Published online Aug 14, 2022. doi: 10.3748/wjg.v28.i30.4120
Peer-review started: November 26, 2021
First decision: January 8, 2022
Revised: January 21, 2022
Accepted: July 18, 2022
Article in press: July 18, 2022
Published online: August 14, 2022
Processing time: 256 Days and 18 Hours
As one of the most common tumors, gastric cancer (GC) has a high mortality rate, since current examination approaches cannot achieve early diagnosis. Fusobacterium nucleatum (Fn) primarily colonized in the oral cavity, has been reported to be involved in the development of gastrointestinal tumor. Until now, little is known about the relationship between salivary Fn and GC.
To determine whether salivary Fn could be a biomarker to diagnose GC and explore the influence of Fn on GC cells.
The abundance of Fn in saliva was quantified by droplet digital polymerase chain reaction in 120 GC patients, 31 atrophic gastritis (AG) patients, 35 non-AG (NAG) patients, 26 gastric polyp (GP) patients, and 20 normal controls (NC) from Qilu Hospital of Shandong University from January 2019 to December 2020. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of Fn as well as traditional serum tumor markers, including carcinoembryonic antigen (CEA), carbohydrate antigen (CA) 19-9, and CA72-4. Transwell assay and wound-healing assay were conducted to assess the influence of Fn infection on GC cells. The expression of epithelial-mesenchymal transition (EMT) markers was detected using western blot assay.
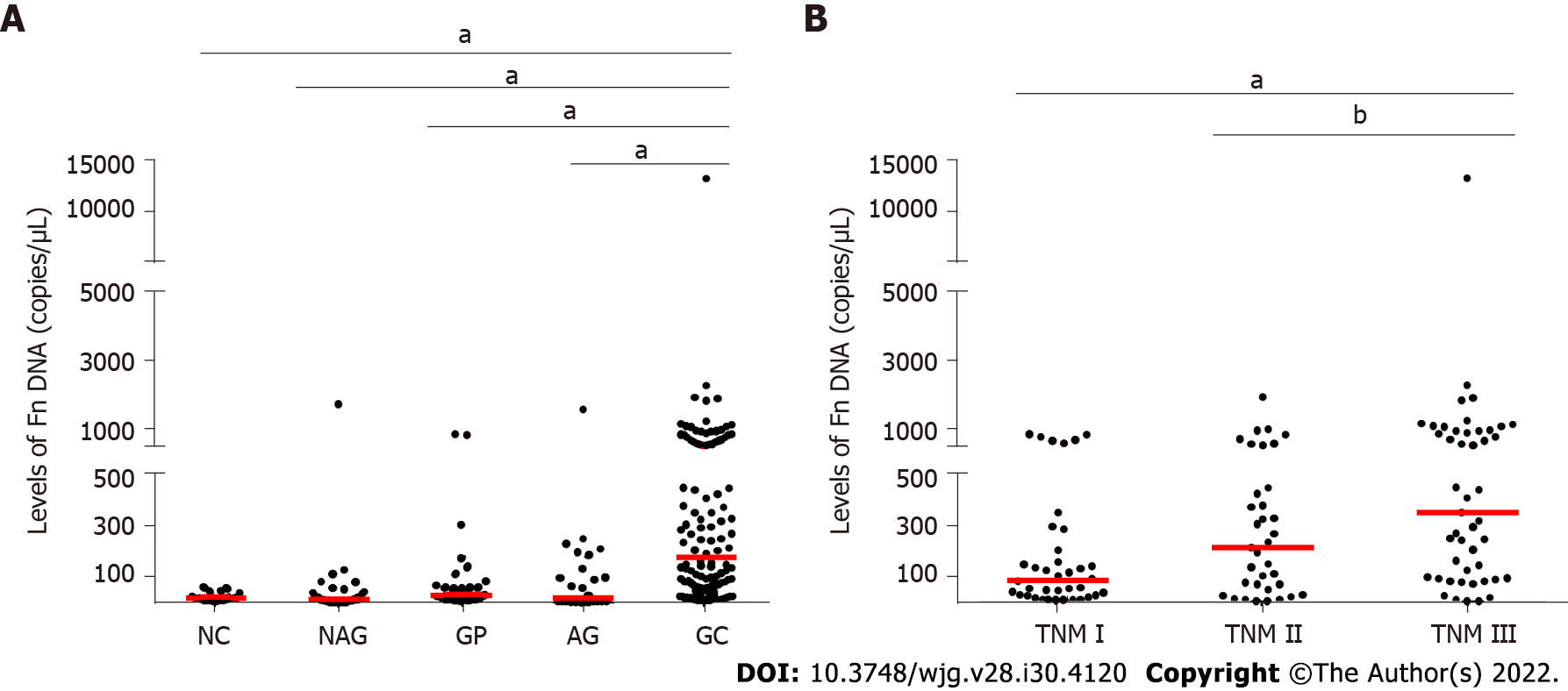
We found that the level of salivary Fn in GC patients was significantly increased compared with those in AG, NAG, and GP patients and NC (P < 0.001). ROC curve analysis showed a favorable capability of Fn (73.33% sensitivity; 82.14% specificity; area under the curve: 0.813) in GC diagnosis, which was superior to that of CEA, CA19-9, CA72-4, ferritin, and sialic acid. The Fn level in saliva of GC patients was increased as the TNM stage increased. GC patients with lymph node metastasis had higher Fn levels than those without metastasis. Both transwell and wound-healing assays indicated that Fn infection promoted the migration and invasion of GC cells. Western blot analysis showed that Fn infection decreased the expression of E-cadherin and increased the expressions of N-cadherin, vimentin, and snail.
Fn abundance in saliva could be used as a promising biomarker to diagnose GC, and Fn infection could promote GC metastasis by accelerating the EMT process.
Core Tip: Fusobacterium nucleatum (Fn), known as a periodontal pathogen, is frequently detected in tissues of gastrointestinal tumor. Here, we found that the level of salivary Fn was increased in gastric cancer (GC) patients, which could be used as a potential biomarker for GC diagnosis, and its diagnostic capability was superior to that of traditional serum tumor markers. We also showed that salivary Fn level was positively associated with the GC TNM stage and lymph node metastasis. Further, experiments in vitro revealed that Fn could promote the migration and invasion of GC cells by promoting the epithelial-mesenchymal transition process. Our findings suggest that salivary Fn is a potential biomarker in diagnosis of GC.
- Citation: Chen WD, Zhang X, Zhang MJ, Zhang YP, Shang ZQ, Xin YW, Zhang Y. Salivary Fusobacterium nucleatum serves as a potential diagnostic biomarker for gastric cancer. World J Gastroenterol 2022; 28(30): 4120-4132
- URL: https://www.wjgnet.com/1007-9327/full/v28/i30/4120.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i30.4120
Gastric cancer (GC) is one of the most common malignant tumors, with morbidity and mortality ranking fifth and third in the world, respectively, posing a great threat to global human health[1]. Although the 5-year survival rate of GC is enhanced with the combination of chemotherapy and surgery, its mortality remains high due to the diagnosis at an advanced stage with a limited therapeutic scheme to choose from. Therefore, effective early diagnosis for GC is especially important.
