Published online Nov 21, 2021. doi: 10.3748/wjg.v27.i43.7530
Peer-review started: May 19, 2021
First decision: June 22, 2021
Revised: June 23, 2021
Accepted: September 15, 2021
Article in press: September 15, 2021
Published online: November 21, 2021
Processing time: 183 Days and 19.7 Hours
Severe acute pancreatitis (SAP) is a deadly inflammatory disease with complex pathogenesis and lack of effective therapeutic options. N6-methyladenosine (m6A) modification of circRNAs plays important roles in physiological and pathological processes. However, the roles of m6A circRNA in the pathological process of SAP remains unknown.
To identify transcriptome-wide map of m6A circRNAs and to determine their biological significance and potential mechanisms in SAP.
The SAP in C57BL/6 mice was induced using 4% sodium taurocholate salt. The transcriptome-wide map of m6A circRNAs was identified by m6A-modified RNA immunoprecipitation sequencing. The biological significance of circRNAs with differentially expressed m6A peaks was evaluated through gene ontology and Kyoto Encyclopedia of Genes and Genomes analysis. The underlying mechanism of m6A circRNAs in SAP was analyzed by constructing of m6A circRNA-microRNA networks. The expression of demethylases was determined by quantitative polymerase chain reaction and western blot to deduce the possible mechanism of reversible m6A process in SAP.
Fifty-seven circRNAs with differentially expressed m6A peaks were identified by m6A-modified RNA immunoprecipitation sequencing, of which 32 were upregulated and 25 downregulated. Functional analysis of these m6A circRNAs in SAP found some important pathways involved in the pathogenesis of SAP, such as regulation of autophagy and protein digestion. In m6A circRNA–miRNA networks, several important miRNAs participated in the occurrence and progression of SAP were found to bind to these m6A circRNAs, such as miR-24-3p, miR-26a, miR-92b, miR-216b, miR-324-5p and miR-762. Notably, the total m6A level of circRNAs was reduced, while the demethylase alkylation repair homolog 5 was upregulated in SAP.
m6A modification of circRNAs may be involved in the pathogenesis of SAP. Our findings may provide novel insights to explore the possible pathogenetic mechanism of SAP and seek new potential therapeutic targets for SAP.
Core Tip: We identified a transcriptome-wide map of N6-methyladenosine (m6A) circRNAs and determined their biological significance and potential mechanisms in severe acute pancreatitis (SAP). The main findings were: (1) Function analysis found that circRNAs with differentially expressed m6A peaks were involved in the key process of SAP; (2) m6A may affect the interplays of circRNAs and microRNAs to participate in the pathogenesis of SAP; and (3) Demethylase alkylation repair homolog 5 may play key roles in dynamic process of m6A to downregulate the total m6A level of circRNAs in SAP. We provided novel insights to explore the possible pathophysiological mechanism of SAP and seek new potential therapeutic targets.
- Citation: Wu J, Yuan XH, Jiang W, Lu YC, Huang QL, Yang Y, Qie HJ, Liu JT, Sun HY, Tang LJ. Genome-wide map of N6-methyladenosine circular RNAs identified in mice model of severe acute pancreatitis. World J Gastroenterol 2021; 27(43): 7530-7545
- URL: https://www.wjgnet.com/1007-9327/full/v27/i43/7530.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i43.7530
Acute pancreatitis (AP) is a pancreatic inflammatory disorder that is associated with substantial morbidity and mortality[1]. Approximately 20% of patients with AP develop into severe AP (SAP)[2]. Due to the extensive pancreatic necrosis, subsequent infection, systemic inflammatory response syndrome and multiple organ failure, the mortality of SAP is up to 30%[2,3]. Previous studies have suggested that some important pathological mechanisms, including premature trypsinogen activation in the acinar cells and macrophages, mitochondrial dysfunction, pathological calcium signaling, endoplasmic reticulum (ER) stress, and impaired autophagy, are involved in the initiation and development of SAP[1]. However, the pathophysiology of SAP is complex and remains unclear, especially the level of gene regulation.
CircRNAs were discovered in the 1970s[4] and were identified as single-stranded covalently closed RNA molecules that lack 5’ caps and 3’ tails[5]. Long after, they were thought to be the byproducts of splicing[6]. In recent years, as high-throughput sequencing developed, thousands of circRNAs were found to be expressed in a wide range of mammalian tissues[7,8], including the pancreas[9], and accumulating studies have demonstrated that circRNAs play vital roles in the whole process and prognosis of many diseases, including cardiovascular diseases[8], cancer[10], neurodevelopmental processes[11], immune responses and immune diseases[12]. The main mechanisms of circRNAs participated in the initiation and development of diseases include the following functions[6,8,10,12]: interplay with RNA-binding proteins, microRNA (miRNA) sponges, regulating the stability of mRNAs, modulating the transcription of parental gene and the templates for protein synthesis. However, the post-transcription modification of circRNAs remains unclear.
N6-methyladenosine (m6A) is the most prevalent internal modification of RNA in eukaryotic cells[13]. In 2017, Zhou et al[14] reported that the m6A modification is widespread in circRNAs and m6A modifications are read and written by the same complexes in circRNAs and mRNAs. The regulatory role of m6A is mainly performed by three homologous factors, namely so-called “writers”, “erasers” and “readers”[13-15]. The writers mainly include methyltransferase-like 3 and 14 proteins (METTL3 and METTL14) and their cofactor WT1-associated protein (WTAP). They form a methyltransferase complex to catalyze the installation of m6A. The erasers, including alkylation repair homolog 5 (ALKBH5) and fat mass and obesity related protein (FTO), can catalyze the oxidative demethylation of N-alkylated nucleic acid bases. The readers are mainly YT521-B homology (YTH) domain containing proteins family, including YTHDC1, YTHDC2, YTHDF1, YTHDF2 and YTHDF3. They can specifically recognize m6A and regulate splicing, localization, degradation and translation of RNAs. Recently, it has been found that the m6A modification of circRNAs plays a key role in innate immunity and tumors though regulating the metabolism and function of circRNAs[15]. In human embryonic stem cells and HeLa cells, m6A circRNAs display cell-type-specific methylation patterns[14]. In colorectal carcinoma, the m6A modification can modulate cytoplasmic export of circNSUN2 and stabilize HMGA2, ultimately enhancing the colorectal liver metastasis[16]. However, the roles of m6A circRNAs in SAP are still unknown.
Here, we investigated the expression profile of m6A circRNAs in SAP through m6A-modified RNA immunoprecipitation sequencing (MeRIP-seq). We evaluated the biological significance of circRNAs with differentially expressed m6A peaks though gene ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and explored their underlying mechanism by construction of m6A circRNA–miRNA networks. In addition, we determined the expression of demethyltransferase, ALKBH5 and FTO, to deduce the possible mechanism of reversible m6A process in SAP.
Male C57BL/6 mice weighing 22-25 g were provided by Chengdu Dashuo Experimental Animal Technology Co. Ltd. All the mice were housed in ventilated plastic cage system and fed with the same food and water for 7 d to adapt to the environment. The entire research protocol was approved by the Institutional Animal Care and Use Committee at the General Hospital of Western Theater Command.
Before the operation, the mice were divided into SAP and control groups randomly (3 mice per group) and fasted for 12 h but had free access to water. Isoflurane (5%) was used to anesthetize mice by induction box prior to surgery. Then, the SAP was induced through 4% sodium taurocholate salt that was slowly retrogradely injected into the biliopancreatic duct with a microinfusion pump. All mice were killed 24 h after the establishment of model, and the blood samples and pancreatic tissues were collected for further analysis.
Pancreatic tissue (0.4 cm × 0.4 cm) was fixed in 4% paraformaldehyde solution. After dehydrating with ethanol, the tissue samples were embedded in paraffin. Then, the samples were cut into about 4-μm-thick sections, and the sections were stained with hematoxylin and eosin. The light microscopy at × 200 magnification was used to examine the slide. The scoring system described previously was used to evaluate the degree of pancreatic injury[17]. The scores were averaged for five different slides that were selected randomly from each pancreas.
The concentrations of lipase and amylase in serum were determined using Lipase Assay kit and Amylase Assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the instructions.
TRIzol reagent (Invitrogen, Carlsbad, CA, United States) was used to extract total RNA from the homogenized pancreatic tissues of the control and SAP groups. The concentration of extracted RNA was measured at OD260 and 280 by NanoDrop ND-2000 instrument (Thermo Fisher Scientific, Waltham, MA, United States). We assessed the integrity of RNA through denaturing agarose gel electrophoresis. The OD A260/A280 ratio between 1.8 and 2.0 was set as the RNA purity standard.
rRNAs in total RNA were removed using Ribo-Zero rRNA Removal Kits (Illumina, San Diego, CA, United States). The removal efficiency of rRNA by the residual determination of 28S and 18S of rRNA using quantitative polymerase chain reaction (qPCR). The fragmented RNA was incubated with the anti-m6A antibody at 4 °C for 2 h in IPP buffer. Then, the mixture was immunoprecipitated by incubation with protein-A beads (Thermo Fisher Scientific) for 2 h at 4 °C. The bound RNA was eluted from the beads with m6A (Berry & Associates) in IPP buffer and then extracted with TRIzol reagent (Thermo Fisher Scientific). The immunoprecipitated RNA and input RNA were used to construct the library using NEBNext® Ultra™ RNA Library Prep Kit and double-ended 150-bp sequencing of the m6A-IP and input samples was performed on an Illumina HiSeq sequencer (performed by Cloudseq Biotech Inc., Shanghai, China).
