Published online Aug 14, 2022. doi: 10.3748/wjg.v28.i30.4221
Peer-review started: July 30, 2021
First decision: August 19, 2021
Revised: August 22, 2021
Accepted: July 18, 2022
Article in press: July 18, 2022
Published online: August 14, 2022
The expression of angiopoietin (ANGPT) 1, ANGPT2, vascular endothelial growth factor (VEGF) A, VEGFB, VEGFC, VEGFD, and placental growth factor (PGF) is significantly higher in tumor tissues than in normal tissues in both unpaired and paired hepatocellular carcinoma (HCC) samples. ANGPT2, VEGFB, VEGFC, and PGF are primarily involved in regulating the activation of the epithelial-mesenchymal transition pathway; ANGPT1 is primarily involved in regulating the activation of the RAS/mitogen-activated protein kinase and receptor tyrosine kinase (RTK) pathways; VEGFA is engaged in regulating the RTK activation pathway; and VEGFD is mainly involved in regulating the activation of the tuberous sclerosis protein/mammalian target of rapamycin pathway. There is a significant difference in overall survival between HCC patients with high and low expression of ANGPT2, PGF, VEGFA, and VEGFD. Disease free survival (DFS) is significantly shorter in HCC patients with high ANGPT2, PGF, and VEGFA expression than in those with low ANGPT2, PGF, and VEGFA expression.
Core Tip: We found that the expression of angiogenesis markers was significantly higher in tumor tissues than in normal tissues in both unpaired and paired hepatocellular carcinoma (HCC) samples. These angiogenesis markers are mainly involved in regulating the activation of the EMT pathway, the RAS/mitogen-activated protein kinase and receptor tyrosine kinase pathways, and the tuberous sclerosis protein/mammalian target of rapamycin pathway. In addition, there was a significant difference in overall survival between HCC patients with high and low expression of angiopoietin-2 (ANGPT2), placental growth factor (PGF), vascular endothelial growth factor A (VEGFA), and VEGFD. Disease free survival was significantly shorter in HCC patients with high ANGPT2, PGF, and VEGFA expression than in those with low ANGPT2, PGF, and VEGFA expression.
- Citation: Miao YD, Tang XL, Wang JT, Mi DH. Prognostic role of expression of angiogenesis markers in hepatocellular carcinoma: A bioinformatics analysis. World J Gastroenterol 2022; 28(30): 4221-4226
- URL: https://www.wjgnet.com/1007-9327/full/v28/i30/4221.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i30.4221
We read with read interest the article by Choi et al[1], in which they initially evaluated plasma levels of angiogenesis biomarkers in hepatocellular carcinoma (HCC) patients, and then assessed their roles in forecasting overall survival (OS) and progression-free survival (PFS), indicating that the plasma level of angiopoietin (ANGPT) 2 was related to tumor stage, liver function, and cancer invasiveness, and that ANGPT2 performed better in predicting OS and PFS than alpha-fetoprotein (AFP), ANGPT1, and vascular endothelial growth factor (VEGF).
We appreciate the authors’ unique perspective in exploring the prognostic role of plasma levels of ANGPT1, ANGPT2, and VEGF in HCC. However, there are some errors in the original text that may cause confusion for readers. For example, the survival curve in figure 3B in the original article should have represented the survival curve between the high and low ANGPT2 expression subgroups, which the authors incorrectly labeled as ANGPT1. Second, it is well known that the VEGF family includes VEGFA, VEGFB, VEGFC, VEGFD, VEGFE, and placental growth factor (PGF)[2,3], so to which VEGF do the authors refer in the text? Usually, VEGF refers to VEGFA, but the authors should have clarified it in the text.
