Published online Sep 21, 2021. doi: 10.3748/wjg.v27.i35.5978
Peer-review started: April 30, 2021
First decision: June 23, 2021
Revised: July 7, 2021
Accepted: August 25, 2021
Article in press: August 25, 2021
Published online: September 21, 2021
The nature of input data is an essential factor when training neural networks. Research concerning magnetic resonance imaging (MRI)-based diagnosis of liver tumors using deep learning has been rapidly advancing. Still, evidence to support the utilization of multi-dimensional and multi-parametric image data is lacking. Due to higher information content, three-dimensional input should presumably result in higher classification precision. Also, the differentiation between focal liver lesions (FLLs) can only be plausible with simultaneous analysis of multi-sequence MRI images.
To compare diagnostic efficiency of two-dimensional (2D) and three-dimensional (3D)-densely connected convolutional neural networks (DenseNet) for FLLs on multi-sequence MRI.
We retrospectively collected T2-weighted, gadoxetate disodium-enhanced arterial phase, portal venous phase, and hepatobiliary phase MRI scans from patients with focal nodular hyperplasia (FNH), hepatocellular carcinomas (HCC) or liver metastases (MET). Our search identified 71 FNH, 69 HCC and 76 MET. After volume registration, the same three most representative axial slices from all sequences were combined into four-channel images to train the 2D-DenseNet264 network. Identical bounding boxes were selected on all scans and stacked into 4D volumes to train the 3D-DenseNet264 model. The test set consisted of 10-10-10 tumors. The performance of the models was compared using area under the receiver operating characteristic curve (AUROC), specificity, sensitivity, positive predictive values (PPV), negative predictive values (NPV), and f1 scores.
The average AUC value of the 2D model (0.98) was slightly higher than that of the 3D model (0.94). Mean PPV, sensitivity, NPV, specificity and f1 scores (0.94, 0.93, 0.97, 0.97, and 0.93) of the 2D model were also superior to metrics of the 3D model (0.84, 0.83, 0.92, 0.92, and 0.83). The classification metrics of FNH were 0.91, 1.00, 1.00, 0.95, and 0.95 using the 2D and 0.90, 0.90, 0.95, 0.95, and 0.90 using the 3D models. The 2D and 3D networks' performance in the diagnosis of HCC were 1.00, 0.80, 0.91, 1.00, and 0.89 and 0.88, 0.70, 0.86, 0.95, and 0.78, respectively; while the evaluation of MET lesions resulted in 0.91, 1.00, 1.00, 0.95, and 0.95 and 0.75, 0.90, 0.94, 0.85, and 0.82 using the 2D and 3D networks, respectively.
Both 2D and 3D-DenseNets can differentiate FNH, HCC and MET with good accuracy when trained on hepatocyte-specific contrast-enhanced multi-sequence MRI volumes.
Core Tip: Our study aimed to assess the performance of two-dimensional (2D) and three-dimensional (3D) densely connected convolutional neural networks (DenseNets) in the classification of focal liver lesions (FLLs) based on multi-parametric magnetic resonance imaging (MRI) with hepatocyte-specific contrast. We used multi-channel data input to train our networks and found that both 2D and 3D-DenseNets can differentiate between focal nodular hyperplasias, hepatocellular carcinomas or liver metastases with excellent accuracy. We conclude that DensNets can reliably classify FLLs based on multi-parametric and hepatocyte-specific post-contrast MRI. Meanwhile, multi-channel input is advantageous when the number of clinical cases available for model training is limited.
- Citation: Stollmayer R, Budai BK, Tóth A, Kalina I, Hartmann E, Szoldán P, Bérczi V, Maurovich-Horvat P, Kaposi PN. Diagnosis of focal liver lesions with deep learning-based multi-channel analysis of hepatocyte-specific contrast-enhanced magnetic resonance imaging. World J Gastroenterol 2021; 27(35): 5978-5988
- URL: https://www.wjgnet.com/1007-9327/full/v27/i35/5978.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i35.5978
Artificial intelligence (AI)-based analysis is one of the fastest evolving fields in medical imaging, thanks to the rapid development of medical physics, electronic engineering, and computer science. The need for computer-aided diagnostics has been further amplified by the continuously increasing demand for imaging studies and the arrival of new modalities that put extra pressure on radiologists while also increasing the probability of diagnostic errors[1]. Meanwhile, deep learning (DL)-based algorithms have started to gain attention among medical researchers, since they provide excellent reproducibility and the ability to quantify aspects of imaging data unobservable to the human eye, resulting in automatically generated statistical reports and predictions, such as the potential of malignancy or metastatic spread and automated volume assessment, among other uses. Nowadays, AI has become compatible with the full spectrum of imaging modalities and has evolved the capacity to diagnose lesions in various organ systems with greater accuracy than a human reader[2]. The processed data often include two-dimensional (2D) slices or three-dimensional (3D) image volumes; moreover, in the case of magnetic resonance imaging (MRI) studies, the different sequences are condensed into a multi-channel input. Due to their efficiency, convolutional neural networks (CNNs) have replaced other machine learning (ML) approaches in most image classification and segmentation tasks[3,4]. Recently, densely connected CNNs (DenseNets) have become more popular than plain CNN architectures. DenseNets use shortcut connections between the convolutional layers to facilitate gradient flow and optimize the number of trainable parameters. In return, these networks yield improved accuracy and efficiency in medical image classification tasks[5].
Focal liver lesions (FLLs) are common incidental findings during imaging studies, and the work-up often requires further diagnostic procedures, such as dynamic contrast-enhanced ultrasound, computed tomography and liver biopsy. Meanwhile, the excellent soft-tissue contrast, volumetric image acquisition and avoidance of ionizing radiation make multi-phase dynamic post-contrast MRI the primary tool for detection and characterization of liver lesions. The use of hepatocyte-specific contrast agents (HSAs), such as gadoxetic acid and gadobenate dimeglumine, further improves the sensitivity and specificity of the diagnosis of FLLs, as the enhancement characteristics of these lesions in the hepatobiliary phase (HBP) correlates with hepatocyte uptake[6,7]. Additionally, HSA-enhanced MRI is capable of detecting lesions smaller than 10 mm, making it an optimal modality for the early detection of liver metastases (METs)[8].
