the most common genotype (56.4%), while the s2m1 genotype was present in only 10.3%[16]. In a similar study conducted in Iran, the most common genotype was s2m2, found in 50% of the isolates[17]. It should be noted that the frequency of the alleles in different populations is influenced by several factors, including evolution, natural selection, mutations, and genetic drift[18].
CagA was identified most frequently together with the VacA genotype s1m1 (P = 0.02), which is considered the most virulent[4]. This result reinforces evidence from a previous study, which found that most VacA s1 strains carry CagA as well[19,20]. Previous studies suggested that the presence of CagA together with certain genotypes of VacA can indicate disease severity. For example, its appearance with the genotype s1m2 or s1m1 was correlated with the appearance of peptic ulcers, while its appearance with s2m2 was correlated with gastritis[17]. In light of the above, it is important to profile the VacA and CagA variants in patients in order to assess disease progression. This may aid in optimizing medical treatment.
Apart from VacA and CagA, urease activity is another indicator of H. pylori virulence. We found a higher urease activity among isolates from the Jewish population as compared to those from Arabs (P < 0.005). No previous study has investigated this issue. As urease activity is influenced by specific food ingredients such as isothiocyanates[10] and essential oils[21], variations in nutrition habits may explain the difference in urease activity between isolates from Arab and Jewish patients. Further studies are needed to confirm our result and investigate its meaning.
Significant differences in urease activity were noted among isolates with specific CagA and VacA genotype combinations. Both CagA (-) s1m1 and CagA (-) s2m2 showed the highest urease activity. This result shows that the absence of CagA may result in increased urease activity, especially when combined with the most virulent alleles of the VacA gene, s1m1, as previously suggested[6].
Given that antibiotic resistance was found to be higher in the Arab population[8], we cautiously suggest that increased urease activity does not coincide with increased antibiotic resistance. In our preliminary analysis, no significant correlation was found between disease severity and urease activity. These results were quite surprising as previous studies did report on such associations[22]. It is possible that this contradiction is due to our relatively small sample size. However, we did find a significant correlation between ethnicity and disease severity (P = 0.002). This finding may be ascribable to differences in health-related lifestyle between different ethnic groups as well as differences in socioeconomic conditions along with cultural and social customs between groups related to their residential environments, which can undoubtedly affect morbidity. Furthermore, it can also be explained by the high antibiotic resistance found among the Arab population[8].
Our study highlighted the importance of incorporating molecular methods for detection of virulence markers of H. pylori in order to tailor optimal treatments for each patient. Further investigation should be performed regarding associations between H. pylori virulence factors and ethnicity.
Helicobacter pylori (H. pylori) is a microaerophilic, Gram-negative bacterium that colonizes the gastric mucosa and infects the stomach epithelium, causing peptic ulcer disease. The genetic variability of H. pylori and its host, combined with environmental factors, have been suggested to affect the clinical outcome. H. pylori pathogenesis is mediated via distinct virulence factors, including the secreted vacuolating cytotoxin A, cytotoxin-associated gene A, and urease.
In recent years, associations between specific virulence markers of H. pylori and gastrointestinal disorders have been suggested.
To investigate the distribution of three virulence factors among isolates from both Arab and Jewish populations and to assess their impact on clinical presentations.
We enrolled 108 patients tested for the presence of vacuolating cytotoxin A and cytotoxin-associated gene A genes and evaluated the urease activity levels. We assessed the clinical state of the patients by hematoxylin and eosin staining of the gastric biopsies from which the bacteria were recovered.
We found associations between disease severity and ethnicity and between some of the virulence factors to ethnicity.
Our study highlighted the importance of incorporating molecular methods for detection of virulence markers of H. pylori in order to tailor optimal treatments for each patient.
Further investigation should be performed regarding associations between H. pylori virulence factors and ethnicity.