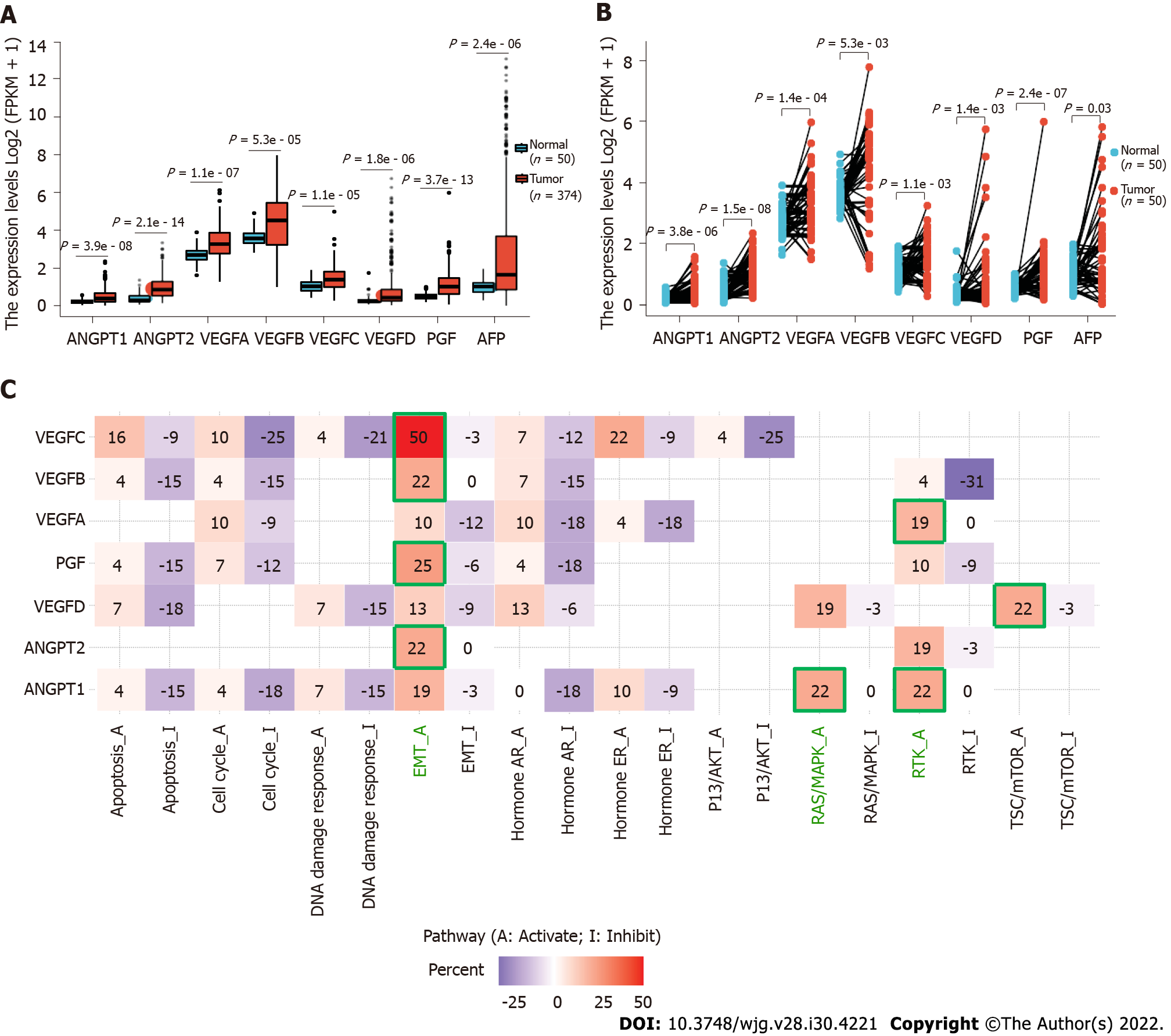
Moreover, it might make the results more significant if the authors could improve the outcome by demonstrating the differential expression of ANGPT1, ANGPT2, and VEGF in normal tissues and HCC tissues as a whole, for example, the analysis of HCC samples in the Cancer Genome Atlas database using bioinformatics. We found that the expression of ANGPT1, ANGPT2, VEGFA, VEGFB, VEGFC, VEGFD, and PGF was significantly higher in cancer samples than in corresponding normal samples in both unpaired and paired HCC samples (Figures 1A and B). Detailed statistical results are reported in Tables 1 and 2.