In the present study, we compared the performance of 2D and 3D-DenseNets in the classification of three types of FLLs, including focal nodular hyperplasia (FNH), hepatocellular carcinoma (HCC) and MET. To guarantee the highest possible prediction rate, we used HSA-enhanced multi-phase dynamic post-contrast MRI scans for the classification task. According to our knowledge, this is the first study to evaluate 2D and 3D-DenseNets for the diagnosis of FLLs and using multi-channel images combining four different MRI sequences. The reporting of this study follows the STROBE Statement checklist of items[9].
In our single-center study, we retrospectively collected multi-phasic MRI studies of patients with FNHs, HCCs or METs, that were acquired using Primovist (gadoxetate disodium), an HSA, from the picture archiving and communication system of the Medical Imaging Centre of our university. As this is a retrospective study, the need for written patient consent was waived by the Institutional Research Ethics Committee. The collected images were acquired between November 2017 and October 2020 using a Philips Ingenia 1.5 T scanner (Cambridge, MA, United States). T2-weighted (T2w) spectral-attenuated inversion recovery (commonly referred to as SPAIR), arterial phase (HAP), portal venous phase (PVP), and HBP scans were collected from each eligible patient for further analysis. Included lesions were either histologically confirmed or exhibited typical characteristics of the given lesion type with MRI. Patients younger than 18 years of age at the time of imaging were excluded from the study. Table 1 contains details of patient demographics, properties of each lesion class, and metastatic lesion origin.
| Patent properties | FNH | HCC | MET | Total |
| Number of patients | 42 | 13 | 14 | 69 |
| Age in years at imaging, mean ± SD | 45 ± 12 | 66 ± 5 | 57 ± 10 | 54 ± 14 |
| Sex | ||||
| Male | 11 | 8 | 8 | 27 |
| Female | 31 | 5 | 6 | 42 |
| Lesion properties | ||||
| Number | 71 | 69 | 76 | 216 |
| Primary type | ||||
| CRC | 21 | |||
| Leiomyosarcoma | 18 | |||
| GI adenocc. or cholangiocc. | 15 | |||
| Breast cc. | 11 | |||
| Pancreas cc. | 7 | |||