The microorganisms in the human body are complex and they play important roles in each part of our body. Over 700 bacterial species have been detected in the human oral cavity, and they interact with each other as well as with the host[2]. The stomach was considered a sterile organ until the discovery of Helicobacter pylori (H. pylori) in 1984[3]. Subsequently, other microorganisms were found in the gastric mucosa[4]. Although the gut microbiota ecosystem is different from that of the oral cavity, there is also a partial similarity between them, since most of the bacteria in the oral cavity enter the stomach when swallowing food and saliva.
Human saliva is a unique biofluid resource with huge clinical diagnostic and risk assessment capacity. Moreover, it is easy to acquire in non-invasive and painless approaches. Most importantly, individual oral environment basically maintains a long-term relatively stable state except for little fluctuation caused by diet, infections, or other factors, and the dominant oral floras in different individuals are host specific[5]. Fusobacterium nucleatum (Fn) is a Gram-negative anaerobic bacterium, which is a normal composition of the oral microenvironment[6,7]. In recent years, due to its increased detection rate in oral infectious diseases, it has been identified as an opportunistic pathogen[8]. Chronic periodontitis is caused by the ecological imbalance of the subgingival plaque biofilm communities, leading to the growth of dominant species, which destroys the host immune response and leads to inflammation[9]. Fn has been proved to be one of the pathogens that grow abnormally before periodontal disease, leading to an imbalance in the composition of oral symbiotic bacteria, which leads to the occurrence of periodontitis[10]. It forms a bridge between the early symbiotic colonizers and the late pathogenic colonizers[11]. In recent years, Fn has also been found to be associated with extra-oral infections, including adverse pregnancy outcomes, rheumatoid arthritis, respiratory tract infections, and gastrointestinal neoplasm[12-14]. Accumulating evidence has shown that Fn is associated with the occurrence, development, and metastasis of colorectal cancer (CRC). However, the information for GC is limited. In the present study, we aimed to detect the Fn abundance in saliva using digital droplet polymerase chain reaction (ddPCR), and establish a new simple and effective diagnostic approach to improve the early diagnosis of GC.
Subjects with GC (n = 120), atrophic gastritis (AG) (n = 31), non-AG (NAG) (n = 35), and gastric polyps (GP) (n = 26) and normal controls (NC) (n = 20) undergoing gastroscopy examination at Qilu Hospital of Shandong University from January 2019 to December 2020 were enrolled in this study. The selection of NC mainly depended on the normal results of gastroscopy examination, biochemical indexes, and tumor marker detection. GC, AG, NAG, and GP were further confirmed based on histopathological analyses. The pathological stage of GC was assessed by the 8th edition TNM staging system. The exclusion criteria were as follows: (1) Subjects who used any antibiotics within the last 4 wk; (2) Subjects with oral inflammation; and (3) Subjects without detailed general clinical data. Whole saliva specimens were collected with Salivette® tube before brushing teeth and eating breakfast in the morning. After collection, samples were immediately transferred to the laboratory with ice gauges and centrifuged at 3000 r/min for 10 min at 4 °C. All saliva samples were stored at -80 °C until further analysis. All study participants provided informed written consent prior to study enrollment. This study was approved by the Ethics Committee of the Qilu Hospital of Shandong University (Approval No. KYLL-2019-2-013).
Total genomic DNA was extracted from 200 μL saliva using the QIAamp Blood Mini Kit (cat. 51106, Qiagen, Hilden, Germany), and purified DNA was finally dissolved in 80 μL AE buffer. The concentration of DNA was determined with a Qubit 3 Fluorometer using an Equalbit 1 × dsDNA HS Assay Kit (cat. EQ121-01, Thermo Fisher Scientific, United States) according to the manufacturer’s instructions.
ddPCR was performed on the bio-digital PCR platform (Turtle Tech Ltd., Shanghai, China) in a 35 μL reaction system consisting of 5 μL genomic DNA, 3.5 μL 10 × dPCR buffer, 1 μL dPCR enzyme, 1 μM primers, and 0.5 μM probe of Fn (forward primer: 5’-TGGTGTCATTCTTCCAAAAATATCA-3’; reverse primer: 5’-AGATCAAGAAGGACAAGTTGCTGAA-3’; probe: 5’-ACTTTAACTCTACCATGTTCA-3’). After the automatic sample loading process was completed, the chips with samples added were transferred into a nucleic acid amplification instrument (Turtle Tech Ltd., Shanghai, China). Briefly, after an initial enzyme activation step at 50 °C for 10 min and then at 90 °C for 10 min, amplification (45 cycles) of Fn DNA was carried out. After amplification, the chips were transferred to the commercial digital PCR platform (BioDigital dPCR System, Turtle Tech. Ltd., Shanghai, China) to detect fluorescence amplitude signals.
The GC cell lines AGS and MKN-28 were purchased from Shanghai Zhongqiao Xinzhou Biotechnology Co., Ltd. (Shanghai, China) and maintained in appropriate medium (Hyclone, United States) supplanted with 10% fetal bovine serum (Hyclone, United States) under the standard conditions (50 mL/L CO2, 37 °C).
Two strains of Fn (ATCC49256 and ATCC25586) used in our assays were purchased from American Type Culture Collection. The strains were first inoculated and cultured on Columbia Blood Agar under the anaerobic condition at 37 °C for 48 h. Then a single colony was transferred to a fluid thioglycolate medium, followed by incubation at 37 °C under anaerobic conditions (90 mL/L N2, 50 mL/L CO2, and 50 mL/L H2) to reach an optical density (OD600) of 0.2 for each bacterial strain.
AGS and MKN-28 cells (2 × 105/per well) were seeded in six-well plates with 2 mL complete RPMI-1640 medium and cultured overnight. The plates were then incubated under an anaerobic condition mentioned above for 24 h. On the next day, the cultured Fn was centrifuged (4400 × g for 5 min at room temperature), and the bottom bacteria were sedimented and then re-suspended with fresh RPMI-1640 medium after the supernatant was discarded. After the OD600 was determined, the resuspended Fn was added to AGS and MKN-28 cells for co-culture. Cell treated with phosphate buffer solution served as a control group. The plates were centrifuged at 250 × g for 5 min and placed back in an anaerobic incubator for 24 h. For all the experiments in this study, Fn bacteria were added to the cells at an multiplicity of infection (MOI) of 100 based on the preliminary experiments, and this value was common in the published articles.