| Gene | Group | Number | Minimum | Maximum | Median | IQR | Lower quartile | Upper quartile | Mean | SD | SE |
| ANGPT1 | Normal | 50 | 0.029 | 0.552 | 0.188 | 0.132 | 0.138 | 0.27 | 0.206 | 0.106 | 0.015 |
| ANGPT1 | Tumor | 374 | 0 | 2.351 | 0.373 | 0.464 | 0.2 | 0.664 | 0.485 | 0.391 | 0.02 |
| ANGPT2 | Normal | 50 | 0.043 | 1.351 | 0.278 | 0.33 | 0.195 | 0.525 | 0.394 | 0.289 | 0.041 |
| ANGPT2 | Tumor | 374 | 0.116 | 3.339 | 0.848 | 0.769 | 0.513 | 1.282 | 0.963 | 0.581 | 0.03 |
| VEGFA | Normal | 50 | 1.616 | 3.901 | 2.687 | 0.473 | 2.439 | 2.911 | 2.717 | 0.445 | 0.063 |
| VEGFA | Tumor | 374 | 1.258 | 6.138 | 3.268 | 1.103 | 2.769 | 3.871 | 3.291 | 0.809 | 0.042 |
| VEGFB | Normal | 50 | 2.816 | 4.919 | 3.568 | 0.523 | 3.325 | 3.848 | 3.636 | 0.444 | 0.063 |
| VEGFB | Tumor | 374 | 0.978 | 8.003 | 4.532 | 2.234 | 3.223 | 5.458 | 4.292 | 1.521 | 0.079 |
| VEGFC | Normal | 50 | 0.408 | 1.901 | 1.019 | 0.453 | 0.787 | 1.239 | 1.057 | 0.355 | 0.05 |
| VEGFC | Tumor | 374 | 0.253 | 4.988 | 1.376 | 0.816 | 0.978 | 1.795 | 1.436 | 0.62 | 0.032 |
| VEGFD | Normal | 50 | 0.054 | 1.74 | 0.236 | 0.151 | 0.164 | 0.316 | 0.307 | 0.28 | 0.04 |
| VEGFD | Tumor | 374 | 0.014 | 6.756 | 0.422 | 0.622 | 0.241 | 0.863 | 0.838 | 1.14 | 0.059 |
| PGF | Normal | 50 | 0.182 | 0.992 | 0.471 | 0.204 | 0.37 | 0.575 | 0.501 | 0.188 | 0.027 |
| PGF | Tumor | 374 | 0.061 | 5.991 | 1.007 | 0.855 | 0.613 | 1.467 | 1.104 | 0.675 | 0.035 |
| AFP | Normal | 50 | 0.266 | 1.969 | 1.016 | 0.507 | 0.714 | 1.221 | 0.992 | 0.416 | 0.059 |
| AFP | Tumor | 374 | 0 | 13.118 | 1.644 | 2.855 | 0.844 | 3.699 | 2.965 | 3.15 | 0.163 |
| Gene | Group | Number | Minimum | Maximum | Median | IQR | Lower quartile | Upper quartile | Mean | SD | SE |
| ANGPT1 | Normal | 50 | 0.029 | 0.552 | 0.188 | 0.132 | 0.138 | 0.27 | 0.206 | 0.106 | 0.015 |
| ANGPT1 | Tumor | 50 | 0.014 | 1.557 | 0.463 | 0.56 | 0.228 | 0.788 | 0.507 | 0.363 | 0.051 |
| ANGPT2 | Normal | 50 | 0.043 | 1.351 | 0.278 | 0.33 | 0.195 | 0.525 | 0.394 | 0.289 | 0.041 |