| Neuroendocrine ileum cc. | 3 | |||
| Papillary thyroid cc. | 1 |
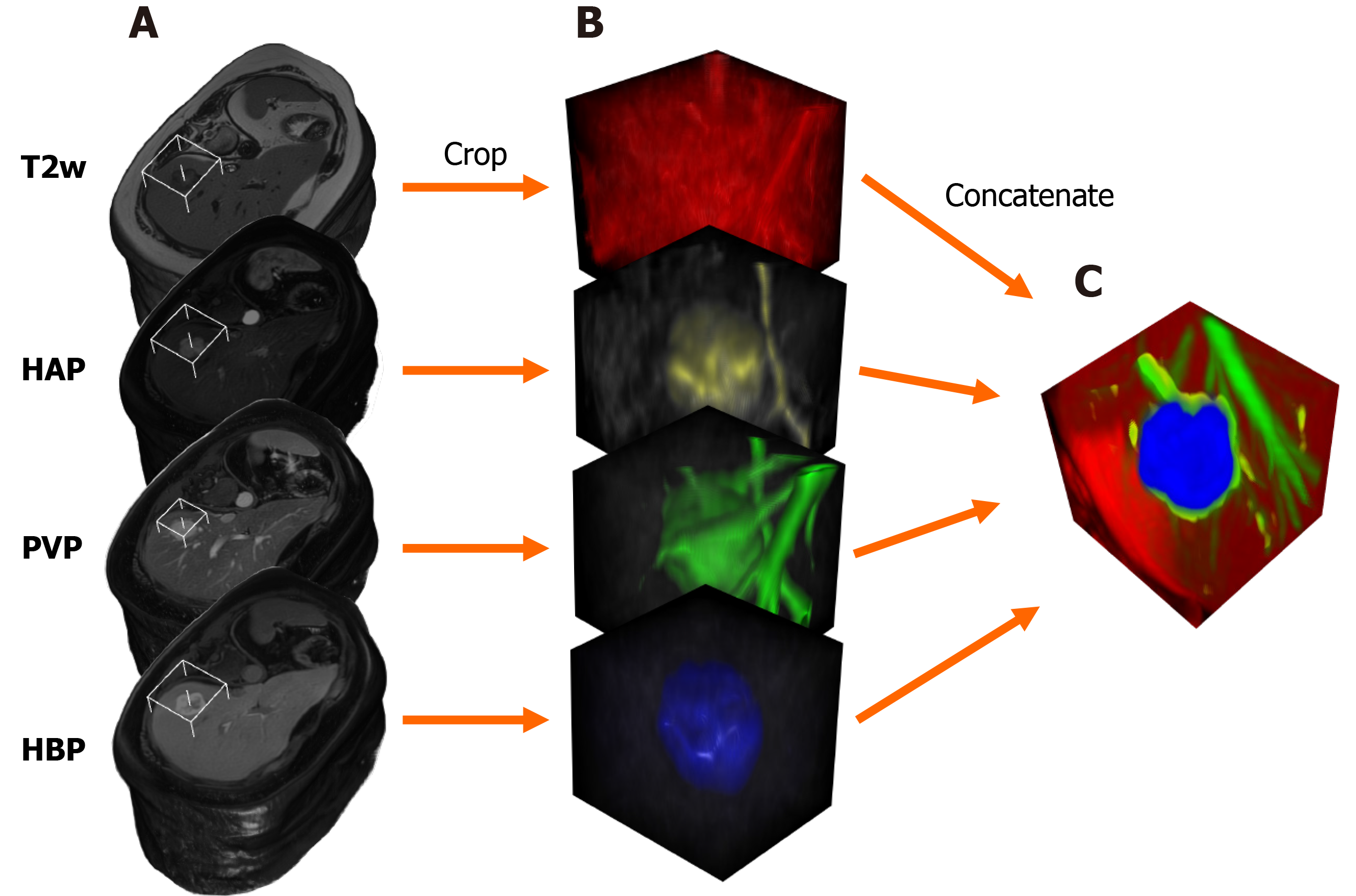
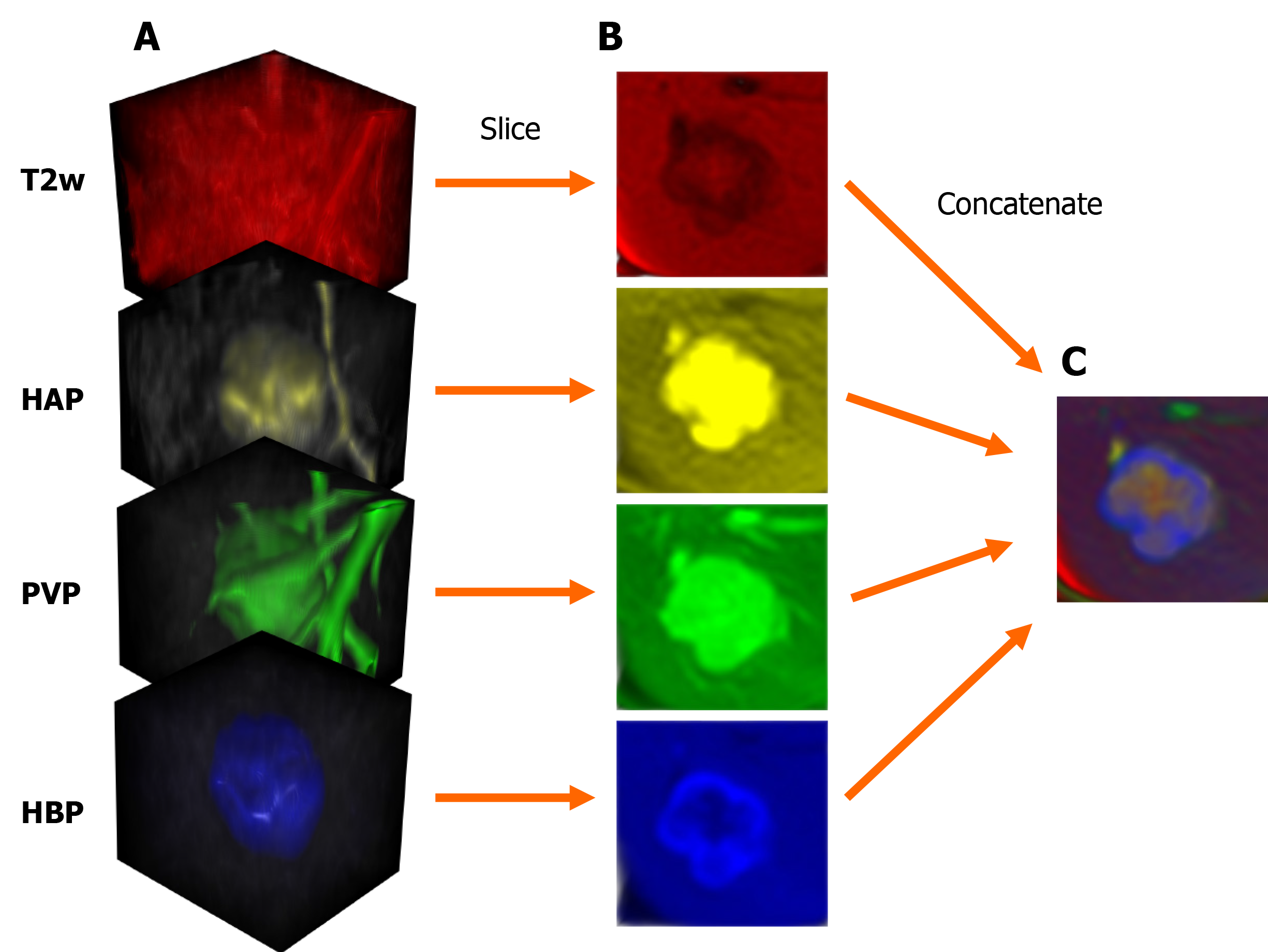
MRI scans were exported as DICOM files, that were then anonymized to remove the patients' social security numbers, birth date, sex, age, body weight, and date of the imaging study. Anonymized PVP and HBP files were resampled and spatially aligned to the corresponding T2w volume using BSpline as a non-rigid registration method via an open-source visualization and medical image computing software, called 3D Slicer (www.slicer.org). 3D Slicer was also used for annotation cropping and file conversion[10,11]. Lesions were annotated by cubic regions of interest (referred to as ROIs). The lesions were then cropped from the aligned HAP, PVP, HBP, and T2w volumes using the same ROI. The cropped volumes were saved as NIfTI files, which were then combined into one four-dimensional (4D) file for each lesion (Figure 1). Cropped lesions were randomly sorted into datasets. After 10-10 lesions were added to the test and validation dataset from each class, the remaining tumors were added to the training dataset. NIfTI files were sliced up into axial PNG images. The resulting T2w, HAP, PVP, and HBP PNG files were concatenated (Figure 2) using a custom-written computer program in Python. The training and validation datasets contained three axial slices of each lesion (i.e. three most representative axial slices of the NIfTI files), while the test set consisted of only one slice from each lesion.
Parameters of concatenated files were modified via transform functions. Image pixel intensity was scaled between -1.0 minimum and 1.0 maximum values. Data augmentation transforms were applied to the training samples, including random rotation (70° range along two axes) and zoom (0.7–1.4 scaling) to enrich training data. PNGs were resized to 64 × 64 resolution. Transformed images were converted to tensors (2D images were converted into 3D tensors, with the additional dimension equaling the number of network input channels), which were then fed to DenseNet264 that used 2D convolutional layers[5].