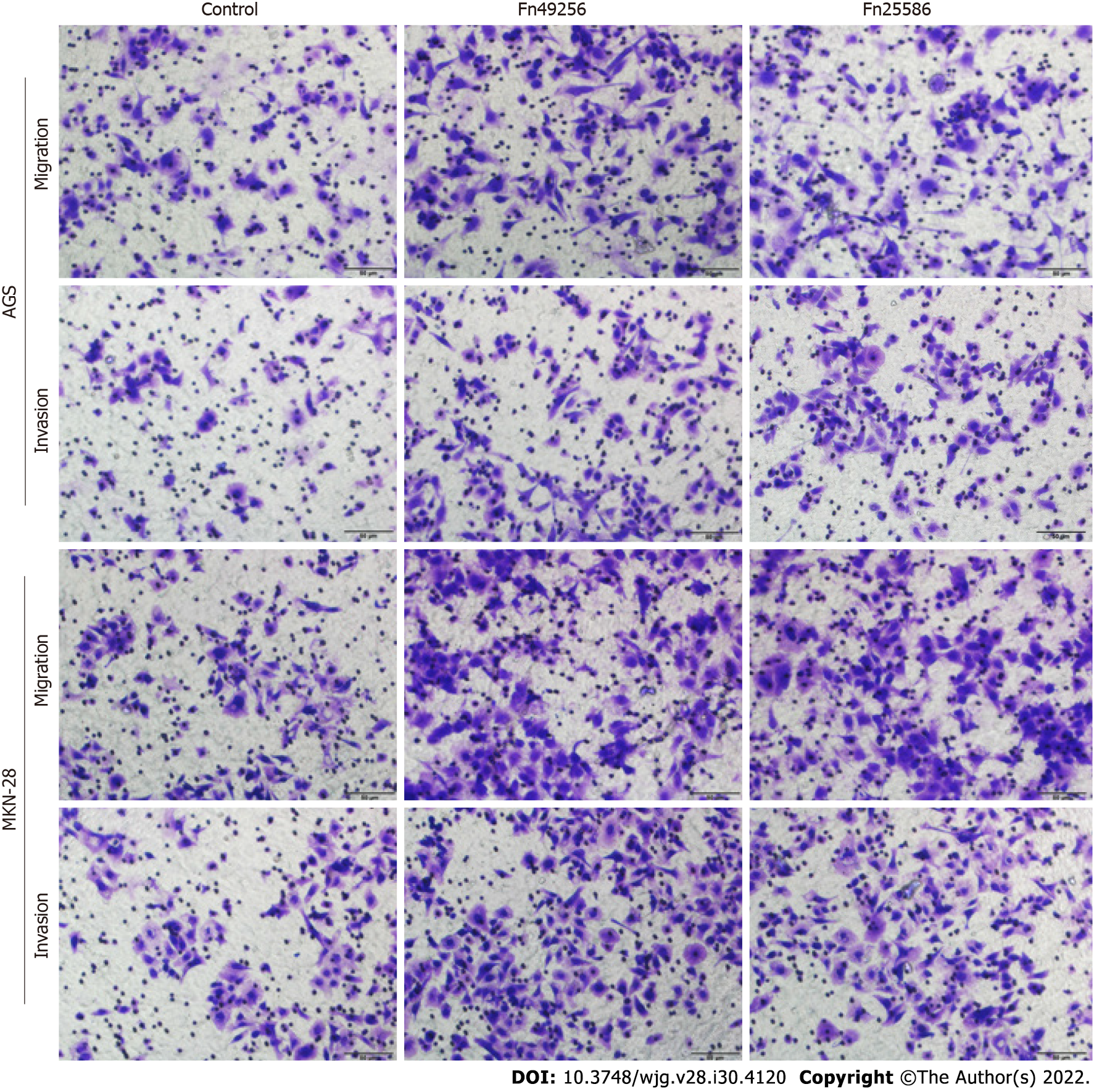
After 24 h of infection, the cells were washed and seeded into the upper chamber of an 8-μm transwell insert (Costar, Corning, New York, United States). Transwell chambers used for invasion assay were pre-coated with Matrigel (Corning), while the chambers used for migration assay were not treated. The upper chamber of the transwell contained 2 × 104 cells in a volume of 200 μL serum-free medium, and 700 μL of medium containing 10% serum was added into the lower chamber. After being cultured for 24 h, the cells in the upper chamber were wiped with the cotton tip, and the adherent cells in the lower chamber were fixed with 40 g/L formaldehyde for 10 min and stained with 0.5% crystal violet for 30 min. The images were captured with an inverted microscope (Olympus, Tokyo, Japan).
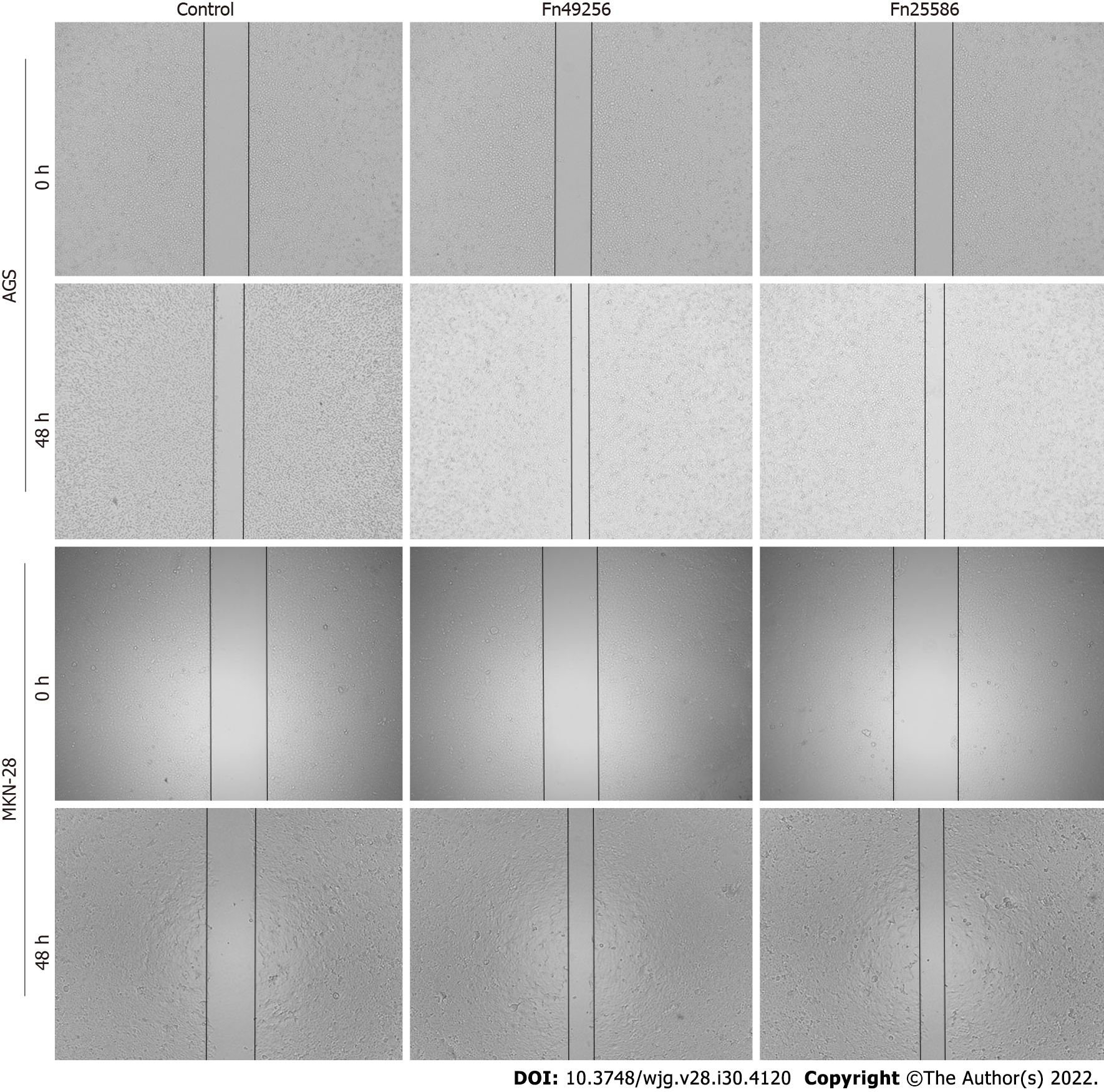
AGS and MKN-28 cells (5 × 105 cells/ per well) were seeded in six-well plates, and the later infection process was the same as the transwell assay above-mentioned. After 24 h of Fn infection, the cells were washed with RPMI-1640 medium (by this time, the cells were grown to 100% confluence, if not, cultured continually). A 200 μL pipette tip was used to make a straight scratch, simulating a wound. Subsequently, the medium and floating cells were removed and replaced with a serum-free culture medium, and the imagines were captured (0 h). After 48 h of cell culture, images were captured again. The cell migration rate was evaluated by comparing the healing degree between 0 h and 48 h.