Paired-end reads were harvested from the Illumina HiSeq 4000 sequencer, and were quality controlled by Q30. To obtain high quality clean reads, 3’ adaptor-trimming and low-quality reads were removed by cutadapt software. The clean reads with high quality of the input library were aligned to the mouse reference genome (UCSC MM10) with STAR software. DCC software was used for detecting and identifying the circRNAs. The identified circRNAs were annotated using the circBase database and Circ2Traits database. For all samples, raw junction reads were normalized to the number of total mapped reads and log2 transformed. The read alignments on the genome were visualized using the tool integrative genomics viewer. The adapter-removal reads were aligned to the reference genome using Hisat2 software. The methylated sites in each sample were identified using MACS software. Differentially methylated sites were identified using diffReps software.
The parent genes of circRNAs with differential m6A peaks were selected to analyze their potential biological roles through GO and KEGG pathway analysis. GO analysis included three parts, namely, biological process (BP) analysis, molecular function (MF) analysis, and cell component (CC) analysis[18]. GO analysis was performed by R topGO package. Fisher’s exact test in Matlab MCR software was applied to calculate the enrichment of each pathway. The bubble plots and column plots were generated using the ggplot2 in R package (https://ggplot2.tidyverse.org).
circRNA containing miRNA-binding sites can bind to miRNA response elements competitively, further regulating the target mRNAs[19]. The top 10 upregulated and top 10 downregulated circRNAs according to the level of m6A were selected to construct circRNA–miRNA networks. The m6A circRNA–miRNA networks were constructed using TargetScan software and miRanda software and the circRNA–miRNA interactions were visualized by Cytoscape.
The top 10 upregulated and top 10 downregulated circRNAs were selected to analyze their homology with human circRNAs. The sequence of human circRNAs was downloaded from circBase database and the sequence of each selected m6A circRNA was blasted against the human circRNAs sequence by the blastn function of Blast software.
The whole pancreatic tissues from SAP and control groups were placed in RIPA lysate buffer with protease inhibitor, phosphatase inhibitor and phenylmethylsulfonyl fluoride inside (Total Protein Extraction Kit; Beijing Solarbio Science and Technology Inc., Beijing, China), and the tissues were homogenized with homogenizer. The tissue homogenate was centrifuged at 12000 g for 30 min at 4 °C, and the supernatant was collected. After protein concentration was measured by BCA Protein Assay Kit (Beyotime Biotechnology, Jiangsu, China), the supernatant was mixed with loading buffer (Beijing Solarbio Science and Technology), boiled at 100 °C for 10 min for protein denaturation, and stored at -80 °C after separation. The target proteins were separated by SDS-PAGE. The proteins were transferred to polyvinylidene difluoride membrane (0.45 μm, IPVH00010; Millipore, Billerica, MA, United States), blocked in 5% nonfat milk for 1 h at room temperature (22 ± 3 °C), and then incubated with primary antibody, FTO (1:1000, D2V1I; Cell Signaling Technology, Danvers, MA, United States), ALKBH5 (1:2000, 16837-1-AP; Proteintech, Rosemont, IL, United States), GAPDH (1:5000, 10494-1-AP; Proteintech) at 4 °C overnight. The membranes were washed with Tris-buffered saline with Tween-20 (TBST) (Beijing Solarbio Science and Technology) three times and incubated with secondary antibody (1:10000, 15015; Proteintech) at room temperature for 1 h. After being washed three times with TBST, the protein bands were visualized by enhanced chemiluminescence (Immobilon Western Chemilum HRP Substrate; Millipore) in a biological imaging system.
The total RNA was extracted from SAP and control groups as described above. qPCR was performed using One Step SYBR® PrimeScript™ RT-PCR kit II (Takara Biotechnology Co., Ltd., Dalian, China) and the primers (ALKBH5: forward 5’-GGCGGTCATCATTCTCAGGAAGAC-3’ and reverse 5’-CTGACAGGCGATCTGAAGCATAGC-3’; FTO: forward 5’-CTCACAGCC TCGGTTTAGTTCCAC-3’ and reverse 5’–CGTCGCCATCGTCTGAGTCATT G-3’; GAPDH: forward 5’-GGTGAAGGTCGGTGTGAACG-3’ and reverse 5’-CTCGCTCCTGGAAGATGGTG-3’) were synthesized by Shanghai Sangon Biotech Co., Ltd.. The outcomes were analyzed by means of 2-ΔΔCT through normalizing the quantity of GAPDH.
GraphPad Prism 8 (La Jolla, CA, United States) and SPSS 22.0 (IBM Corp., Armonk, NY, United States) were used for performing statistical analyses. Student’s t test was used for estimating statistically significance between two groups. The results were evaluated through Spearman’s correlation coefficient test. All values are shown as mean ± SE of the mean; P < 0.05 was regarded as statistically significant.
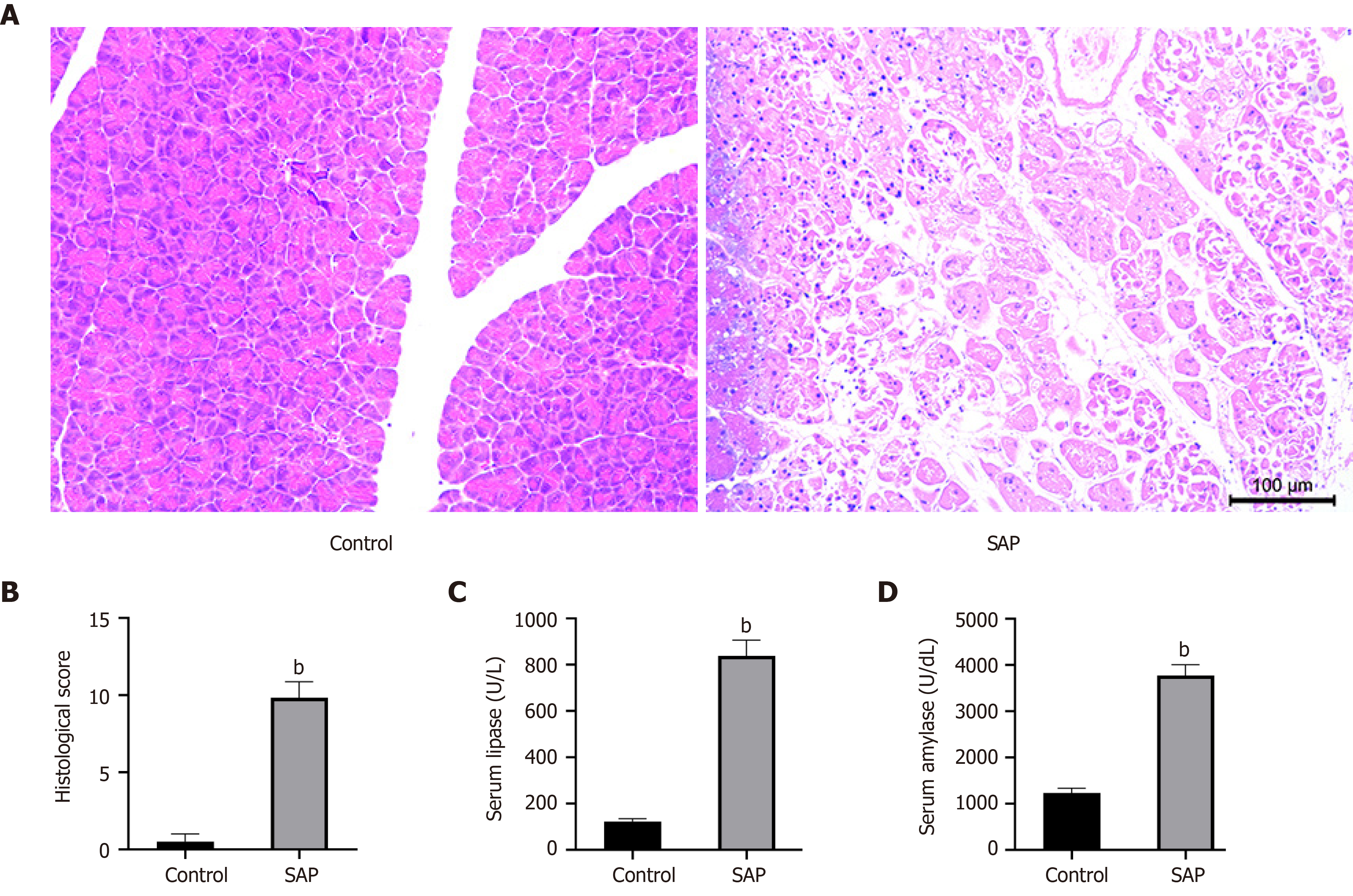
Twenty-four hours after treatment with sodium taurocholate salt, the staining of hematoxylin and eosin on the pancreatic tissues from the SAP group showed typical histopathological changes, including pancreatic lobular edema, extensive acinar cell necrosis, focal expansion of the pancreatic interlobular septum and granulocyte infiltration (Figure 1A). By contrast, under light microscopy, the pancreases from the control group had a complete normal structure. Figure 1B showed the corresponding histopathological scores. At the same time, considering that the levels of serum lipase and amylase are as one of the diagnostic criteria of AP[20], we determined their concentrations in serum. As a result, the serum lipase and amylase levels in the SAP group were also markedly higher than those in the control group (P < 0.05; Figure 1C and 1D). These results confirmed the successful establishment of the SAP mice model.