| ANGPT2 | Tumor | 50 | 0.193 | 2.324 | 1.056 | 0.77 | 0.747 | 1.517 | 1.111 | 0.517 | 0.073 |
| VEGFA | Normal | 50 | 1.616 | 3.901 | 2.687 | 0.473 | 2.439 | 2.911 | 2.717 | 0.445 | 0.063 |
| VEGFA | Tumor | 50 | 1.471 | 5.974 | 3.102 | 1.087 | 2.801 | 3.888 | 3.287 | 0.902 | 0.128 |
| VEGFB | Normal | 50 | 2.816 | 4.919 | 3.568 | 0.523 | 3.325 | 3.848 | 3.636 | 0.444 | 0.063 |
| VEGFB | Tumor | 50 | 1.164 | 7.789 | 4.833 | 1.993 | 3.323 | 5.317 | 4.328 | 1.575 | 0.223 |
| VEGFC | Normal | 50 | 0.408 | 1.901 | 1.019 | 0.453 | 0.787 | 1.239 | 1.057 | 0.355 | 0.05 |
| VEGFC | Tumor | 50 | 0.261 | 3.233 | 1.398 | 0.819 | 1.013 | 1.831 | 1.459 | 0.633 | 0.09 |
| VEGFD | Normal | 50 | 0.054 | 1.74 | 0.236 | 0.151 | 0.164 | 0.316 | 0.307 | 0.28 | 0.04 |
| VEGFD | Tumor | 50 | 0.014 | 5.746 | 0.367 | 0.562 | 0.231 | 0.793 | 0.832 | 1.207 | 0.171 |
| PGF | Normal | 50 | 0.182 | 0.992 | 0.471 | 0.204 | 0.37 | 0.575 | 0.501 | 0.188 | 0.027 |
| PGF | Tumor | 50 | 0.144 | 5.991 | 1.072 | 0.833 | 0.67 | 1.503 | 1.16 | 0.859 | 0.121 |
| AFP | Normal | 50 | 0.266 | 1.969 | 1.016 | 0.507 | 0.714 | 1.221 | 0.992 | 0.416 | 0.059 |
| AFP | Tumor | 50 | 0 | 5.824 | 1.033 | 1.31 | 0.725 | 2.036 | 1.62 | 1.383 | 0.196 |
We also found that ANGPT2, VEGFB, VEGFC, and PGF are mainly involved in regulating the activation of the EMT pathway; ANGPT1 is prominently involved in regulating the activation of the RAS/mitogen-activated protein kinase and receptor tyrosine kinase (RTK) pathways; VEGFA is engaged in regulating the activation of the RTK pathway; and VEGFD is mainly involved in regulating the activation of the tuberous sclerosis protein/mammalian target of rapamycin pathway (Figure 1C). These results are consistent with those of previously reported studies[4-7]. Our findings could be a supplement to Choi et al’s study[1]. In the future, the roles of ANGPT1, ANGPT2, and VEGF in the development of HCC should be further explored.