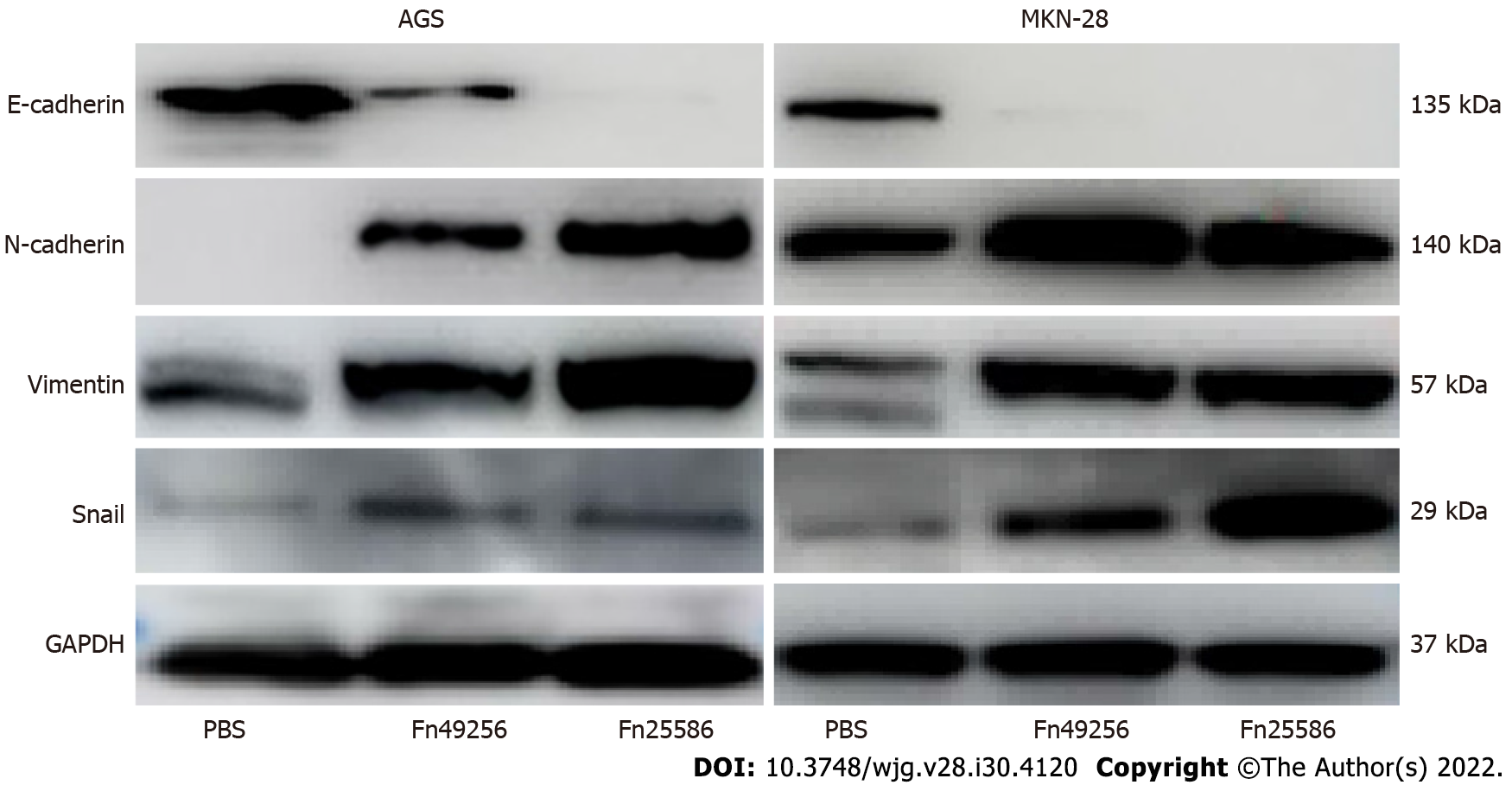
The AGS and MKN-28 cells with or without Fn infection were lysed in radioimmune precipitation assay buffer for 30 min on ice. Then the cell lysates were centrifuged at 12000 r/min for 10 min at 4 °C, and the supernatant was collected. The bicinchoninic acid protein assay kit (Pierce, Rockford, United States) was used to quantify protein concentrations. Subsequently, equal amounts of proteins (30 μg) were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis, and then electrophoretically transferred onto polyvinylidene fluoride membranes. The blots were blocked with 5% skim milk in Tris-buffered saline with tween (TBST) (BD Biosciences, San Jose, CA, United States) containing 0.1% tween-20 (Bio-Rad, Richmond, CA, United States) at room temperature for 1 h. The membranes were then incubated overnight at 4 °C with the primary antibodies against E-cadherin (1:1000, Cell Signaling, #3195S, Danvers, MA, United States), N-cadherin (1:1000, Cell Signaling, #13116T), vimentin (1:1000, Cell Signaling, #5741S, Danvers, MA, United States), Snail (1:1000, Cell Signaling, #3879S), and GAPDH (1:1000, Cell Signaling, #5174S). The membranes were washed three times with 1 × TBST buffer, followed by incubation with anti-rabbit HRP-conjugated secondary antibody (1:5000; Huaan, Hangzhou, China) at room temperature for 1.5 h. The immunoreactive bands were visualized using an Amersham Imager 680 (GE, Boston, United States). GAPDH served as a loading control.