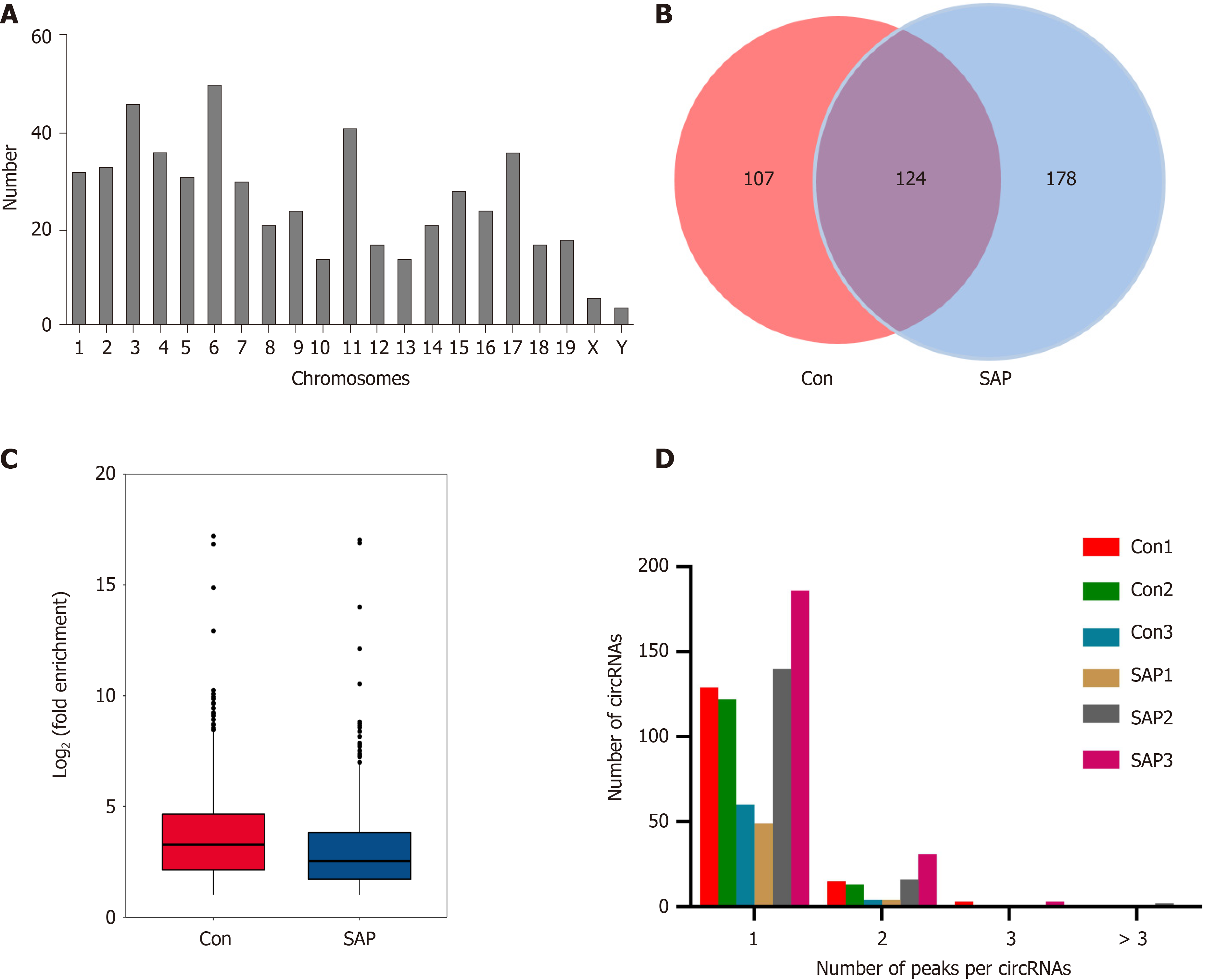
We used MeRIP-seq to investigate the expression of m6A circRNAs in pancreatic tissues from the control and SAP groups. We had submitted the data to the online repository, which can be found at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173298. Before performing MeRIP-seq, the residual determination of 28S and 18S of rRNA showed that the rRNAs in total RNA were removed effectively (Supplementary Figure 1). In general, a total of 409 m6A circRNAs were identified in all chromosomes (Figure 2A). Among these, 178 were specifically expressed in the SAP group, 107 in the control group, and 124 were shared in both groups (Figure 2B). m6A level in total circRNAs from the SAP group was lower than that from the control group (Figure 2C). Besides, > 80% of circRNAs contained only one m6A peak in both SAP and control groups (Figure 2D).
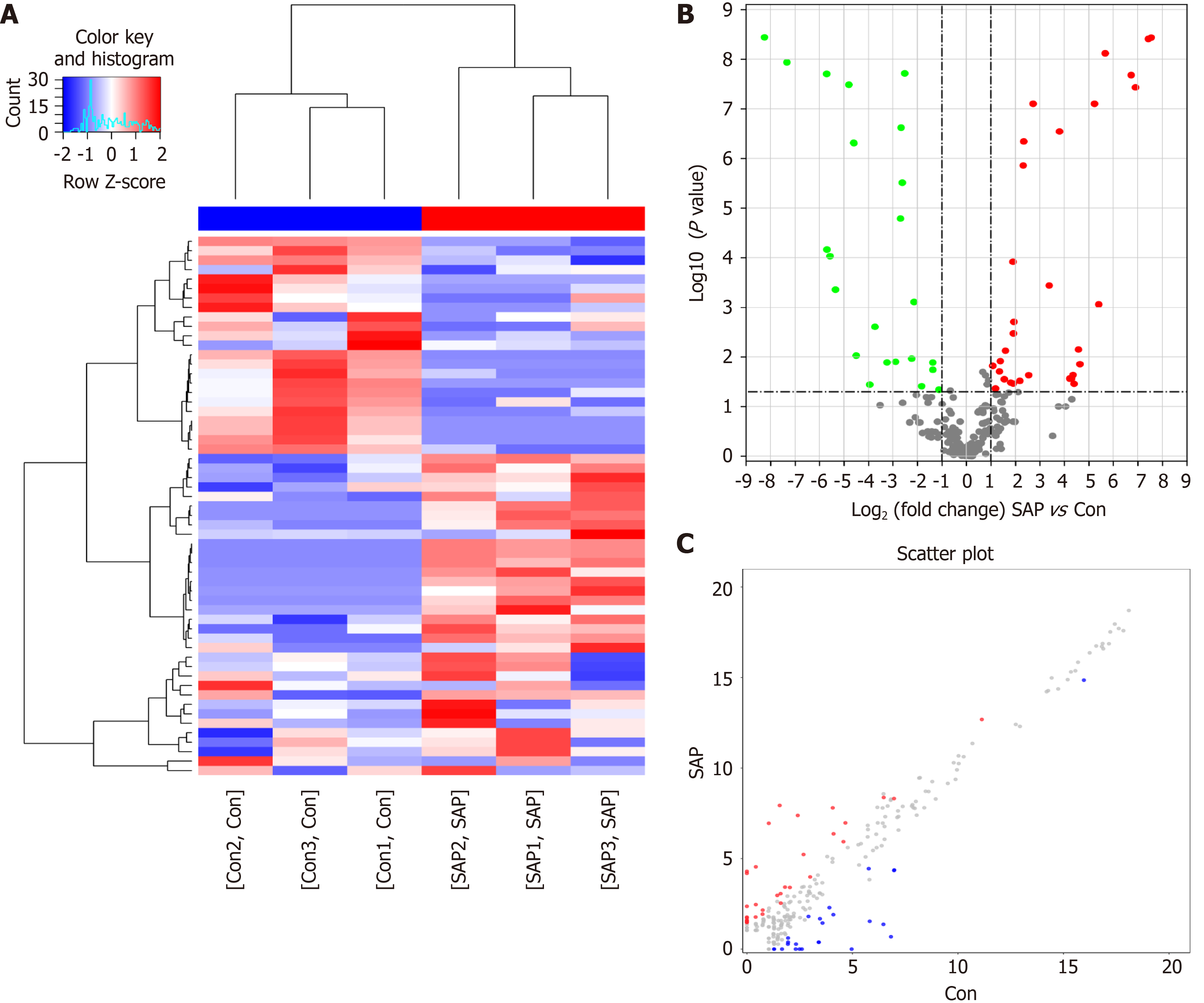
To understand the biological role of m6A modification of circRNAs in SAP, the circRNAs with differentially expressed (DE) m6A peaks were further analyzed. Significant differential expression was defined as fold-change > 2 and P < 0.05. Compared with the control group, 57 circRNAs with DE m6A peaks were identified; 32 were upregulated and 25 downregulated in the SAP group. Table 1 presents the top 10 methylated m6A sites that were up- and downregulated within circRNAs. Figure 3A shows the m6A circRNAs expression profile in the SAP and control groups though hierarchical cluster analysis. The scatter plot exhibits the variation of DE m6A circRNAs between the SAP and control groups (Figure 3B). The volcano plot depicted DE m6A circRNAs between the two groups (Figure 3C).