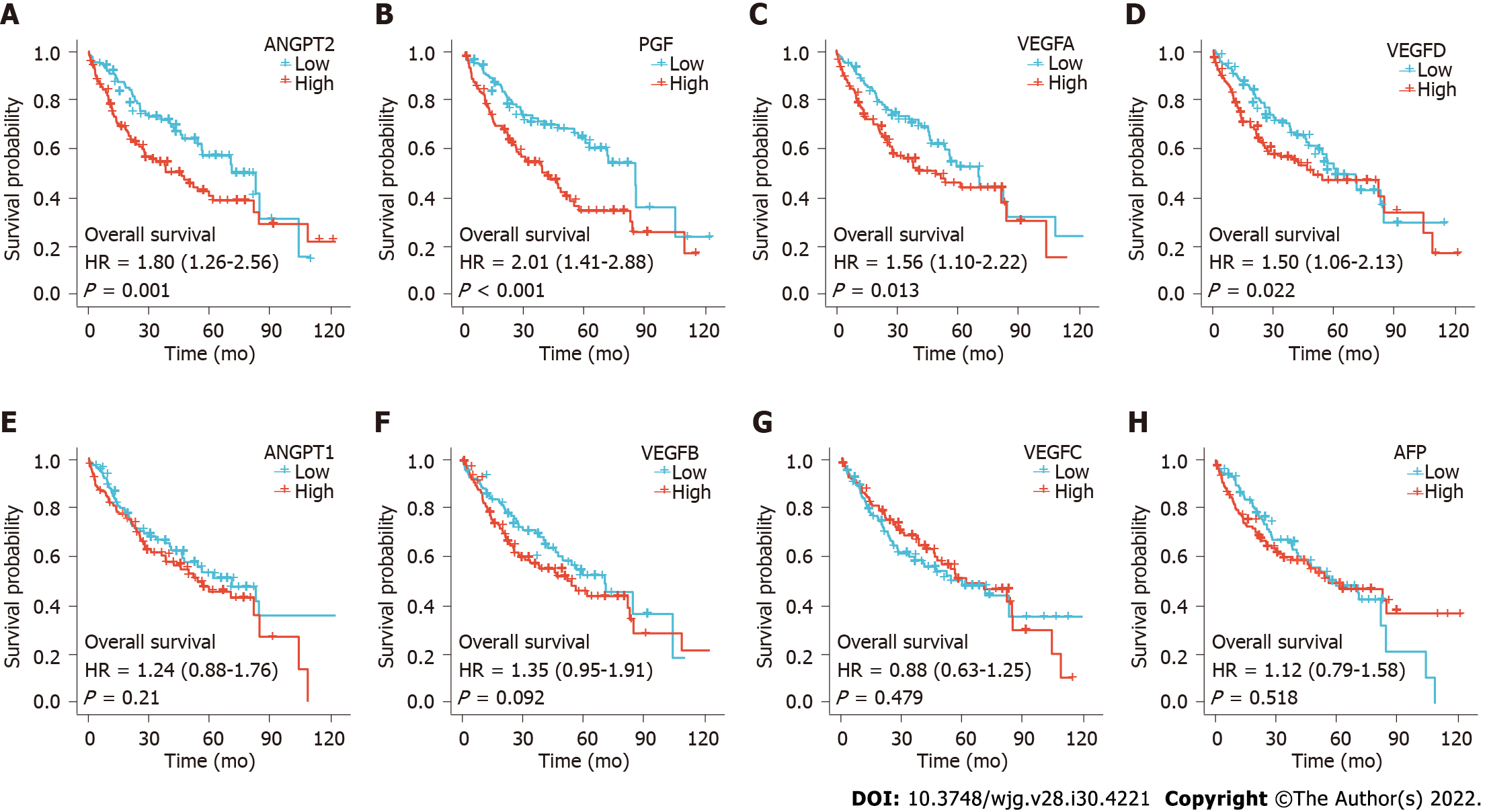
Choi et al[1] found that OS was significantly shorter in the high ANGPT2 and high AFP subgroups than in the low ANGPT2 and AFP subgroups, respectively, though the differences in OS rates were not significant between the high and low ANGPT1 subgroups or between the high and low VEGF subgroups. Our study found that OS was significantly shorter in patients with high ANGPT2, PGF, VEGFA, or VEGFD expression than in those with low expression, respectively (Figures 2A-D; P < 0.05). However, there was no significant difference in survival time between patients with high and low expression of ANGPT1, VEGFB, VEGFC, and AFP (Figures 2E-H; P > 0.05). Prognostic data for HCC came from Liu et al[8].