Statistical analyses were conducted using SPSS software (SPSS Inc., Chicago, IL, United States) and graphs were prepared using GraphPad Prism 8.0 (La Jolla, CA). Statistical review was performed by a biomedical statistician. Fn abundance in different groups is expressed as the median and interquartile range. A χ2 test was used for the comparison of categorical variables. A Kruskal-Wallis test was performed for the global comparison of multiple groups and post-hoc multiple comparisons were performed using the Mann-Whitney U test. Receiver operating characteristic (ROC) curve analysis was conducted via MedCalc15.2.2 to evaluate the diagnostic value of Fn as well as other serum tumor markers, and the best cut-off values were calculated by the Youden index. All statistical tests were two-tailed, and P < 0.05 was considered statistically significant.
Our ddPCR results showed a significant difference in salivary Fn among the patients with GC, AG, NAG, and GP, and NC. And the Fn level was significantly increased in GC patients compared with patients with AG, NAG, and GP and NC while there was no difference among AG, NAG, and GP patients and NC (Figure 1A). Moreover, the Fn level was increased as the TNM stage increased (Figure 1B). Further clinical characteristic analysis showed that GC patients with lymph node metastasis had higher Fn levels than those without (Table 1). However, the Fn level in GC was not significantly associated with age, gender, tumor size, pathological differentiation, or invasion depth.
| Feature | Number of cases | Fusobacterium nucleatum level | P value |
| Age (yr) | 0.780 | ||
| < 61 | 56 | 153.00 (39.20-542.00) | |
| ≥ 61 | 64 | 204.00 (57.30-566.00) | |
| Gender | 0.961 | ||
| Male | 99 | 161.00 (53.52-620.60) | |
| Female | 21 | 213.60 (46.63-335.90) | |
| Tumor size (cm) | 0.450 | ||
| < 5 | 76 | 152.90 (52.21-443.80) | |
| ≥ 5 | 44 | 239.90 (47.31-846.80) | |
| Pathological differentiation | 0.658 | ||
| Well | 77 | 147.50 (33.98-543.90) | |
| Moderate | 33 | 155.80 (84.94-754.60) | |
| Poor | 10 | 280.80 (78.46-518.70) | |
| Invasion depth | 0.227 | ||
| T1 | 29 | 124.60 (32.21-415.80) | |
| T2 | 60 | 298.30 (76.58-666.90) | |
| T3 | 21 | 102.10 (68.62-385.70) | |
| T4 | 10 | 223.00 (66.99-790.30) | |
| Lymph nodes metastasis | < 0.001 | ||
| N0 | 43 | 80.14 (23.98-241.80) | |
| N1 | 25 | 324.50 (72.43-549.10) | |
| N2 | 22 | 239.60 (79.36-501.70) | |
| N3 | 30 | 478.00 (96.09-1054.00) |
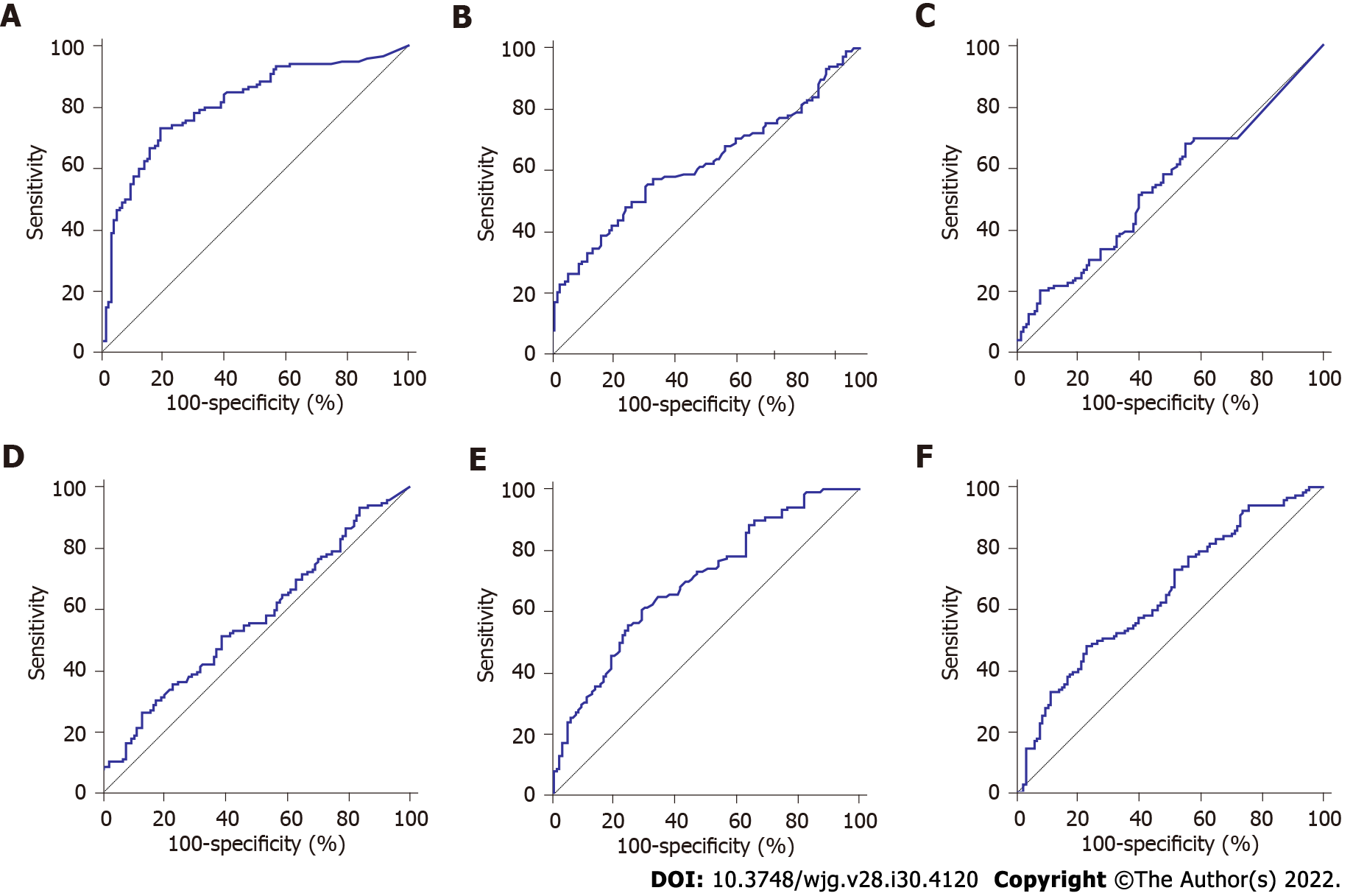
ROC analysis indicated some diagnostic power with a fitted area under the curve (AUC) of 0.813 (95% confidence interval: 0.756-0.861) (Figure 2A). When the Youden index was used to calculate the optimal cut-off value of the Fn copy number as 67.58 copies/μL, the sensitivity and specificity were 73.33% and 82.14%, respectively. To better evaluate the diagnostic value of Fn in saliva for GC, the routine clinical test records of traditional serum tumor markers, including CEA, CA19-9, CA72-4, ferritin, and sialic acid were assessed. The upper cut-off values of these markers were determined according to the manufacturer’s directions. Table 2 shows that the Fn level was significantly higher in GC patients compared with AG, NAG, and GP patients and NC, while CEA, CA19-9, CA72-4, and ferritin did not significantly different between GC patients and the other four groups. Although sialic acid was also significantly different among the five groups, its diagnostic value was still lower than that of Fn, according to the ROC curves.