| PeakStart | PeakEnd | circRNA | Regulation | Fold-change | P value | |
| chr15 | 98658229 | 98658320 | chr15:98656602-98658435- | Up | 187.2 | 3.67392E-09 |
| chr11 | 74929241 | 74929540 | chr11:74928993-74990215+ | Up | 172.8 | 3.92116E-09 |
| chr9 | 108248361 | 108248660 | chr9:108207543-108263690- | Up | 120.034482 | 3.70238E-08 |
| chr2 | 153763381 | 153763760 | chr2:153756037-153769786+ | Up | 106.330434 | 2.09239E-08 |
| chr18 | 30281961 | 30282053 | chr18:30276981-30282053+ | Up | 50.9545454 | 7.61781E-09 |
| chr19 | 40346381 | 40346760 | chr19:40314443-40373578- | Up | 42.4 | 0.026929988 |
| chr16 | 94641481 | 94641740 | chr16:94611419-94694141+ | Up | 37.6772727 | 7.9549E-08 |
| chr10 | 60144412 | 60144720 | chr10:60144413-60144723- | Up | 24.9 | 0.014047401 |
| chr11 | 44652781 | 44652825 | chr11:44651797-44652825+ | Up | 23.9 | 0.007108941 |
| chr7 | 63895821 | 63896100 | chr7:63891679-63938495- | Up | 21.1 | 0.034908475 |
| chr9 | 107852341 | 107852720 | chr9:107847268-107860459- | Down | 302.6 | 3.63198E-09 |
| chr8 | 104143561 | 104143760 | chr8:104143031-104143793+ | Down | 160.728571 | 1.15908E-08 |
| chr1 | 150426881 | 150427260 | chr1:150413021-150442180+ | Down | 52.095238 | 1.98141E-08 |
| chr1 | 13312381 | 13312680 | chr1:13298706-13325802- | Down | 51.7 | 6.86061E-05 |
| chr6 | 119970581 | 119970800 | chr6:119951703-120038640- | Down | 47.4 | 9.37286E-05 |
| chr11 | 23271132 | 23271205 | chr11:23261835-23271205+ | Down | 40.6 | 0.000442849 |
| chr4 | 108499346 | 108499398 | chr4:108486454-108508433+ | Down | 27.79 | 3.27262E-08 |
| chr9 | 102619691 | 102619760 | chr9:102618811-102619760- | Down | 24.25 | 4.83147E-07 |
| chr11 | 32296401 | 32296600 | chr11:32283981-32297161+ | Down | 24.1 | 4.96449E-07 |
| chr9 | 69414201 | 69414580 | chr9:69408311-69432615+ | Down | 22.6 | 0.00943617 |
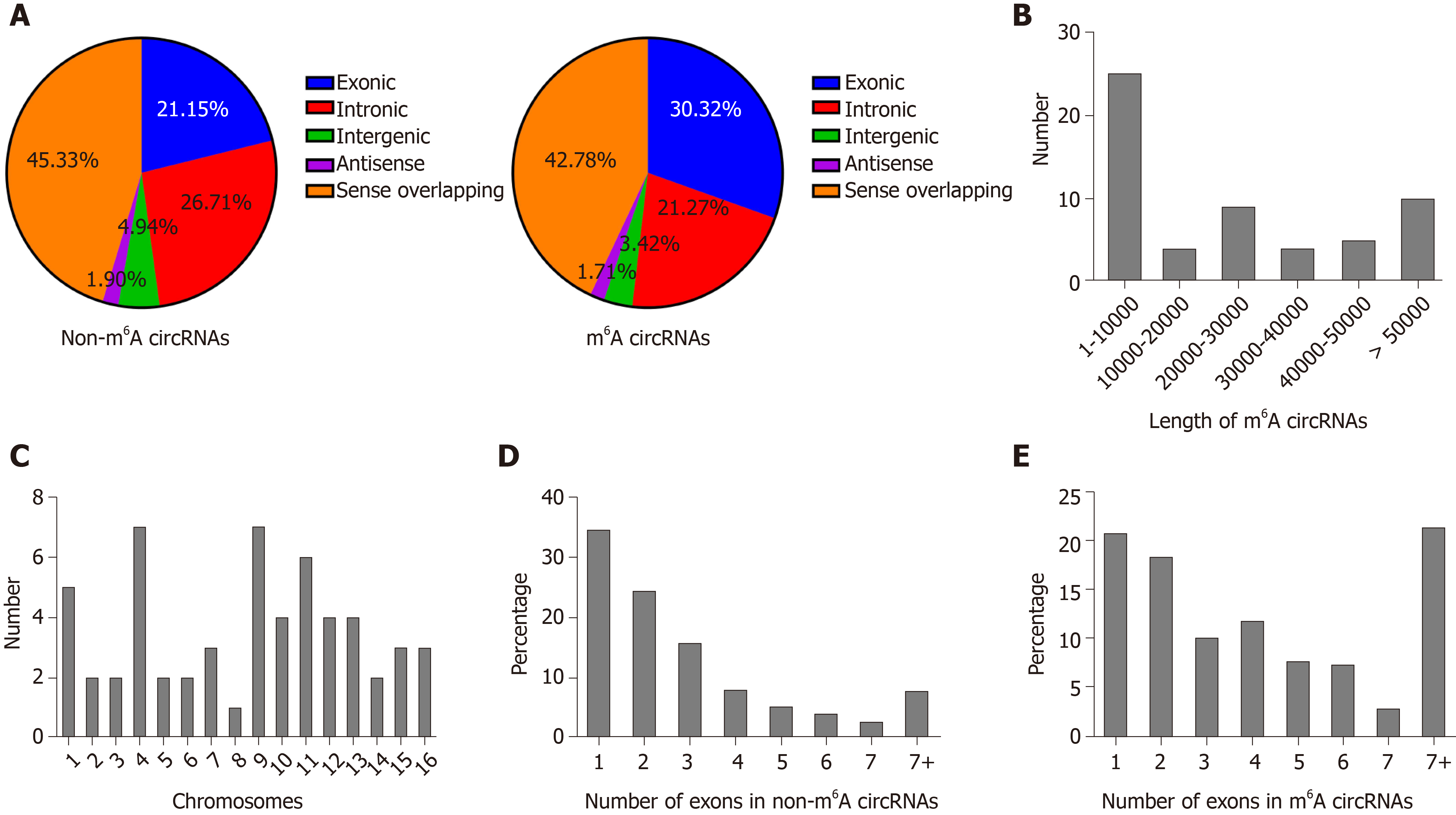
We identified 903 m6A peaks distributed on 781 circRNAs and it is reported that circRNAs can be generated from any region of the genome[21]. Therefore, we firstly analyzed the genomic distribution of m6A and non-m6A circRNAs according to their genomic origins to explore their distribution features. As a results, in non-m6A circRNAs, 45.33% were sense overlapping, 21.15% exonic, 26.71% intronic, 4.94% intergenic and a few antisense; in m6A circRNAs, 42.78% were sense overlapping, 30.32% exonic, 21.27% intronic, 3.42% intergenic and a few antisense (Figure 4A). These results indicated that the majority of m6A and non-m6A circRNAs were commonly encoded by sense overlapping sequences and the number of circRNAs that generated from protein-coding genes in m6A circRNAs was more than those in non-m6A circRNAs.
We further analyzed the distribution of circRNAs with DE m6A peaks. The length of DE m6A circRNAs was mainly enriched in 1–10 000 base pairs (Figure 4B). Although the host genes of m6A circRNAs located in all chromosomes, the dysregulated parts mostly located in chromosomes 4, 9 and 11 (Figure 4C). A previous study reported that most circRNAs that derived from protein-coding genes spanned two or three exons[14]. In this study, the majority of circRNAs from protein-coding genes spanned one or two exons (Figure 4D). Similarly, the majority of m6A circRNAs and non-m6A circRNAs were more commonly encoded by a single or two exons (Figure 4E).