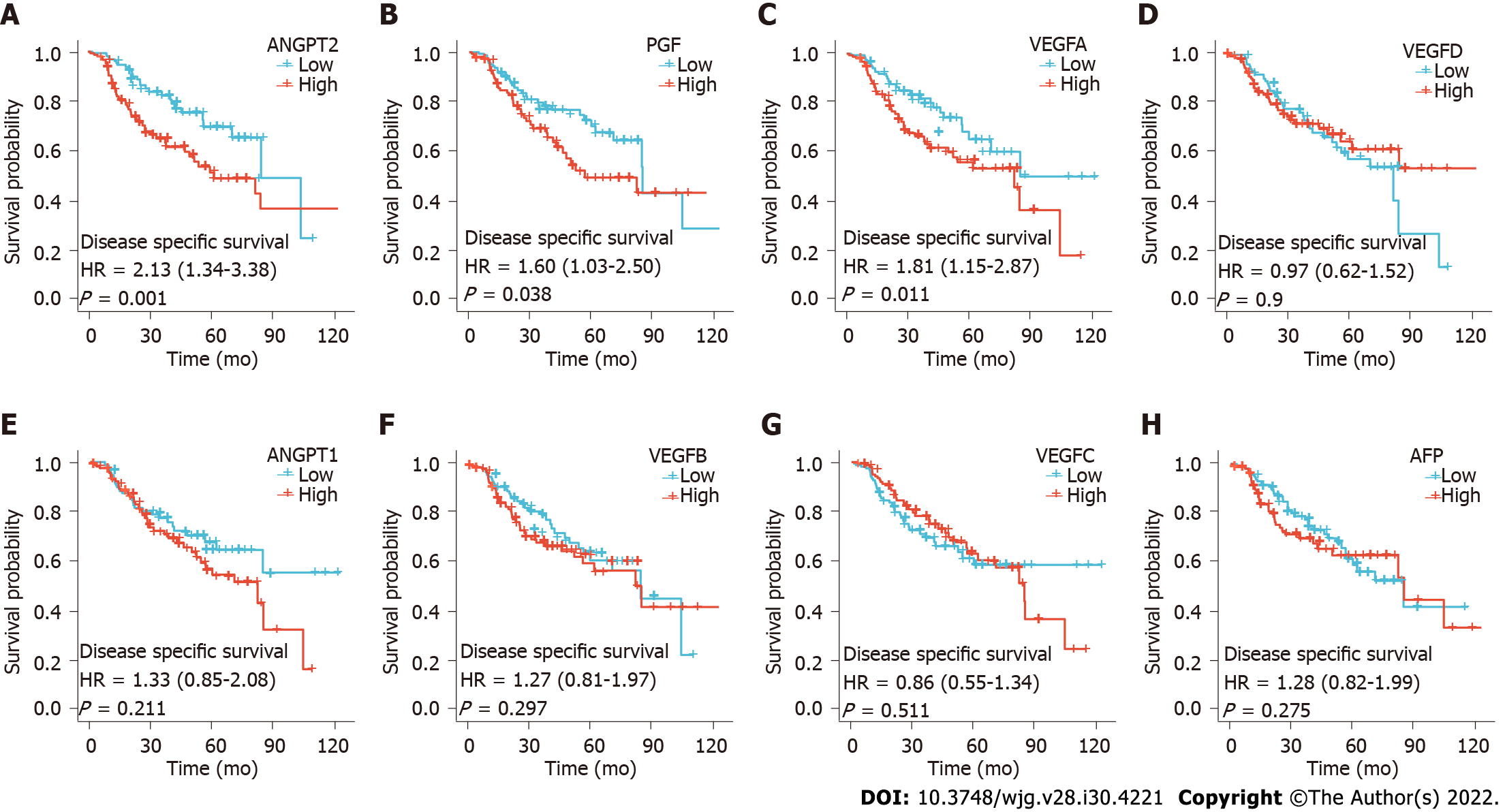
In addition, we also analyzed the differences in disease free survival (DFS) between patients with high and low angiogenesis marker expression. We found that DFS was significantly shorter in the high ANGPT2, PGF, and VEGFA groups than in the low ANGPT2, PGF, and VEGFA groups, respectively (Figures 3A, B and C; P < 0.05). However, there was no significantly difference in DFS between groups with high and low expression of AFP, ANGPT1, VEGFB, VEGFC, and VEGFD (Figures 3D-H; P > 0.05). The above results confirm that the study performed by Choi et al[1] is of great value and that our discovery could be a supplement to their research.