In the case of the 3D-DenseNet264 network, NIfTI voxels were resampled to isovolumetric shape, voxel intensities were rescaled between -1.0 minimum and 1.0 maximum value and NIfTI files were resized to 64 × 64 × 64 spatial resolution. The four NIfTI files were concatenated (T2w, HAP, PVP, HBP) to be used as multi-channel input for the 3D CNN. We used random 90° rotation (along two spatial axes), random 60° rotation (along x and y axes), random zoom (between 0.8 and 1.35), and random flipping on the training samples. MR volumes were converted to 4D tensors (number of channels, x-, y- and z-dimensions) that were used as network input. We used DenseNet264 models through the Pytorch-based open-source Medical Open Network For Artificial Intelligence (i.e. MONAI) framework[12]. We used categorical cross-entropy loss to measure the prediction error of the network during training and an Adam optimizer to update model parameters[13]. Networks were trained for 70 epochs. Using a Tesla T4 graphical processing unit, the 2D network was trained for 18 min, while the 3D CNN was trained for 41 min. Validation set area under the receiver operating characteristic curve (AUROC) values were calculated after each epoch, and the model with the highest average AUC value was saved as the final model.
The trained models were used to make predictions on an independent test dataset consisting of 10 lesions from each class. The tumor type with the highest probability, according to the last softmax layer of the convolutional networks, was chosen as the predicted lesion type via an argmax function, encoding the predicted diagnosis as 1, while the predicted incorrect classes as 0. Specificity, sensitivity, f1 score, positive predictive value (PPV), negative predictive value (NPV) were calculated for each class based on these outputs.
Classification performance was also measured using AUC values of each class, calculated from the softmax layer probability outputs. DeLong’s test was used to determine the statistical significance between the test performance of the 2D and 3D classifiers[14].
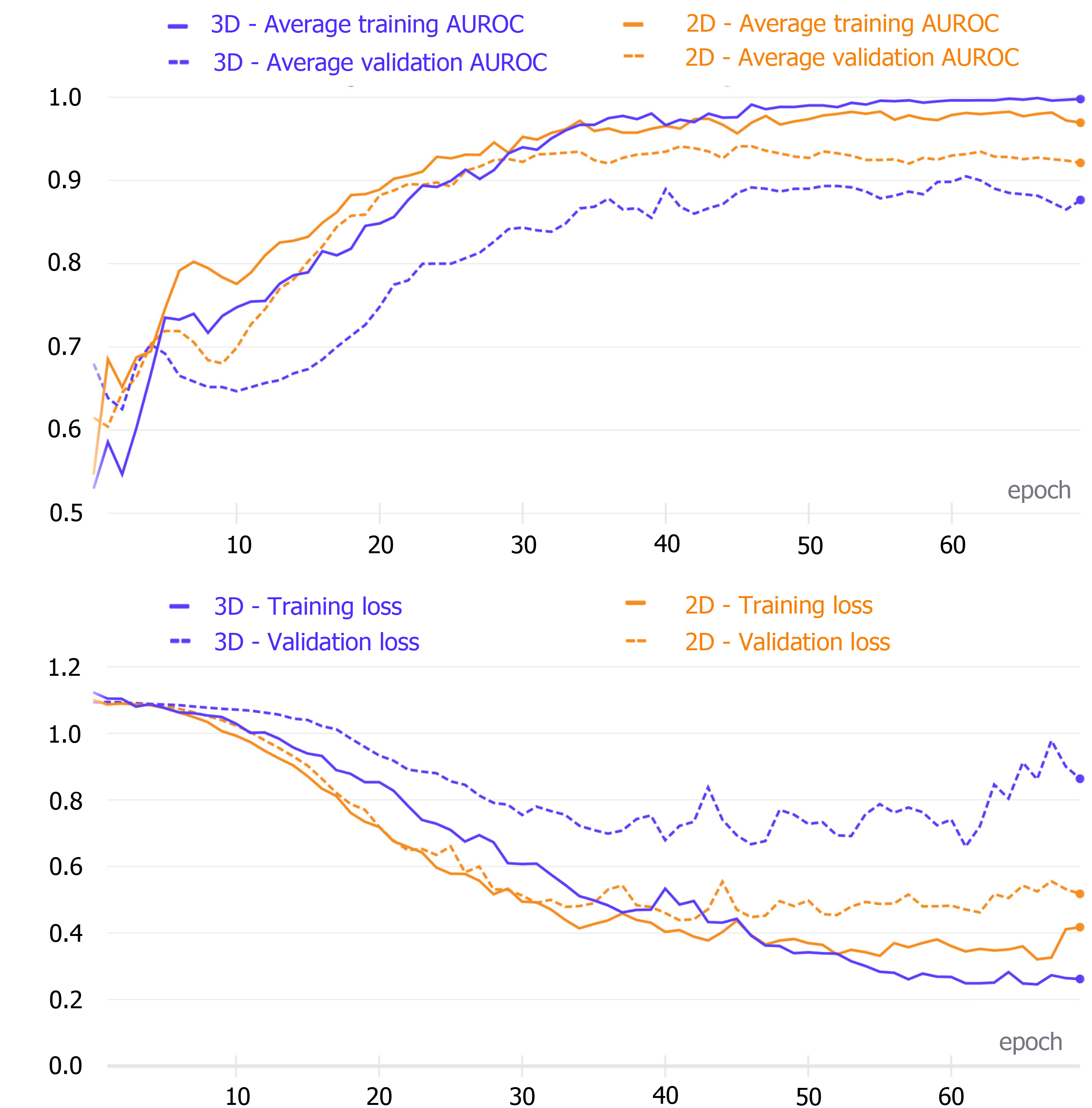
The 2D model achieved the highest average validation set AUC after the 46th epoch, while the best average AUC value of the 3D network was reached after the 62nd epoch. These models were saved and then used to make test set predictions (Figure 3).