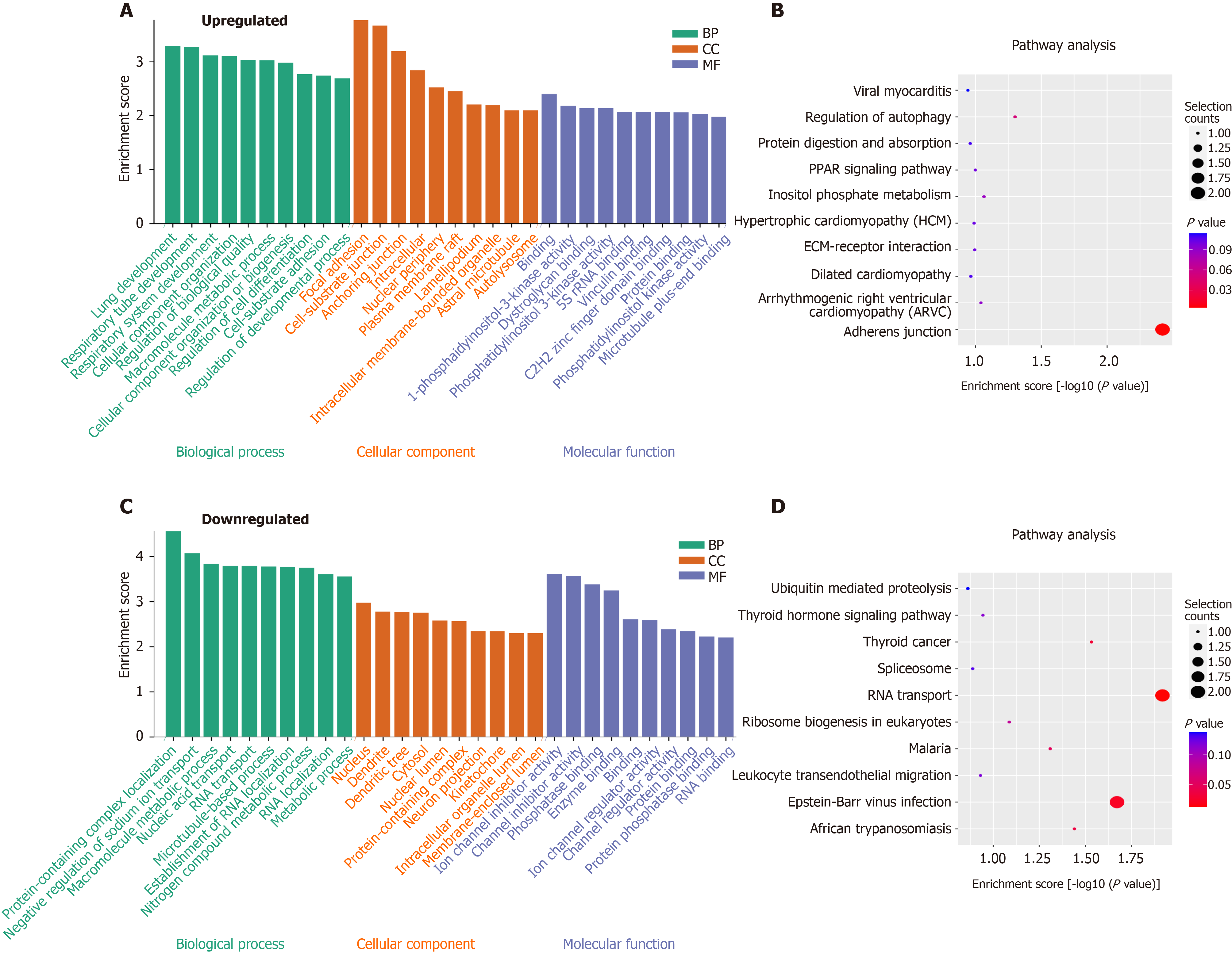
To explore the function of m6A circRNAs in SAP, GO analysis and KEGG pathway analysis of circRNAs with the DE m6A peaks were performed. Figure 5A presented the top 10 GO terms of circRNAs with upregulated m6A peaks from the three aspects: BP, CC and MF. For BP, the most enriched and meaningful GO terms were cellular component organization, macromolecule metabolic process and regulation of developmental process. For CC, the top three terms were focal adhesion, cell–substrate junction and anchoring junction. For MF, the main represented GO terms were C2H2 zinc finger domain binding and protein binding. The top 10 pathways from KEGG pathway analysis for circRNAs with upregulated m6A peaks were selected and presented in a bubble chart (Figure 5B). Among them, protein digestion and absorption and regulation of autophagy were the major signaling pathways associated with the SAP progression.
The GO terms of circRNAs with downregulated m6A peaks are presented in Figure 5C. For BP, protein-containing complex localization, RNA transport and macromolecule metabolic process were the most enriched and meaningful GO terms. For CC, nucleus, dendrite and dendritic tree were the top three terms. For MF, the main represented GO terms were channel regulator activity, RNA, enzyme and protein binding. As for the KEGG pathway analysis of circRNAs with downregulated m6A peaks, RNA transport was the main pathway (Figure 5D).
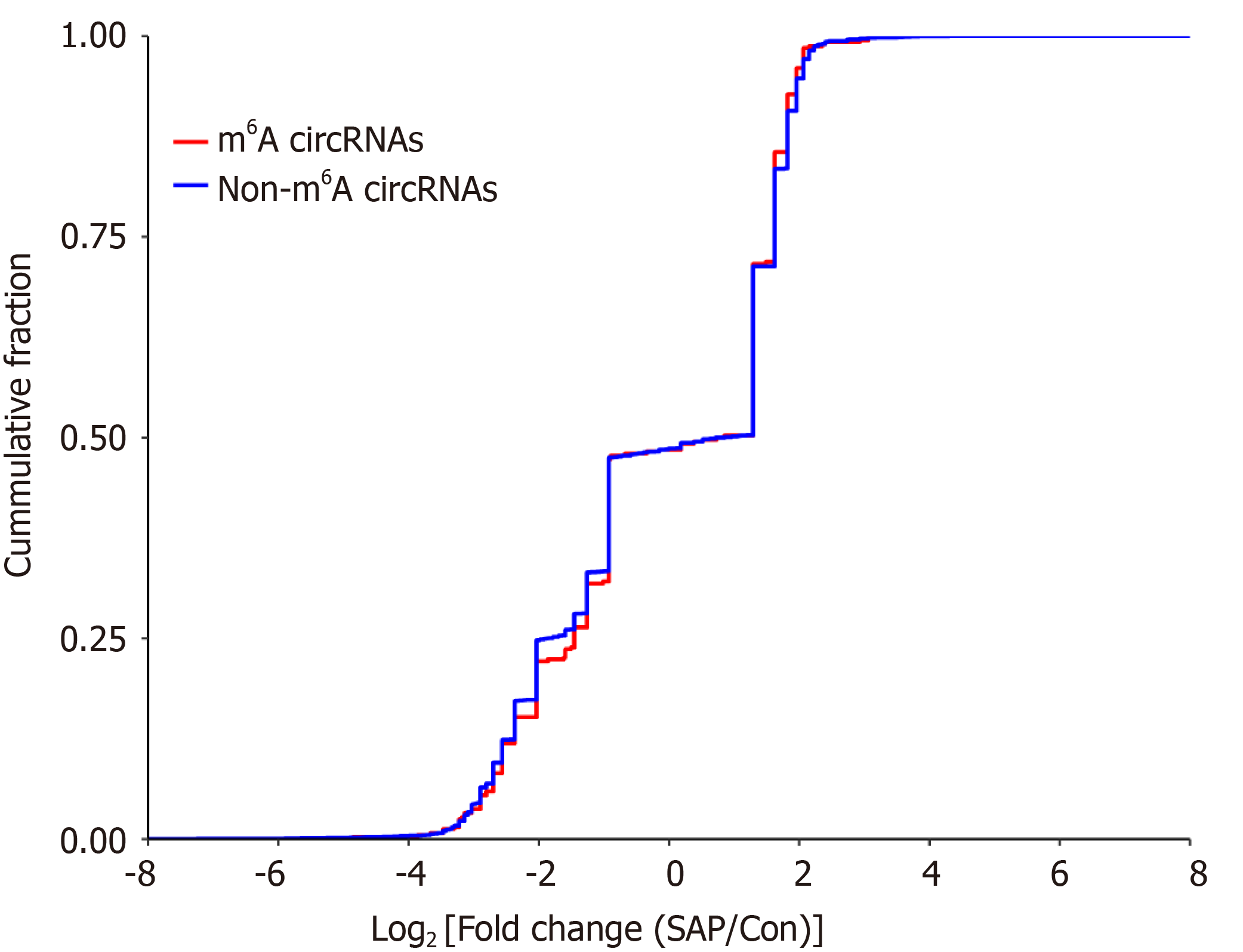
To explore whether m6A modification could affect the expression of circRNAs, we analyzed the expression of m6A circRNAs. The expression level of these circRNAs with DE m6A peaks did not have significant differences (fold-change < 2 or P > 0.05; Supplementary Table 1), indicating that m6A modification of circRNAs did not influence the expression of circRNAs. To verify this result further, we analyzed the cumulative distribution of circRNA expression between the control and SAP groups for m6A and non-m6A circRNAs (Figure 6). This was consistent with the above result.