| Normal control | Atrophic gastritis | Non-atrophic gastritis | Gastric polyps | Gastric cancer | |
| Cases | 20 | 31 | 35 | 26 | 120 |
| Gender (male/female) | 13/7 | 17/14 | 19/16 | 15/11 | 73/47 |
| Age (yr)1 | 56.90 ± 16.02 | 62.13 ± 10.56 | 60.80 ± 14.54 | 58.17 ± 15.26 | 61.38 ± 10.31 |
| CEA (mg/L)2,3 | 2.03 (1.22-2.56) | 1.95 (1.49-53.37) | 1.72 (1.44-2.60) | 1.36 (1.10-1.85) | 2.46 (1.38-4.63) |
| CA19-9 (pg/mL)2,3 | 6.38 (3.61-11.39) | 10.67 (8.68-16.35) | 10.30 (7.09-15.07) | 9.20 (5.68-12.09) | 11.46 (6.75-20.28) |
| CA72-4 (U/mL)2,3 | 2.86 (0.98-6.48) | 0 (0-3.18) | 2.41 (1.50-4.65) | 1.71 (0.15-4.63) | 2.58 (0-5.69) |
| Ferritin (ng/mL)2,3 | 143.15 (97.04-273.98) | 96.98 (69.33-206.55) | 126.70 (74.49-245.30) | 182.95 (101.13-263.88) | 86.20 (25.99-165.75) |
| Sialic acid (g/L)2,4 | 54.30 (51.45-56.75) | 52.50 (46.50-55.30) | 53.70 (48.25-60.55) | 56.2 (50.53-59.65) | 59.55 (53.28-65.33) |
| Fusobacterium nucleatum (copies/μL)2,4 | 16.70 (2.4-26.10) | 28.611 (5.99-61.50) | 12.18 (0.74-35.55) | 16.32 (1.59-122.234) | 177.09 (53.09-547.11) |
The ROC curves presented that the AUC of CEA, CA19-9, CA72-4, ferritin, and sialic acid was 0.620, 0.570, 0.541, 0.648, and 0.693, respectively (Figures 2B-F). In addition, the sensitivity of Fn was higher compared with the other traditional serum markers. As for the specificity, the value of Fn was higher than those of CEA, CA72-4, ferritin, and sialic acid, but lower than that of CA19-9. Of note, although the specificity of CA19-9 was high, its sensitivity was relatively low (Table 3). These results indicated that the diagnostic capability of Fn was superior to that of CEA, CA19-9, CA72-4, ferritin, and sialic acid.
| Index | AUC | Cut-off value | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
| Fn | 0.813 | 67.58 copies/μL | 73.33 | 82.14 | 81.48 | 74.19 |
| CEA | 0.620 | 5 ng/mL | 55.00 | 69.64 | 66.00 | 59.09 |
| CA19-9 | 0.570 | 69.2 pg/mL | 26.67 | 86.61 | 68.09 | 52.43 |
| CA72-4 | 0.541 | 6.9 U/mL | 67.50 | 44.64 | 56.64 | 56.18 |
| Ferritin | 0.648 | 400 ng/mL | 51.67 | 23.21 | 41.89 | 30.95 |
| Sialic acid | 0.693 | 75.4 mg/dL | 60.83 | 70.54 | 68.87 | 62.70 |
To evaluate the effect of Fn infection in vitro, we infected AGS and MKN-28 cells with Fn. The transwell assay indicated that Fn infection significantly enhanced the invasive and migratory capacities of AGS and MKN-28 cells (Figure 3). Consistent with these results, the wound-healing assay showed that Fn infection promoted the migration of these cells (Figure 4). Since epithelial-mesenchymal transition (EMT) is an important process of metastasis, we examined the effect of Fn infection on the expression of proteins involved in the EMT process. The western blot analysis revealed that Fn infection decreased the expression of epithelial markers, such as E-cadherin, but increased the expression of mesenchymal phenotype-associated molecules, such as N-cadherin, vimentin, and snail (Figure 5).
In the present study, we reported the relationship between the Fn level in saliva and GC development for the first time. First, a more sensitive ddPCR approach was used to determine the amount of Fn DNA in saliva, and the results showed that a higher number of Fn existed in the saliva of GC patients compared with NC and patients with benign disease. Importantly, salivary Fn had good diagnostic value for GC, which was superior to traditional serum tumor markers, such as CEA, CA19-9, CA72-4, ferritin, and sialic acid, providing a new direction for the diagnosis of GC. We also found that the Fn level was positively associated with TNM stage and lymph node metastasis. Furthermore, transwell and wound-healing assays verified the role of Fn in promoting the invasion and migration of GC cells. Subsequently, we indicated that Fn infection promoted GC metastasis by accelerating the EMT process.
It is essential to diagnose GC at an early stage. Currently, gastroscopy in combination with biopsy is considered the most accurate method to diagnose GC. However, early asymptomatic patients have poor compliance with gastroscopy due to its invasiveness, which directly reduces the rate of early diagnosis of GC. Serum tumor markers are another approach to screen GC. For example, CEA, CA19-9, and CA72-4 are widely used in clinical practice. However, serum tumor markers are not recommended for routine clinical screening due to the lack of specificity and sensitivity[15]. In a cohort consisting of 587 early GC patients, the positive rate of CA19-9, CA125, and CEA was 4.8%, 1.9%, and 4.3%, respectively[16]. For Fn in saliva, it is not only easy to obtain, but also shows a better diagnostic capacity for GC. Moreover, salivary Fn has a pleasurable tissue specificity as a diagnostic marker for GC.