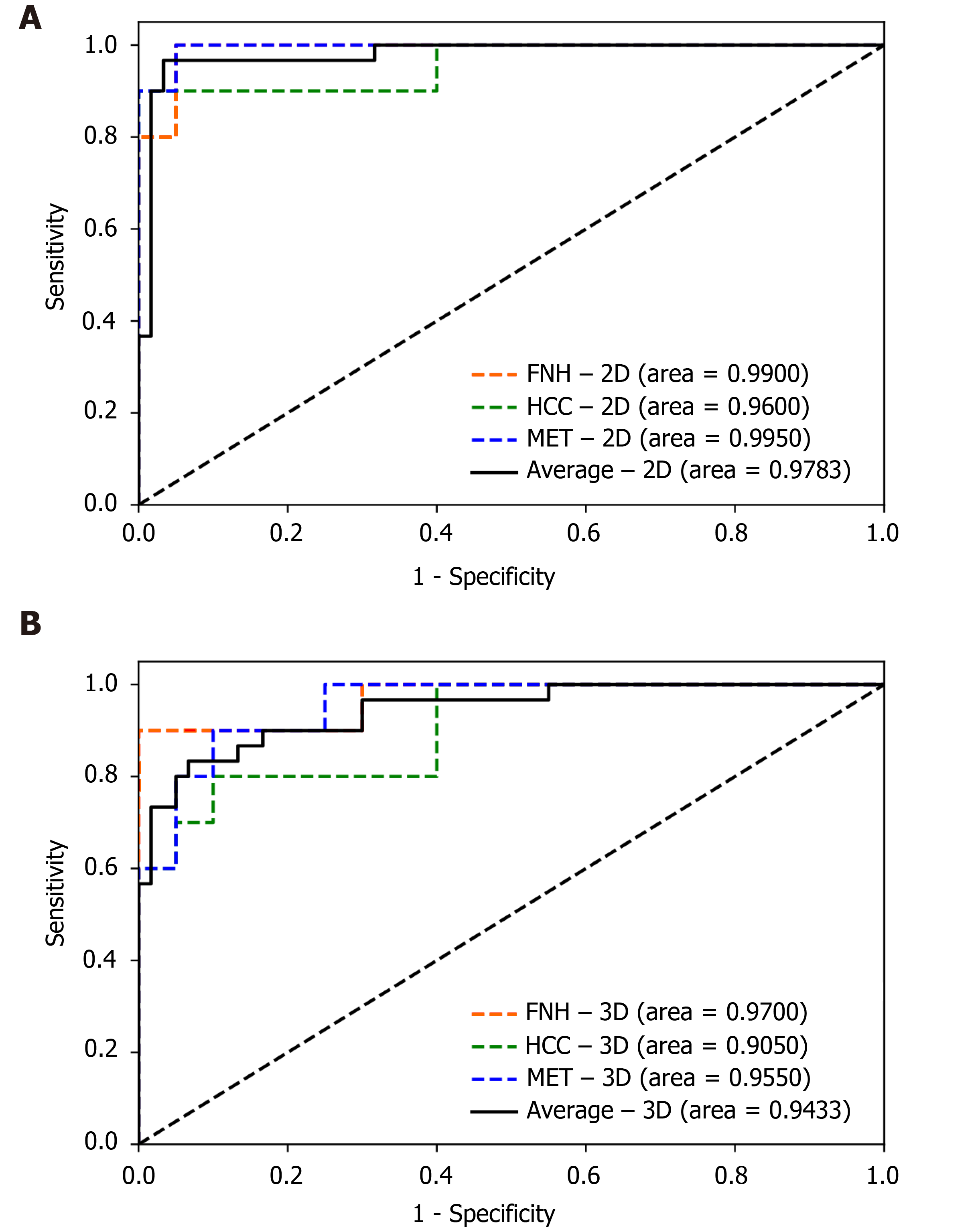
The finalized 2D and 3D networks were evaluated on the same independent test set, consisting of 10 lesions from each tumor type. On the independent test set, the finalized 2D model achieved 0.9900 [95% confidence interval (CI): 0.9664–1.0000], 0.9600 (95%CI: 0.8786–1.0000) and 0.9950 (95%CI: 0.9811–1.0000) AUC values for FNH, HCC and MET respectively, with an average AUC of 0.9783 (95%CI: 0.9492–1.0000). The finalized 3D model achieved 0.9700 (95%CI: 0.9077–1.0000), 0.9050 (95%CI: 0.7889–1.0000) and 0.9550 (95%CI: 0.8890–1.0000) AUC values for FNH, HCC and MET diagnosis, and an average AUC value of 0.9433 (95%CI: 0.8942–0.9924) on the test dataset (Figure 4). No statistically significant difference was found between the diagnostic performance of the 2D and 3D classifiers based on the ROC curve comparison for the three classes (Z = 0.7007, P = 0.4835 for FNH; Z = 0.7812, P = 0.4347 for HCC; Z = 1.3069, P = 0.1913 for MET). The 2D input data achieved excellent results in the distinction between all three lesion classes, similar to the 3D network (Table 2). Both networks achieved excellent PPV, sensitivity, f1 score, NPV, and specificity values for all three classes. The highest diagnostic accuracy was achieved by both networks for FNH and MET, while both networks demonstrated lower AUC values for HCC (Table 2). PPV, sensitivity, f1 score, specificity and an NPV of 0.9091, 1.0000, 0.9524, 0.9500, 1.000 values were achieved by the 2D model for FNH diagnosis. The 3D network performed FNH classification with similar PPV (0.9000), sensitivity (0.9000), f1 score (0.9000), specificity (0.9500) and NPV (0.9500) values as the 2D network. During HCC classification both the 2D and 3D models reached acceptable metrics with PPVs of 1.000 and 0.8750, sensitivities of 0.8000 and 0.7000, f1 scores of 0.8889 and 0.7778, specificities of 1.000 and 0.9500, lastly NPVs of 0.9091 and 0.8636. For the differentiation of METs from FNHs and HCCs the use of the 2D DenseNet resulted in a PPV of 0.9091, sensitivity of 1.000, f1 score of 0.9524, specificity of 0.9500 and NPV of 1.000, while the 3D DenseNet achieved values of 0.7500, 0.9000, 0.8182, 0.8500 and 0.9444 for PPV, sensitivity, f1 score, specificity and NPV respectively. On average, both the 2D and 3D trained models could distinguish FNHs, HCCs and METs reliably with PPVs of 0.9394 and 0.8417, sensitivities of 0.9333 and 0.8333, f1 scores of 0.9312 and 0.8320, specificities of 0.9667 and 0.9167, NPVs of 0.9697 and 0.9194.