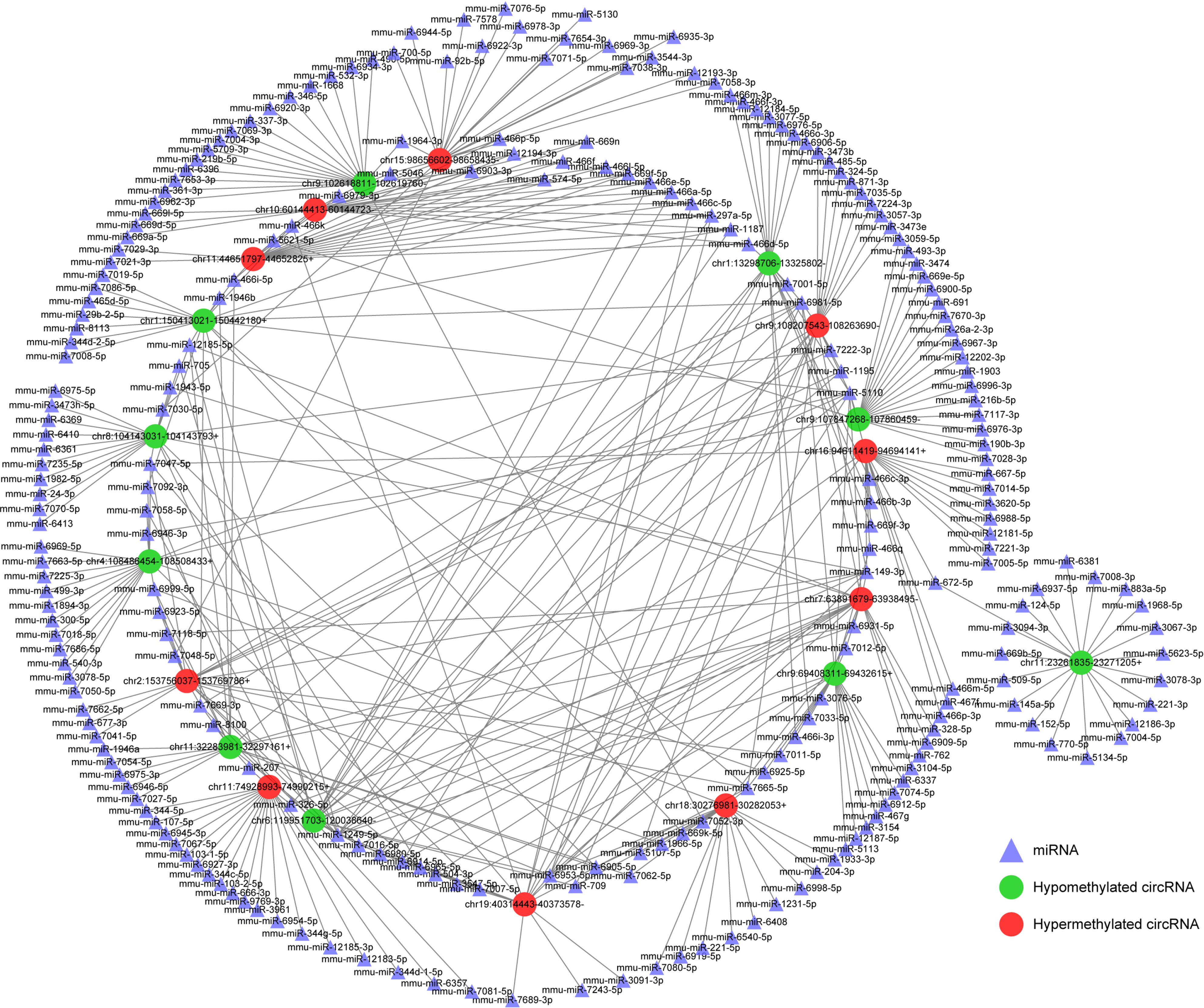
Given the importance of circRNA–miRNA interaction[22] and to further explore the underlying mechanism of these circRNAs with DE m6A peaks, the top 10 upregulated and top 10 downregulated circRNAs according to the level of m6A were selected to construct circRNA–miRNA networks. In this network map, several important miRNAs participated in the occurrence and development of SAP were found to bind to these m6A circRNAs (Figure 7), such as miR-24-3p, miR-26a, miR-92b, miR-216b, miR-324-5p and miR-762. These data suggest that these circRNAs with DE m6A peaks might play a role in the pathological process of SAP.
To explore whether the circRNAs with DE m6A peaks identified in mouse SAP may have similar roles in human SAP, we performed the conservation analysis of the sequence of the top 10 upregulated and top 10 downregulated circRNAs preliminarily. Through aligning with the sequence of human circRNAs that downloaded from circBase database, we found that 15/20 of the selected circRNAs that have highly similar sequences to human circRNAs (sequence identity > 80%), as shown in the Table 2. These results suggested that these circRNAs may have similar roles in human SAP.
| Human circRNA | |||||
| Mouse circRNA | Human circRNA | Hg19 location | Transcript | Parent gene | Sequence identity, % |
| chr15:98656602-98658435- | hsa_circ_0026065 | chr12:49223538-49245957- | NM_004818 | DDX23 | 88.75 |
| chr11:74928993-74990215+ | hsa_circ_0041387 | chr17:2139785-2203958- | NM_001170957 | SMG6 | 86.32 |
| chr9:108207543-108263690- | hsa_circ_0124055 | chr3:49514281-49548252+ | NM_001177634 | DAG1 | 85.36 |
| chr2:153756037-153769786+ | hsa_circ_0059811 | chr20:31436477-31438211+ | NM_012325 | MAPRE1 | 84.18 |
| chr19:40314443-40373578- | hsa_circ_0094611 | chr10:97110965-97114724- | ENST00000371247.2 | SORBS1 | 93.52 |
| chr16:94611419-94694141+ | hsa_circ_0115989 | chr21:38792600-38888974+ | ENST00000338785.3 | DYRK1A | 91.34 |
| chr7:63891679-63938495- | hsa_circ_0034321 | chr15:31619082-31670102+ | NM_015995 | KLF13 | 85.89 |
| chr9:107847268-107860459- | hsa_circ_0065768 | chr3:50000008-50114685+ | NM_005777 | RBM6 | 90.84 |
| chr1:150413021-150442180+ | hsa_circ_0111511 | chr1:186294895-186325581- | NM_003292 | TPR | 87.91 |
| chr1:13298706-13325802- | hsa_circ_0113369 | chr1:42166586-42254891- | ENST00000247584.5 | HIVEP3 | 91.30 |
| chr6:119951703-120038640- | hsa_circ_0024963 | chr12:939168-990955+ | NM_001184985 | WNK1 | 92.02 |
| chr11:23261835-23271205+ | hsa_circ_0120688 | chr2:61749745-61764803- | ENST00000404992.2 | XPO1 | 95.11 |
| chr4:108486454-108508433+ | hsa_circ_0012539 | chr1:52927184-53018762- | NM_001009881 | ZCCHC11 | 92.38 |
| chr11:32283981-32297161+ | hsa_circ_0118668 | chr2:202780266-202790202- | None | None | 91.48 |
| chr9:69408311-69432615+ | hsa_circ_0035568 | chr15:60720627-60748993- | NM_024611 | NARG2 | 87.08 |
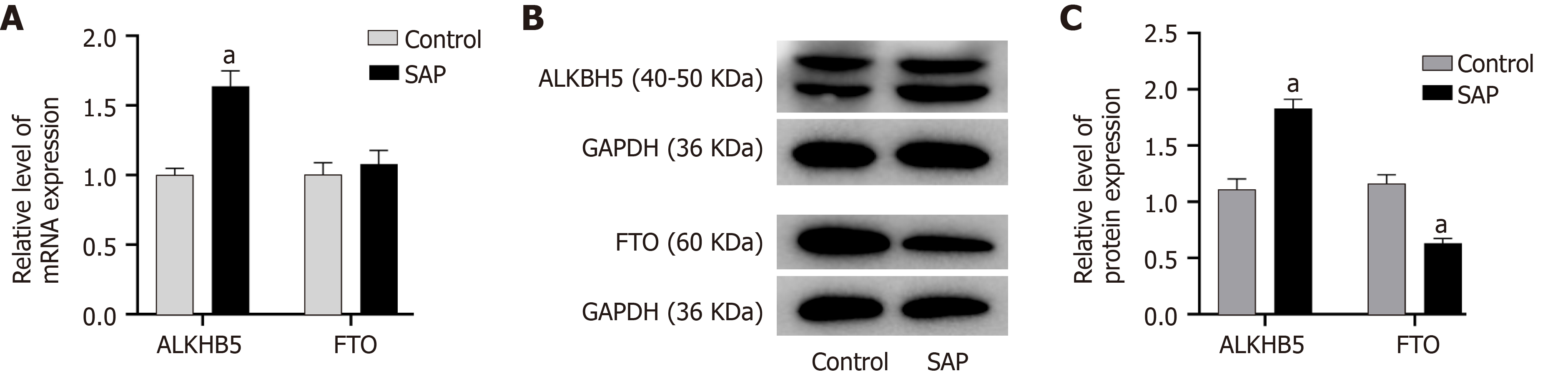
Given that the total m6A level of circRNAs was reduced in SAP and to explore how the m6A level was regulated in SAP, we detected the protein and mRNA expression of two demethyltransferases (ALKBH5 and FTO). FTO was reduced at the level of protein, but ALKBH5 was increased in SAP at both the level of mRNA and protein (Figure 8). These results indicated that ALKBH5 might be related to the dynamic process of m6A in SAP.