We thank Mr. Wadie Abu Dahoud for the statistical analysis.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: Israel
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Netto ERA, Brazil; Selvin ST, United States; Zhang J, China; Zojaji M, Iran S-Editor: Chen YL L-Editor: Filipodia P-Editor: Chen YL
| 1. | Moreno-Ochoa MF, Valencia ME, Morales-Figueroa GG, Moya-Camarena SY. Association of cagA+ Helicobacter pylori strains with high urease activity and dyspepsia in Mexican adults. Rev Gastroenterol Mex (Engl Ed). 2020;85:404-409. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 2. | Panayotopoulou EG, Sgouras DN, Papadakos KS, Petraki K, Breurec S, Michopoulos S, Mantzaris G, Papatheodoridis G, Mentis A, Archimandritis A. CagA and VacA polymorphisms are associated with distinct pathological features in Helicobacter pylori-infected adults with peptic ulcer and non-peptic ulcer disease. J Clin Microbiol. 2010;48:2237-2239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Palframan SL, Kwok T, Gabriel K. Vacuolating cytotoxin A (VacA), a key toxin for Helicobacter pylori pathogenesis. Front Cell Infect Microbiol. 2012;2:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 200] [Article Influence: 15.4] [Reference Citation Analysis (2)] |
| 4. | Sheikh AF, Yadyad MJ, Goodarzi H, Hashemi SJ, Aslani S, Assarzadegan MA, Ranjbar R. CagA and vacA allelic combination of Helicobacter pylori in gastroduodenal disorders. Microb Pathog. 2018;122:144-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Hatakeyama M, Higashi H. Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis. Cancer Sci. 2005;96:835-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 183] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 6. | Ghalehnoei H, Ahmadzadeh A, Farzi N, Alebouyeh M, Aghdaei HA, Azimzadeh P, Molaei M, Zali MR. Relationship between ureB Sequence Diversity, Urease Activity and Genotypic Variations of Different Helicobacter pylori Strains in Patients with Gastric Disorders. Pol J Microbiol. 2016;65:153-159. [PubMed] |
| 7. | Shimoyama T, Fukuda S, Liu Q, Nakaji S, Fukuda Y, Sugawara K. Helicobacter pylori water soluble surface proteins prime human neutrophils for enhanced production of reactive oxygen species and stimulate chemokine production. J Clin Pathol. 2003;56:348-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Yeganeh M, Paritsky M, On A, Azrad M, Roshrosh H, Moalem R, Peretz A. Characteristics of Antibiotic Resistance of Helicobacter pylori Among Adult Arab and Jewish Populations in Northern Israel. Microb Drug Resist. 2019;25:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015;6:791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 675] [Cited by in RCA: 895] [Article Influence: 89.5] [Reference Citation Analysis (0)] |
| 10. | Fahey JW, Stephenson KK, Wade KL, Talalay P. Urease from Helicobacter pylori is inactivated by sulforaphane and other isothiocyanates. Biochem Biophys Res Commun. 2013;435:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 11. | Perez-Perez GI. Accurate diagnosis of Helicobacter pylori. Culture, including transport. Gastroenterol Clin North Am. 2000;29:879-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 177] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 13. | Akeel M, Shehata A, Elhafey A, Elmakki E, Aboshouk T, Ageely H, Mahfouz M. Helicobacter pylori vacA, cagA and iceA genotypes in dyspeptic patients from southwestern region, Saudi Arabia: distribution and association with clinical outcomes and histopathological changes. BMC Gastroenterol. 2019;19:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 14. | Muhsen K, Barak M, Shifnaidel L, Nir A, Bassal R, Cohen D. Helicobacter pylori infection is associated with low serum ferritin levels in Israeli Arab children: a seroepidemiologic study. J Pediatr Gastroenterol Nutr. 2009;49:262-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Muhsen K, Sinnereich R, Beer-Davidson G, Nassar H, Abu Ahmed W, Cohen D, Kark JD. Correlates of infection with Helicobacter pylori positive and negative cytotoxin-associated gene A phenotypes among Arab and Jewish residents of Jerusalem. Epidemiol Infect. 2019;147:e276. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Idowu A, Mzukwa A, Harrison U, Palamides P, Haas R, Mbao M, Mamdoo R, Bolon J, Jolaiya T, Smith S, Ally R, Clarke A, Njom H. Detection of Helicobacter pylori and its virulence genes (cagA, dupA, and vacA) among patients with gastroduodenal diseases in Chris Hani Baragwanath Academic Hospital, South Africa. BMC Gastroenterol. 2019;19:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (2)] |
| 17. | Pajavand H, Alvandi A, Mohajeri P, Bakhtyari S, Bashiri H, Kalali B, Gerhard M, Najafi F, Abiri R. High Frequency of vacA s1m2 Genotypes Among Helicobacter pylori Isolates From Patients With Gastroduodenal Disorders in Kermanshah, Iran. Jundishapur J Microbiol. 2015;8:e25425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Chen N, Juric I, Cosgrove EJ, Bowman R, Fitzpatrick JW, Schoech SJ, Clark AG, Coop G. Allele frequency dynamics in a pedigreed natural population. Proc Natl Acad Sci U S A. 2019;116:2158-2164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 19. | Atherton JC, Cao P, Peek RM Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771-17777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1078] [Cited by in RCA: 1108] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 20. | Homan M, Luzar B, Kocjan BJ, Orel R, Mocilnik T, Shrestha M, Kveder M, Poljak M. Prevalence and clinical relevance of cagA, vacA, and iceA genotypes of Helicobacter pylori isolated from Slovenian children. J Pediatr Gastroenterol Nutr. 2009;49:289-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Korona-Glowniak I, Glowniak-Lipa A, Ludwiczuk A, Baj T, Malm A. The In Vitro Activity of Essential Oils against Helicobacter Pylori Growth and Urease Activity. Molecules. 2020;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 22. | Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, Maciejewski R. Helicobacter pylori Virulence Factors-Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment. Cells. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 213] [Article Influence: 42.6] [Reference Citation Analysis (0)] |