| Input data | PPV | Sensitivity | F1 score | Specificity | NPV |
| FNH 2D | 0.9091 | 1.0000 | 0.9524 | 0.9500 | 1.0000 |
| 3D | 0.9000 | 0.9000 | 0.9000 | 0.9500 | 0.9500 |
| HCC 2D | 1.0000 | 0.8000 | 0.8889 | 1.0000 | 0.9091 |
| 3D | 0.8750 | 0.7000 | 0.7778 | 0.9500 | 0.8636 |
| MET 2D | 0.9091 | 1.0000 | 0.9524 | 0.9500 | 1.0000 |
| 3D | 0.7500 | 0.9000 | 0.8182 | 0.8500 | 0.9444 |
| Mean 2D | 0.9394 | 0.9333 | 0.9312 | 0.9667 | 0.9697 |
| 3D | 0.8417 | 0.8333 | 0.8320 | 0.9167 | 0.9194 |
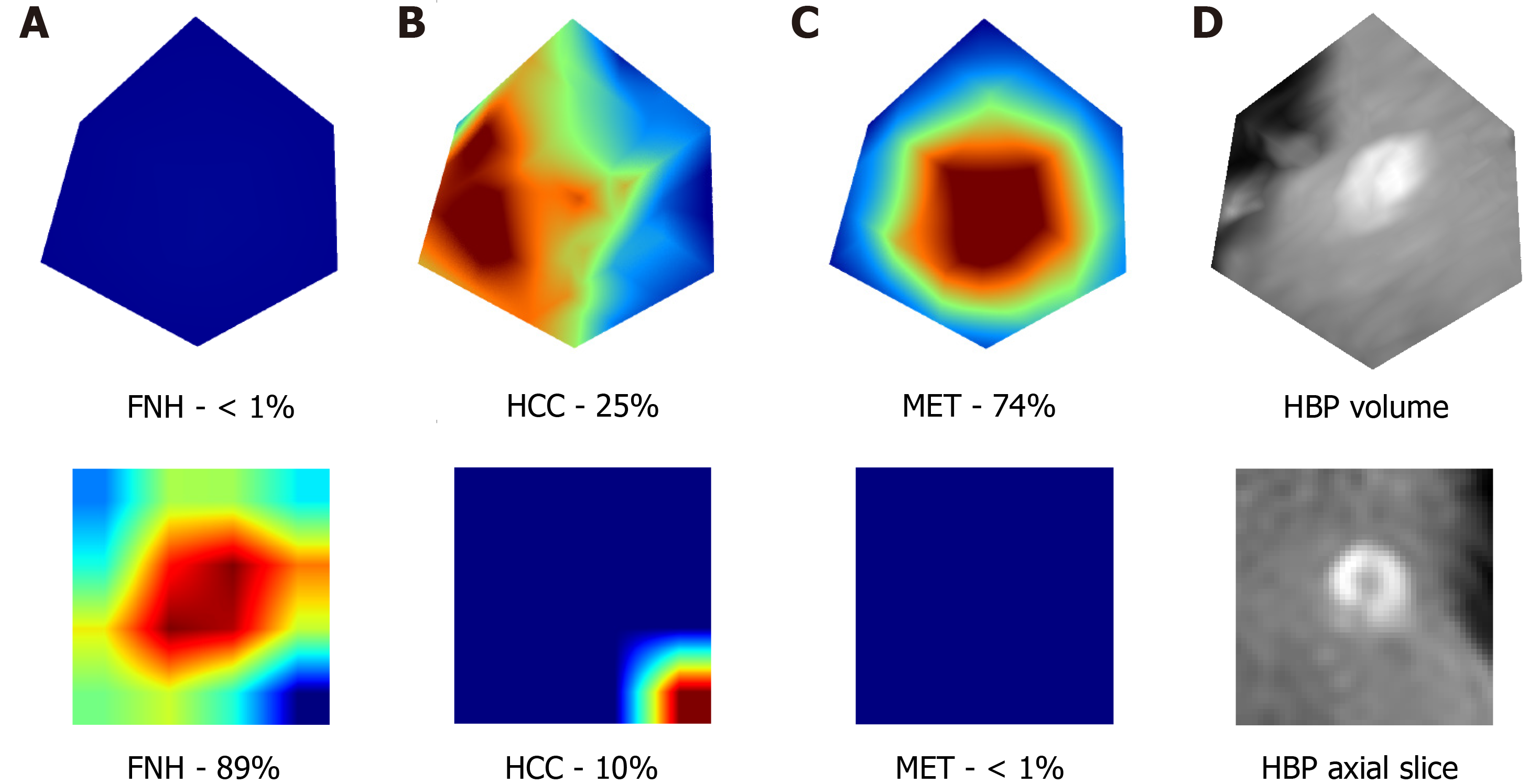
In addition, these results are supported by the extraction of attention maps from the trained models using test set images. We implemented an open-source software (M3d-CAM) to visualize the most important regions for diagnosis-making[15]. The extracted attention maps may correlate with the certainty with which a model classifies FLLs. By marking the areas within images, based on which the model makes a decision, attention maps form optimal bases of training dataset tailoring for certain radiological or other medical computer vision tasks by focusing on image regions that are difficult to analyze for the trained neural network (Figure 5).
FLLs are common findings during liver imaging, and the differentiation of benign and malignant types of FLLs is a significant diagnostic challenge, as imaging signs may overlap between different pathologies which can substantially alter the therapeutic decision. Therefore, precise and reproducible differential diagnosis of FLLs is critical for optimal patient management.