In the present study, we identified transcriptome-wide map of m6A circRNAs and determined their biological significance and potential mechanisms for the first time in SAP. The main findings are: (1) We identified 57 circRNAs with DE m6A peaks and found these DE m6A circRNAs were involved in the key process of SAP by GO and KEGG analysis, such as protein digestion and regulation of autophagy; (2) In m6A circRNA-miRNA networks, several important miRNAs participated in the initiation and development of SAP were found to bind to these m6A circRNAs potentially, suggesting that m6A may affect the interplays with miRNAs; and (3) The total m6A level was reduced in SAP, and the demethylase ALKBH5 was found to be upregulated in SAP, indicating that ALKBH5 may be related to dynamic process of m6A in SAP. These results suggested that m6A modification on circRNAs may be involved in the pathophysiology of SAP, which may provide novel insights to explore the possible pathophysiological mechanism of SAP and seek new potential therapeutic targets.
To find effective therapeutic targets for SAP, many studies have explored the underlying molecular mechanisms of SAP. Our previous study found that many circRNAs are expressed in mice with SAP[9] and these circRNAs play an important role in the pathogenetic mechanism of SAP[9,23]. In recent years, m6A modification of circRNAs was found to be widespread[14] and gained widespread attention in epigenetics. Several important studies have investigated the roles of m6A modification in circRNA metabolism and found that m6A circRNAs play key roles in some diseases[16,24-28]. In circRNA metabolism, m6A modifications can regulate its translation through recognition by YTHDF3 and eIF4G2, and this progress of translation can be enhanced by METTL3/14 and inhibited by FTO[24,25]. In addition, m6A circRNAs associate with YTHDF2 in an HRSP12-dependent manner and are selectively downregulated by RNase P/MRP[26]. In innate immunity, Chen et al[27] found that unmodified circRNA adjuvant induces antigen-specific T and B cell responses, but m6A modification could abrogate circRNA immunity though YTHDF2-mediated suppression. In male germ cells, the back splicing tends to occur mainly at m6A-enriched sites, which are usually located around the start and stop codons in linear mRNAs, resulting in about half of circRNAs containing large open reading frames. This potential mechanism could ensure long-lasting and stable protein production for specific physiological processes when lacking the corresponding linear mRNAs[28]. These findings showed the important roles of m6A in circRNAs during disease progress. Therefore, it is essential to explore the roles of m6A circRNAs in SAP.
In the present study, the function analysis of DE m6A circRNAs in SAP found that two important pathways were involved in the pathogenesis of SAP, including protein digestion and regulation of autophagy. As an important pathological cellular event, the activation of premature trypsinogen can result in acinar cell necrosis[1]. Many pancreatic injury factors, such as trauma, obstruction of the pancreatic duct and alcohol, can initiate the fusion of lysosomes with zymogen in acinar cells, leading to the activation of trypsinogen through cathepsin B to trypsin. Once trypsin is released, it can cause self-digestion in and outside the acinar cells, and the release of cathepsin B can cause necroptosis. As a cytoprotective mechanism, autophagy can process and recycle various aged, defective or damaged cytoplasmic contents[29]. Selective macroautophagy is a biological process during which specific damaged organelles and misfolded proteins are processed and recycled. Autophagy is accomplished via a series of steps, which start with the enucleation of cytoplasmic inclusions in the open double membrane formed by the ER, Golgi apparatus and plasma membrane[30]. Knocking out ATG7 genes (which are important to form autophagosome) and LAMP genes could lead to pancreatitis with extensive inflammation in mice[29,31]. Importantly, impaired autophagy leads to trypsinogen activation, ER stress and mitochondrial dysfunction. These events can together make acinar cells become more susceptible to other insults and cellular death[1]. In addition, RNA transport is enriched in GO terms of note, and Chen et al[16] found that m6A modification can modulate the export of circNSUN2 to the cytoplasm, suggesting that m6A modification regulates transport of circRNAs in SAP. These results were consisted with the hypothesis that m6A modification of circRNAs participated in the progression of SAP.
m6A modification of mRNA can influence its expression by regulating transcription, splicing and degradation[32]. In circRNAs, Zhou et al[14] and Su et al[33] reported that m6A levels are correlated with expression levels of circRNAs in HeLa cells and a rat model of hypoxia-mediated pulmonary hypertension. However, in SAP, we found m6A modification in circRNAs was not associated with expression of circRNAs, suggesting that m6A circRNAs function in SAP though other mechanisms, such as miRNA sponges. It is worth mentioning that more direct evidence is currently needed to support that m6A can affect circRNA expression.
miRNA sponges is an important function of circRNAs. Cytoplasmic circRNAs can prevent miRNAs from binding to target mRNAs by competitive binding to miRNA response elements, further playing a key role in diseases[8,34]. For instance, in lung squamous cell carcinoma, circTP63 can competitively bind to miR-873-3p and prevent miR-873-3p from decreasing the level of FOXM1. The FOXM1 can upregulate the expression of CENPA and CENPB, ultimately facilitating cell cycle progression[35]. In SAP, circHIPK3 can enhance pyroptosis via regulating the miR-193a-5p/GSDMD axis in acinar cells, ultimately aggravating this disease[36]. In our previous study, we found that circZFP644 could sponge miR-21-3p, thereby participating in the pathogenesis of SAP[9]. Recently, Su et al[33] found that m6A modification of circRNAs could influence the interactions between circRNAs and miRNAs. Therefore, analysis of m6A circRNA–miRNA networks was performed in this study. Several important miRNAs participated in the pathological process of SAP were found to bind to these m6A circRNAs, such as miR-24-3p, miR-26a, miR-92b, miR-216b, miR-324-5p and miR-762. For example, in caerulein-stimulated AR42J cells, expression of miR-92b-3p was decreased, while overexpression of miR-92b-3p could downregulate the expression of TRAF3 and inhibit the MKK3-p38 pathway, attenuating inflammatory response and autophagy[37]. These results suggest that m6A modification of circRNAs functions by influencing the interactions between circRNAs and miRNAs.
m6A modification is a reversible process that occurs by methyltransferase complex consisting of METTL3, METTL14 and WTAP, and is “erased” by ALKBH5 and FTO[13,15]. In pancreatic cancer, ALKBH5 could regulate the post-transcriptional activation of PER1 through m6A abolishment, thereby inhibiting the cancer[38]. In hepatocellular carcinoma, ALKBH5 could attenuate expression of LYPD1 by an m6A-dependent manner and act as a tumor suppressor[39]. Overall, this evidence has suggested that ALKBH5 plays an essential role in m6A modification. In this study, we found that expression level of ALKBH5 was upregulated in SAP. Consistent with this result, total m6A level of circRNAs in SAP was reduced, indicating that ALKBH5 may play a role in the dynamic process of m6A in SAP.
However, there are still limitations in our study. Firstly, further in vivo and in vitro experiments are needed to further explore the m6A circRNA-mediated precise regulatory mechanisms in SAP. Secondly, the conservation analysis of the m6A circRNAs showed that these circRNAs may have similar roles in human SAP. However, their clinical significance and the results should be investigated further in SAP patients. Additionally, the precise mechanism of ALKBH5 in m6A circRNAs during SAP needs to be studied. Actually, these are in our next plans to explore the roles of m6A circRNAs in SAP.
In conclusion, our study identified the transcriptome-wide profiling of m6A circRNAs in SAP and predicted their biological significance and possible potential mechanisms, providing new insights to explore the possible pathophysiological mechanism of SAP and seek new potential therapeutic targets.