H. pylori is a well-known risk factor for GC. However, other bacterial genera are also detected in the gastric mucosa, such as Prevotella, Rothia, Fusobacterium, and Klebsiella[17,18]. Fn, a resident colony of bacteria in the oral cavity, is detected significantly higher in the GC tissue compared with the para-cancerous tissue[19]. There is a microbial succession hypothesis to explain the decrease of H. pylori and elevation of the non-dominant flora in GC[18]. The hypothesis proposes that H. pylori can produce urease through the mucus layer, enhancing pH value to modify the microenvironment and triggering a strong inflammatory response. In such a process, gastric mucosal integrity is destroyed, and other secondary bacteria invade the mucus layer, causing more aggressive tumor occurrence. Therefore, Fn in the oral cavity can easily reach the stomach and colonize through the gastric mucosa barrier as the swallowing of saliva, forming an invasion focus. Thus, we think that Fn cooperates with H. pylori to promote GC development. Hsieh et al[20] have shown that Fn colonization leads to worse prognosis in GC patients with H. pylori positivity.
As a high-frequency periodontal pathogen, Fn has been confirmed to promote the initiation, proliferation, invasion, and chemoresistance of CRC in recent years. There are several suggested mechanisms, such as enhancing adhesion and evasion ability, modulating host immune evasion, and improving autophagy to cause chemoresistance[21-23]. It is worth noting that Fn is considered to play a more important role in promoting the formation of tumor, rather than its development[24]. Consistent with these findings, Fn has been confirmed to have key pathogenic characteristics, such as virulence factor FadA and characteristic adhesion protein Fap2[25-27]. In addition to promoting the occurrence and development of CRC, accumulating evidence has shown that Fn also plays an important role in promoting CRC metastasis[28,29]. In our present study, a high level of Fn was found in GC patients with lymph node metastasis. Although its impact in promoting tumor metastasis has been recognized, the role of Fn in such a process and its potential mechanism remain largely unclear. Chen et al[30] have detected Fn colonization in metastatic lung lesions of nude mice, and they further verified that Fn infection up-regulates KRT7 (type II cytokeratin, which plays a role in maintaining the structural integrity of the cells as well as promoting motile activities) to promote CRC metastasis. Besides, Chen et al[31] have identified that Fn activates autophagy to promote CRC metastasis. It is worth mentioning that EMT is the most well-validated method by which Fn promotes CRC metastasis, and it has been discovered that the effect is enhanced when the MOI is increased[32-34]. EMT is a classical pathway promoting metastasis. It is a special program that enables settled epithelial cells to gain the ability to migrate as single cells, which can enhance mobility, invasion, and resistance to apoptosis, conferring metastatic properties of cancer cells[35]. The examinations conducted in CRC both in vivo and in vitro have verified that Fn can promote the EMT process[28]. We supplemented the evidence that Fn promoted the EMT process in GC cells. Moreover, Fn infection significantly decreased the expression of cell adhesion molecules and increased the expression of mesenchymal phenotype-associated molecules, eventually resulting in GC metastasis. Taken together, we identified that Fn promoted GC metastasis by facilitating the EMT process.
Targeting Fn may reduce the progression of related cancers to ascertain the origin of Fn in cancers, and determining the route by which it reaches the tumor is of therapeutic significance. It has been found for a long time that part of Fn detected in CRC tissue is originated from the oral cavity, while how Fn in the oral cavity is transported to other body sites is also a hot topic. Initially, Fn strains with similar sequences were found in human primary CRC tissues and paired liver metastases, suggesting that these bacteria can migrate to distant sites with CRC cells[29]. Therefore, some scholars believe that Fn enters tumor tissue via blood circulation. The existing data have shown that Fn is detected at higher loads in GC tissue[19], and our data suggested that Fn abundance was increased in the saliva of GC patients. Could this indicate that Fn could directly enter the gastrointestinal tract, and then colonize epithelial cells? For Fn can produce a variety of adhesion factors as well as invasive factors, it is easy to adhere to the gastrointestinal mucosa, and form an inflammatory microenvironment, which is conducive to the formation of tumor. However, some evidence has also shown that Fn in the blood is more successful in CRC colonization than being gavaged, indicating that Fn the in blood (from a bleeding wound in the gum or some other way) plays a more important role in forming colonization than the ones in the digestive tract, even if there is a disadvantage in quantity[36]. Which type of Fn transmission is dominant in tumor formation (or the two work together) remains to be further studied.
There are also some limitations in our study. First, the small sample size caused by high cost led to selection bias. In follow-up studies, larger, multicenter studies are required to consolidate the findings. Second, the flora in the oral cavity is diverse. Whether other floras have an impact on the occurrence and development of GC, or whether they have interacted with Fn needs to be further explored.
Collectively, Fn in saliva exhibits a good predictive ability and represents a promising diagnostic marker for GC. Given the tumorigenicity and metastasis-promoting properties of Fn, we suggest that the elimination of Fn could reduce the incidence of GC and improve the treatment outcomes, which is similar to triple or quadruple anti-H. pylori therapy.
Gastric cancer (GC) is a common malignant tumor in the digestive tract. Although the 5-year survival rate of GC is enhanced with the combination of chemotherapy and surgery, its mortality remains high due to the diagnosis at an advanced stage. It is essential to develop a new method to diagnose GC at an early stage.
Fusobacterium nucleatum (Fn) primarily colonized in the oral cavity, has been detected in tissues of GC in recent years. We wondered whether there is a correlation between salivary Fn and GC.
The research purpose was to find a new simple and effective biomarker to diagnose GC.
The abundance of Fn in saliva was quantified by droplet digital polymerase chain reaction in 120 GC patients, 31 atrophic gastritis (AG) patients, 35 non-AG (NAG) patients, 26 gastric polyp patients, and 20 normal controls (NC). The diagnostic value of Fn was evaluated and compared with traditional serum tumor markers, including carcinoembryonic antigen (CEA), carbohydrate antigen (CA)19-9, CA72-4, ferritin, and sialic acid. Transwell and wound-healing assays were conducted to assess the influence of Fn infection on GC cells. Western blot analysis was performed to detect the expression of epithelial-mesenchymal transition (EMT) markers.
Fn had favorable diagnostic value in classifying GC from NC and benign diseases, which was superior to those traditional serum tumor markers, such as CEA, CA19-9, CA72-4, ferritin, and sialic acid. Salivary Fn level was positively associated with the GC TNM stage and lymph node metastasis. Further, in vitro experiments revealed that Fn could promote the migration and invasion of GC cells. Western blot analysis indicated that Fn infection decreased the expression of epithelial marker such as E-cadherin, and increased the expression of mesenchymal markers such as N-cadherin, vimentin, and snail.
Fn in saliva could be used as a promising biomarker to diagnose GC, and Fn infection could promote GC metastasis by accelerating the EMT process.