Today, the most accurate imaging modality to diagnose FLLs is multi-phase dynamic contrast-enhanced MRI. Extracellular contrast agents (ECAs) are commonly used to perform multi-phase dynamic post-contrast MRI studies to differentiate between lesions based on their distinct contrast enhancement patterns, such as HAP hyper-enhancement or washout in the PVP[16]. In comparison to ECAs, HSAs are taken up by hepatocytes and (in part) excreted through the biliary tract; thus, they can better differentiate between those lesions that consist of functionally active and impaired hepatocytes or those that are extrahepatic in origin[7]. This behavior of HSAs is utilized for making a distinction between FNH and hepatocellular adenoma, or to detect small foci of HCC and MET within the surrounding liver parenchyma[17,18].
In the current study, we evaluated different AI models on liver MRI images for the prediction of 216 FLLs compiled from three different types of lesions, namely FNHs, HCCs and METs. To ensure that the models could achieve the highest possible prediction rate, we narrowed down our data collection to only those four MRI sequences that provided the highest tissue contrast compared to the neighboring parenchyma or depicted distinctive imaging features of the lesion types. For the same reason, we used only HSA-enhanced scans for the analysis. We collected post-contrast images from HAP, PVP and HBP, and a T2w SPAIR image in the case of each lesion. A similar image analysis strategy was used by Hamm et al[19], who predicted 494 FLLs from six categories, including simple cyst, cavernous hemangioma, FNH, HCC, intrahepatic cholangiocarcinoma, and colorectal cancer METs using a 3D CNN model. The authors used HAP, PVP and delayed venous phase MRI images for the classification of the FLLs. They reported that the CNN model demonstrated 0.92 accuracy, 0.92 sensitivity and 0.98 specificity. The disadvantage of this study compared to ours was that it did not include HBP images, with only ECA images used for the MRI scans.
There are a handful of studies that included conventional ML methods and achieved reasonably good results. Wu et al[20], for example, extracted radiomics features from non-enhanced multi-parametric MRI images of FLLs and used them in ML models to differentiate between hepatic haemangioma and HCC. The final classifier achieved an AUC of 0.89, a sensitivity of 0.822 and a specificity of 0.714. Jansen et al[21], in their 2019 paper, used traditional ML methods for the same problem achieving an average accuracy of 0.77 for five major FLL types.
Our models' performance in the test set was comparable to or even surpassed those from previous publications, as the AUC, sensitivity and specificity were excellent for both the 2D (0.9783, 0.9333 and 0.9667 respectively) and 3D (0.9433, 0.8333 and 0.9167 respectively) architectures, which demonstrates the robustness of our data collection and analysis.
The quality and quantity of input data are pivotal when training neural networks. MRI liver tumor analysis using DL methods has steeply increased, but there is evidence lacking to support the use of 2D or 3D data. The additional dimension in 3D network inputs makes them computationally more demanding and the different data augmentation methods and hyperparameters must be well chosen to avoid artifacts. The 2D neural networks have the advantage of pretraining, which may improve classification accuracy[16,22,23]. Our study supports the results of Wang et al[24] and Hamm et al[19], emphasizing the need for multi-channel input volumes in order to achieve better accuracy. In contrast to these approaches, we have also utilized HBP images, thereby increasing the number of input channels to four in order to improve accuracy and additionally trained 2D CNNs, proving them to be just as effective classifiers as 3D models.
The selected architecture of the DL model can substantially alter classification accuracy. It is a novelty of our analysis that compared to previous examinations we utilized a DenseNet architecture. DenseNets contain multiple dense blocks, where each layer is connected with the residuals from previous layers. DenseNets require fewer trainable parameters at the same depth than conventional CNNs, as newly learned features are shared through all layers[5]. Our results are among the first to indicate that this highly efficient network design can enhance the performance of AI models for the classification of multi-parametric MRI images of FLLs.
Our study's limitations are the low number of patients involved, the retrospective nature of the study, and that it was conducted within a single institute. Further improvement may be achieved by additional data collection (including additional lesion classes) and the use of more MRI volumes and different data augmentation methods as well as the use of pre-trained networks.
Based on our study, we can say that routinely acquired radiological image materials can be used for analysis with AI methods, such as CNNs. According to our results, densely connected CNNs trained on multi-sequence MRI scans can be promising new alternatives to single-phase approaches; furthermore, the use of multi-dimensional input volumes can help the AI-based diagnosis of FLLs. According to our results, 3D and 2D DenseNets can reach similar performance in the differentiation of FLLs based on a small dataset of MRI images. The use of gadoxetate disodium-enhanced MRI scans can also enhance the diagnostic performance of MRI-based hepatic lesion classification.
Interest in medical applications of artificial intelligence (AI) has steeply risen in the last few years. As one of the most obvious beneficiaries of the advances in computer vision, radiology research has also put AI in a prominent position. Convolutional neural networks are the state-of-the-art methods used in computer vision. Focal liver lesions (FLLs) are common findings during imaging, which can best be evaluated via hepatocyte-specific contrast-enhanced magnetic resonance imaging (MRI).
Though convolutional neural networks are widely used for medical image research purposes, the effect of input, such as data dimensionality and the effect of multiple input channels, has not yet been widely examined in this area. MRI volumes presumably hold more complex information about each lesion; as such, three-dimensional inputs may be more difficult to process and properly use for classification tasks in comparison to two-dimensional axial slices. The combination of multiple MRI sequences in addition to the use of hepatocyte-specific contrast agents (HSAs) may also affect diagnostic accuracy.
Our research aimed to compare two- and three-dimensional DenseNets264 networks for the multi-phasic hepatocyte-specific contrast-enhanced MRI-based classification of FLLs.
T2-weighted, arterial phase, portal venous phase, and hepatobiliary phase volumes of focal nodular hyperplasias, hepatocellular carcinomas and liver metastases were used to train the two models. Diagnostic performance was evaluated on an independent test set, based on area under the curve, positive and negative predictive values (NPVs), sensitivity, specificity and f1 score.