Severe acute pancreatitis (SAP) is a lethal inflammatory disease with mortality up to 30%. But the genetic pathological mechanism of SAP remains unclear and SAP is still lack of effective therapeutic options. N6-methyladenosine (m6A) modification of circular (circ)RNAs plays a key role in many diseases and physiological processes through regulating the metabolism and function of circRNAs. However, the role of m6A circRNA in SAP has been unexplored yet.
The pathophysiology of SAP at the level of gene regulation is complex and remains unclear. circRNAs are found to participate in many physiological processes and play key roles in pathological processes during SAP. m6A modification can affect the “fate” of m6A modified circRNAs, thereby participating in the regulation of diseases. Therefore, we want to explore whether the m6A modification of circRNAs is related to the pathophysiological mechanism of SAP, and determine their biological significance and potential mechanisms.
The present study aims to determine the transcriptome-wide map of m6A circRNAs and explore their biological significance and its possible mechanisms in SAP.
The SAP C57BL/6 mice model was induced by retrograde injection of 4% sodium taurocholate salt. m6A-modified RNA immunoprecipitation sequencing was used to determine the transcriptome-wide map of m6A circRNAs. The biological significance of circRNAs with differentially expressed m6A peaks was identified by GO and KEGG analysis. m6A circRNA-microRNA networks was constructed to explore the underlying mechanism of m6A circRNAs in SAP. The expression of demethylases was measured by western blot and qPCR. H&E staining and measurement of serum lipase and amylase were performed to assess the establishment of SAP mice model.
In the identified transcriptome-wide map of m6A circRNAs, there were 57 circRNAs with differentially expressed m6A peaks; among which, 32 were upregulated and 25 downregulated. Important pathways in the pathogenetic process during SAP were found by functional analysis of these m6A circRNAs, such as protein digestion and regulation of autophagy. m6A circRNA–miRNA networks showed that several important miRNAs in pathogenesis of SAP were bind to these m6A circRNAs, such as miR-24-3p, miR-26a, miR-92b, miR-216b, miR-324-5p and miR-762. To be note, the total m6A level of circRNAs was reduced in SAP, accompanied by the upregulated demethylase ALKBH5.
The transcriptome-wide profiling of m6A circRNAs in SAP was identified, and the biological significance and possible potential mechanisms of m6A circRNAs in SAP were predicted, providing new insights into exploring the possible pathophysiological mechanism of SAP and new potential therapeutic targets.
This present study for the first time identified transcriptome-wide map of m6A circRNAs and determined their biological significance and potential mechanisms. However, the m6A circRNA-mediated precise regulatory mechanisms are need to be explore further in vivo and vitro experiments. What’s more, further studies are needed to reveal the precise mechanism of ALKBH5 in m6A circRNAs during SAP. In the future, we will explore them and investigate these m6A circRNAs in SAP patients.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Kontos CK, Surbatovic M S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Xing YX
| 1. | Lee PJ, Papachristou GI. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol. 2019;16:479-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 516] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 2. | Trikudanathan G, Wolbrink DRJ, van Santvoort HC, Mallery S, Freeman M, Besselink MG. Current Concepts in Severe Acute and Necrotizing Pancreatitis: An Evidence-Based Approach. Gastroenterology. 2019;156:1994-2007.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 248] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 3. | Garg PK, Singh VP. Organ Failure Due to Systemic Injury in Acute Pancreatitis. Gastroenterology. 2019;156:2008-2023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 371] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 4. | Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 591] [Cited by in RCA: 811] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 5. | Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18:547-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 135] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 6. | Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 126] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 7. | Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838-1847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 664] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 8. | Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16:503-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 264] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 9. | Yang Y, Ren J, Huang Q, Wu J, Yuan X, Jiang W, Wen Y, Tang L, Sun H. CircRNA Expression Profiles and the Potential Role of CircZFP644 in Mice With Severe Acute Pancreatitis via Sponging miR-21-3p. Front Genet. 2020;11:206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 10. | Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 833] [Cited by in RCA: 1157] [Article Influence: 144.6] [Reference Citation Analysis (0)] |
| 11. | Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, Gentsch J, Wang F; Dominantly Inherited Alzheimer Network (DIAN), Salloway S, Masters CL, Lee JH, Graff-Radford NR, Chhatwal JP, Bateman RJ, Morris JC, Karch CM, Harari O, Cruchaga C. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22:1903-1912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 248] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 12. | Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, Yang A, Wang T. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588-607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 213] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 13. | Shulman Z, Stern-Ginossar N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat Immunol. 2020;21:501-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 280] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 14. | Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017;20:2262-2276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 341] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 15. | Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, Liu J, Sun Z. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol Cancer. 2020;19:105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 223] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 16. | Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, Wang F, Ma NF, Guan X, Yun JP, Wang FW, Xu RH, Dan Xie. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 518] [Article Influence: 86.3] [Reference Citation Analysis (0)] |
| 17. | Schmidt J, Rattner DW, Lewandrowski K, Compton CC, Mandavilli U, Knoefel WT, Warshaw AL. A better model of acute pancreatitis for evaluating therapy. Ann Surg. 1992;215:44-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 673] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 18. | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29963] [Cited by in RCA: 28883] [Article Influence: 1155.3] [Reference Citation Analysis (1)] |
| 19. | Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1116] [Cited by in RCA: 1589] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 20. | Crockett SD, Wani S, Gardner TB, Falck-Ytter Y, Barkun AN; American Gastroenterological Association Institute Clinical Guidelines Committee. American Gastroenterological Association Institute Guideline on Initial Management of Acute Pancreatitis. Gastroenterology. 2018;154:1096-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 556] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 21. | Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 325] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 22. | Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 800] [Article Influence: 114.3] [Reference Citation Analysis (0)] |
| 23. | Wang B, Wu J, Huang Q, Yuan X, Yang Y, Jiang W, Wen Y, Tang L, Sun H. Comprehensive Analysis of Differentially Expressed lncRNA, circRNA and mRNA and Their ceRNA Networks in Mice With Severe Acute Pancreatitis. Front Genet. 2021;12:625846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 24. | Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, Incarnato D, Oliviero S, Fatica A, Morlando M, Bozzoni I. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020;31:107641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 245] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 25. | Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, Wong CC, Xiao X, Wang Z. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 960] [Cited by in RCA: 1397] [Article Influence: 174.6] [Reference Citation Analysis (0)] |
| 26. | Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol Cell. 2019;74:494-507.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 425] [Article Influence: 70.8] [Reference Citation Analysis (0)] |
| 27. | Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, Hur S, Chang HY. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol Cell. 2019;76:96-109.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 405] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 28. | Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ, Tang Y, Zhang X, Qin W, Zhang Y, Song G, Zheng W, Wang J, Chen W, Wei X, Xie Z, Klukovich R, Zheng H, Quilici DR, Yan W. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30:211-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 149] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 29. | Antonucci L, Fagman JB, Kim JY, Todoric J, Gukovsky I, Mackey M, Ellisman MH, Karin M. Basal autophagy maintains pancreatic acinar cell homeostasis and protein synthesis and prevents ER stress. Proc Natl Acad Sci U S A. 2015;112:E6166-E6174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 30. | Gukovskaya AS, Gukovsky I, Algül H, Habtezion A. Autophagy, Inflammation, and Immune Dysfunction in the Pathogenesis of Pancreatitis. Gastroenterology. 2017;153:1212-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 256] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 31. | Mareninova OA, Hermann K, French SW, O'Konski MS, Pandol SJ, Webster P, Erickson AH, Katunuma N, Gorelick FS, Gukovsky I, Gukovskaya AS. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009;119:3340-3355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 32. | Zhu ZM, Huo FC, Pei DS. Function and evolution of RNA N6-methyladenosine modification. Int J Biol Sci. 2020;16:1929-1940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 33. | Su H, Wang G, Wu L, Ma X, Ying K, Zhang R. Transcriptome-wide map of m6A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics. 2020;21:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 34. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5550] [Article Influence: 396.4] [Reference Citation Analysis (0)] |
| 35. | Cheng Z, Yu C, Cui S, Wang H, Jin H, Wang C, Li B, Qin M, Yang C, He J, Zuo Q, Wang S, Liu J, Ye W, Lv Y, Zhao F, Yao M, Jiang L, Qin W. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat Commun. 2019;10:3200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 284] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 36. | Wang J, Li X, Liu Y, Peng C, Zhu H, Tu G, Yu X, Li Z. CircHIPK3 Promotes Pyroptosis in Acinar Cells Through Regulation of the miR-193a-5p/GSDMD Axis. Front Med (Lausanne). 2020;7:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 37. | Sun H, Tian J, Li J. MiR-92b-3p ameliorates inflammation and autophagy by targeting TRAF3 and suppressing MKK3-p38 pathway in caerulein-induced AR42J cells. Int Immunopharmacol. 2020;88:106691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 38. | Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q, Wan R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 2020;19:91. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 261] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 39. | Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R, Cheng Q, Yang B, Feng X, Lu Y, Xie H, Zhou L, Wu J, Zheng S. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m6A-guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 199] [Article Influence: 39.8] [Reference Citation Analysis (0)] |