We utilized R (version 4.0.3) to perform statistical analyses and display the results. The differential expression analysis of angiogenesis markers between HCC tissues and corresponding normal tissues was performed using the Wilcoxon rank-sum test, and the results are presented by using R-package “ggplot2”[9]. Survival analysis was completed through log-rank test and COX regression. Pathway analysis was performed based on the online database GSCALite (http://bioinfo.life.hust.edu.cn/web/GSCALite/)[10].
We are grateful to the professors at the School of Foreign Languages of Lanzhou University for their help in the language polish of this manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhong C, China S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Choi GH, Jang ES, Kim JW, Jeong SH. Prognostic role of plasma level of angiopoietin-1, angiopoietin-2, and vascular endothelial growth factor in hepatocellular carcinoma. World J Gastroenterol. 2021;27:4453-4467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 2. | Heloterä H, Alitalo K. The VEGF family, the inside story. Cell. 2007;130:591-592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Thomas JL, Eichmann A. The power of VEGF (vascular endothelial growth factor) family molecules. Cell Mol Life Sci. 2013;70:1673-1674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Kong D, Zhou H, Neelakantan D, Hughes CJ, Hsu JY, Srinivasan RR, Lewis MT, Ford HL. VEGF-C mediates tumor growth and metastasis through promoting EMT-epithelial breast cancer cell crosstalk. Oncogene. 2021;40:964-979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 5. | Wang X, Xing Z, Xu H, Yang H, Xing T. Development and validation of epithelial mesenchymal transition-related prognostic model for hepatocellular carcinoma. Aging (Albany NY). 2021;13:13822-13845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | Bi X, Niu J, Ding W, Zhang M, Yang M, Gu Y. Angiopoietin-1 attenuates angiotensin II-induced ER stress in glomerular endothelial cells via a Tie2 receptor/ERK1/2-p38 MAPK-dependent mechanism. Mol Cell Endocrinol. 2016;428:118-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 7. | Chen H, Guan R, Lei Y, Chen J, Ge Q, Zhang X, Dou R, Chen H, Liu H, Qi X, Zhou X, Chen C. Lymphangiogenesis in gastric cancer regulated through Akt/mTOR-VEGF-C/VEGF-D axis. BMC Cancer. 2015;15:103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 8. | Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V; Cancer Genome Atlas Research Network, Hu H. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173:400-416.e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1998] [Cited by in F6Publishing: 1819] [Article Influence: 303.2] [Reference Citation Analysis (0)] |
| 9. | Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912-2914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 792] [Cited by in F6Publishing: 984] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 10. | Liu CJ, Hu FF, Xia MX, Han L, Zhang Q, Guo AY. GSCALite: a web server for gene set cancer analysis. Bioinformatics. 2018;34:3771-3772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 361] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 11. | Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, Schmidt H, Amstutz P, Craft B, Goldman M, Rosenbloom K, Cline M, O'Connor B, Hanna M, Birger C, Kent WJ, Patterson DA, Joseph AD, Zhu J, Zaranek S, Getz G, Haussler D, Paten B. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35:314-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 471] [Cited by in F6Publishing: 663] [Article Influence: 110.5] [Reference Citation Analysis (0)] |