The study found that via the use of either two- or three-dimensional convolutional neural networks and the combination of multiple MRI sequences, the average area under the curve, sensitivity, specificity, NPV, positive predictive value and f1 scores of comparable level can be achieved.
According to our findings, two- and three-dimensional networks can both be used for highly accurate differentiation of multiple classes of FLLs by combining multiple MRI phases and using HSAs.
This study’s findings can help to clarify the potential applicability of two- and three-dimensional multi-channel MRI images for the convolutional neural network-based classification of FLLs using HSAs.
The authors would like to express their gratitude to Dr. Endre Szabó, mathematician from the Alfréd Rényi Institute of Mathematics of The Hungarian Academy of Sciences, for expert review of the manuscript and discussion of the AI analysis and statistical methods. The authors also thank Tamás Wentzel and Lilla Petovsky, technicians of the MRI unit of the Medical Imaging Center, Semmelweis University, for their enthusiasm for the current research and professionalism during patient examinations.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Hungary
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bork U S-Editor: Fan JR L-Editor: A P-Editor: Xing YX
| 1. | Bhargavan M, Kaye AH, Forman HP, Sunshine JH. Workload of radiologists in United States in 2006-2007 and trends since 1991-1992. Radiology. 2009;252:458-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 2. | Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts HJWL. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin. 2019;69:127-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 848] [Cited by in F6Publishing: 561] [Article Influence: 112.2] [Reference Citation Analysis (2)] |
| 3. | Kim J, Min JH, Kim SK, Shin SY, Lee MW. Detection of Hepatocellular Carcinoma in Contrast-Enhanced Magnetic Resonance Imaging Using Deep Learning Classifier: A Multi-Center Retrospective Study. Sci Rep. 2020;10:9458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 4. | Singh SP, Wang L, Gupta S, Goli H, Padmanabhan P, Gulyás B. 3D Deep Learning on Medical Images: A Review. Sensors (Basel). 2020;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 143] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 5. | Huang G, Liu Z, Pleiss G, Van Der Maaten L, Weinberger K. Convolutional Networks with Dense Connectivity. IEEE Trans Pattern Anal Mach Intell. 2019;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 103] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 6. | Kim YY, Park MS, Aljoqiman KS, Choi JY, Kim MJ. Gadoxetic acid-enhanced magnetic resonance imaging: Hepatocellular carcinoma and mimickers. Clin Mol Hepatol. 2019;25:223-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Thian YL, Riddell AM, Koh DM. Liver-specific agents for contrast-enhanced MRI: role in oncological imaging. Cancer Imaging. 2013;13:567-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Coenegrachts K. Magnetic resonance imaging of the liver: New imaging strategies for evaluating focal liver lesions. World J Radiol. 2009;1:72-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.7] [Reference Citation Analysis (1)] |
| 9. | von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61:344-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5754] [Cited by in F6Publishing: 7128] [Article Influence: 445.5] [Reference Citation Analysis (0)] |
| 10. | Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323-1341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4857] [Cited by in F6Publishing: 3988] [Article Influence: 332.3] [Reference Citation Analysis (0)] |
| 11. | Klein S, Staring M, Pluim JP. Evaluation of optimization methods for nonrigid medical image registration using mutual information and B-splines. IEEE Trans Image Process. 2007;16:2879-2890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 195] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 12. | Zenodo. The MONAI Consortium. Project MONAI. [cited 10 March 2021]. Available from: https://www.f6publishing.com/Forms/Manuscript/Editorial/ReviewAndEditProcess.aspx?id=WJG-27-5978. [Cited in This Article: ] |
| 13. | Kingma DP, Ba J. Adam: A method for stochastic optimization. 2014 Preprint. Available from: arXiv:1412.6980v9. [Cited in This Article: ] |
| 14. | DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837-845. [PubMed] [Cited in This Article: ] |
| 15. | Gotkowski K, Gonzalez C, Bucher A, Mukhopadhyay A. M3d-CAM: A PyTorch library to generate 3D data attention maps for medical deep learning. 2020 Preprint. Available from: arXiv:2007.00453v1. [Cited in This Article: ] |
| 16. | Matos AP, Velloni F, Ramalho M, AlObaidy M, Rajapaksha A, Semelka RC. Focal liver lesions: Practical magnetic resonance imaging approach. World J Hepatol. 2015;7:1987-2008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 43] [Article Influence: 4.8] [Reference Citation Analysis (2)] |
| 17. | Granata V, Fusco R, de Lutio di Castelguidone E, Avallone A, Palaia R, Delrio P, Tatangelo F, Botti G, Grassi R, Izzo F, Petrillo A. Diagnostic performance of gadoxetic acid-enhanced liver MRI vs multidetector CT in the assessment of colorectal liver metastases compared to hepatic resection. BMC Gastroenterol. 2019;19:129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 18. | Grieser C, Steffen IG, Kramme IB, Bläker H, Kilic E, Perez Fernandez CM, Seehofer D, Schott E, Hamm B, Denecke T. Gadoxetic acid enhanced MRI for differentiation of FNH and HCA: a single centre experience. Eur Radiol. 2014;24:1339-1348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 19. | Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin M, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. 2019;29:3338-3347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 152] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 20. | Wu J, Liu A, Cui J, Chen A, Song Q, Xie L. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med Imaging. 2019;19:23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 21. | Jansen MJA, Kuijf HJ, Veldhuis WB, Wessels FJ, Viergever MA, Pluim JPW. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS One. 2019;14:e0217053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Liu Z, Tang H, Lin Y, Han SJapa. Point-voxel cnn for efficient 3d deep learning. 2020 Preprint. Available from: arXiv:1907.03739v2. [Cited in This Article: ] |
| 23. | Morid MA, Borjali A, Del Fiol G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput Biol Med. 2021;128:104115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 117] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 24. | Wang C, Hamm C, Letzen B, Duncan J. A probabilistic approach for interpretable deep learning in liver cancer diagnosis. SPIE Medical Imaging. United States, California, San Diego: SPIE, 2019. [Cited in This Article: ] |