Human saliva is a unique bio-fluid resource with huge clinical diagnostic and risk assessment capacity. Salivary Fn has promising value for diagnosing GC.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Imai Y, Japan; Mincione G, Italy; Mishra TS, India; Rahimi-Movaghar E, Iran S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 55842] [Article Influence: 7977.4] [Reference Citation Analysis (132)] |
| 2. | Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721-5732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1867] [Cited by in RCA: 2022] [Article Influence: 101.1] [Reference Citation Analysis (0)] |
| 3. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3302] [Cited by in RCA: 3265] [Article Influence: 79.6] [Reference Citation Analysis (1)] |
| 4. | Alarcón T, Llorca L, Perez-Perez G. Impact of the Microbiota and Gastric Disease Development by Helicobacter pylori. Curr Top Microbiol Immunol. 2017;400:253-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Rasiah IA, Wong L, Anderson SA, Sissons CH. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Arch Oral Biol. 2005;50:779-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Kabwe M, Brown TL, Dashper S, Speirs L, Ku H, Petrovski S, Chan HT, Lock P, Tucci J. Genomic, morphological and functional characterisation of novel bacteriophage FNU1 capable of disrupting Fusobacterium nucleatum biofilms. Sci Rep. 2019;9:9107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 7. | Habib AM, Islam MS, Sohel M, Mazumder MH, Sikder MO, Shahik SM. Mining the Proteome of Fusobacterium nucleatum subsp. nucleatum ATCC 25586 for Potential Therapeutics Discovery: An In Silico Approach. Genomics Inform. 2016;14:255-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 723] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 9. | Fragkioudakis I, Riggio MP, Apatzidou DA. Understanding the microbial components of periodontal diseases and periodontal treatment-induced microbiological shifts. J Med Microbiol. 2021;70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 10. | Nozawa A, Oshima H, Togawa N, Nozaki T, Murakami S. Development of Oral Care Chip, a novel device for quantitative detection of the oral microbiota associated with periodontal disease. PLoS One. 2020;15:e0229485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Baek K, Ji S, Choi Y. Complex Intratissue Microbiota Forms Biofilms in Periodontal Lesions. J Dent Res. 2018;97:192-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 555] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 13. | Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM, Redline RW. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet Gynecol. 2010;115:442-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 210] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 14. | Wang X, Buhimschi CS, Temoin S, Bhandari V, Han YW, Buhimschi IA. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One. 2013;8:e56131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 15. | Sekiguchi M, Matsuda T. Limited usefulness of serum carcinoembryonic antigen and carbohydrate antigen 19-9 levels for gastrointestinal and whole-body cancer screening. Sci Rep. 2020;10:18202. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 16. | Feng F, Tian Y, Xu G, Liu Z, Liu S, Zheng G, Guo M, Lian X, Fan D, Zhang H. Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 267] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 17. | Ianiro G, Molina-Infante J, Gasbarrini A. Gastric Microbiota. Helicobacter. 2015;20 Suppl 1:68-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 18. | Hutton ML, Kaparakis-Liaskos M, Turner L, Cardona A, Kwok T, Ferrero RL. Helicobacter pylori exploits cholesterol-rich microdomains for induction of NF-kappaB-dependent responses and peptidoglycan delivery in epithelial cells. Infect Immun. 2010;78:4523-4531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW, Lin YJ, Deng YF, Hsu WT, Wu CS, Li C. Increased Abundance of Clostridium and Fusobacterium in Gastric Microbiota of Patients with Gastric Cancer in Taiwan. Sci Rep. 2018;8:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 197] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 20. | Hsieh YY, Tung SY, Pan HY, Chang TS, Wei KL, Chen WM, Deng YF, Lu CK, Lai YH, Wu CS, Li C. Fusobacterium nucleatum colonization is associated with decreased survival of helicobacter pylori-positive gastric cancer patients. World J Gastroenterol. 2021;27:7311-7323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 21. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 814] [Cited by in RCA: 1478] [Article Influence: 184.8] [Reference Citation Analysis (0)] |
| 22. | Zhang S, Yang Y, Weng W, Guo B, Cai G, Ma Y, Cai S. Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J Exp Clin Cancer Res. 2019;38:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 23. | Dong G, Wang M, Gu G, Li S, Sun X, Li Z, Cai H, Zhu Z. MACC1 and HGF are associated with survival in patients with gastric cancer. Oncol Lett. 2018;15:3207-3213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Shang FM, Liu HL. Fusobacterium nucleatum and colorectal cancer: A review. World J Gastrointest Oncol. 2018;10:71-81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 192] [Cited by in RCA: 196] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 25. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1511] [Cited by in RCA: 1669] [Article Influence: 139.1] [Reference Citation Analysis (1)] |
| 26. | Fardini Y, Wang X, Témoin S, Nithianantham S, Lee D, Shoham M, Han YW. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol. 2011;82:1468-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 27. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 978] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 28. | Yan X, Liu L, Li H, Qin H, Sun Z. Clinical significance of Fusobacterium nucleatum, epithelial-mesenchymal transition, and cancer stem cell markers in stage III/IV colorectal cancer patients. Onco Targets Ther. 2017;10:5031-5046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 29. | Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 1064] [Article Influence: 133.0] [Reference Citation Analysis (0)] |
| 30. | Chen S, Su T, Zhang Y, Lee A, He J, Ge Q, Wang L, Si J, Zhuo W. Fusobacterium nucleatum promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes. 2020;11:511-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 162] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 31. | Chen Y, Chen Y, Zhang J, Cao P, Su W, Deng Y, Zhan N, Fu X, Huang Y, Dong W. Fusobacterium nucleatum Promotes Metastasis in Colorectal Cancer by Activating Autophagy Signaling via the Upregulation of CARD3 Expression. Theranostics. 2020;10:323-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 32. | Wang Q, Yu C, Yue C, Liu X. Fusobacterium nucleatum produces cancer stem cell characteristics via EMT-resembling variations. Int J Clin Exp Pathol. 2020;13:1819-1828. [PubMed] |
| 33. | Yu MR, Kim HJ, Park HR. Fusobacterium nucleatum Accelerates the Progression of Colitis-Associated Colorectal Cancer by Promoting EMT. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 34. | Duan C, Tang X, Wang W, Qian W, Fu X, Deng X, Han C, Hou X. L-fucose ameliorates the carcinogenic properties of Fusobacterium nucleatum in colorectal cancer. Oncol Lett. 2021;21:143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 35. | Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 523] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 36. | Abed J, Maalouf N, Manson AL, Earl AM, Parhi L, Emgård JEM, Klutstein M, Tayeb S, Almogy G, Atlan KA, Chaushu S, Israeli E, Mandelboim O, Garrett WS, Bachrach G. Colon Cancer-Associated Fusobacterium nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front Cell Infect Microbiol. 2020;10:400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 155] [Article Influence: 31.0] [Reference Citation Analysis (0)] |