Published online Aug 21, 2021. doi: 10.3748/wjg.v27.i31.5171
Peer-review started: January 25, 2021
First decision: March 29, 2021
Revised: April 11, 2021
Accepted: August 3, 2021
Article in press: August 3, 2021
Published online: August 21, 2021
Processing time: 204 Days and 9.6 Hours
Pancreatic ductal adenocarcinoma (PDAC) represents a challenging pathology with very poor outcomes and is increasing in incidence within the general population. The majority of patients are diagnosed incidentally with insidious symptoms and hence present late in the disease process. This significantly affects patient outcomes: the only cure is surgical resection but only up to 20% of patients present with resectable disease at the time of clinical presentation. The use of “omic” technology is expanding rapidly in the field of personalised medicine - using genomic, proteomic and metabolomic approaches allows researchers and clinicians to delve deep into the core molecular processes of this difficult disease. This review gives an overview of the current findings in PDAC using these “omic” approaches and summarises useful markers in aiding clinicians treating PDAC. Future strategies incorporating these findings and potential application of these methods are presented in this review article.
Core Tip: Treatment for pancreatic ductal adenocarcinoma is limited by the severity of the pathology, limited biomarkers and late presentation of patients. Utilising genomic, proteomic and metabolomic research into pancreatic ductal adenocarcinoma has provided insight into understanding the disease process as well as providing suitable markers of diagnosis and treatment to improve clinical outcomes.
- Citation: Rajesh S, Cox MJ, Runau F. Molecular advances in pancreatic cancer: A genomic, proteomic and metabolomic approach . World J Gastroenterol 2021; 27(31): 5171-5180
- URL: https://www.wjgnet.com/1007-9327/full/v27/i31/5171.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i31.5171
Pancreatic adenocarcinoma or pancreatic ductal adenocarcinoma (PDAC) is an aggre
The field of “omics” research refers to the use of high throughput technologies to globally analyse a biological system at the molecular level. This takes place on mul
The first of these fields to emerge was genomics, with the goal of characterising the genome and the variations of its structure and expression leading to pathogenesis[5]. Cancer being essentially a disease of the genome, occurring through the accumulation of genetic mutations, the insights gleaned from such analysis is invaluable for cancer research[6].
With the advent of next-generation sequencing [whole-genome sequencing (WGS), whole-exome sequencing (WES) and RNA sequencing], this can now be done quicker and more accurately than traditional methods[7]. WES, which sequences all protein coding exons, is more abundantly utilised as it is more accurate and relatively less expensive compared to WGS, which also has the issue of complex data analysis and interpretation[6].
At the protein level, proteomic studies identify and quantify the proteome of a biological system, their interactions and post-translational modifications[5]. Proteo
Metabolomics involves the study of all the low molecular weight metabolites within a sample that gives a comprehensive reflection of the sample’s phenotype at a given time[10]. Identification and quantification of these metabolites can indicate the meta
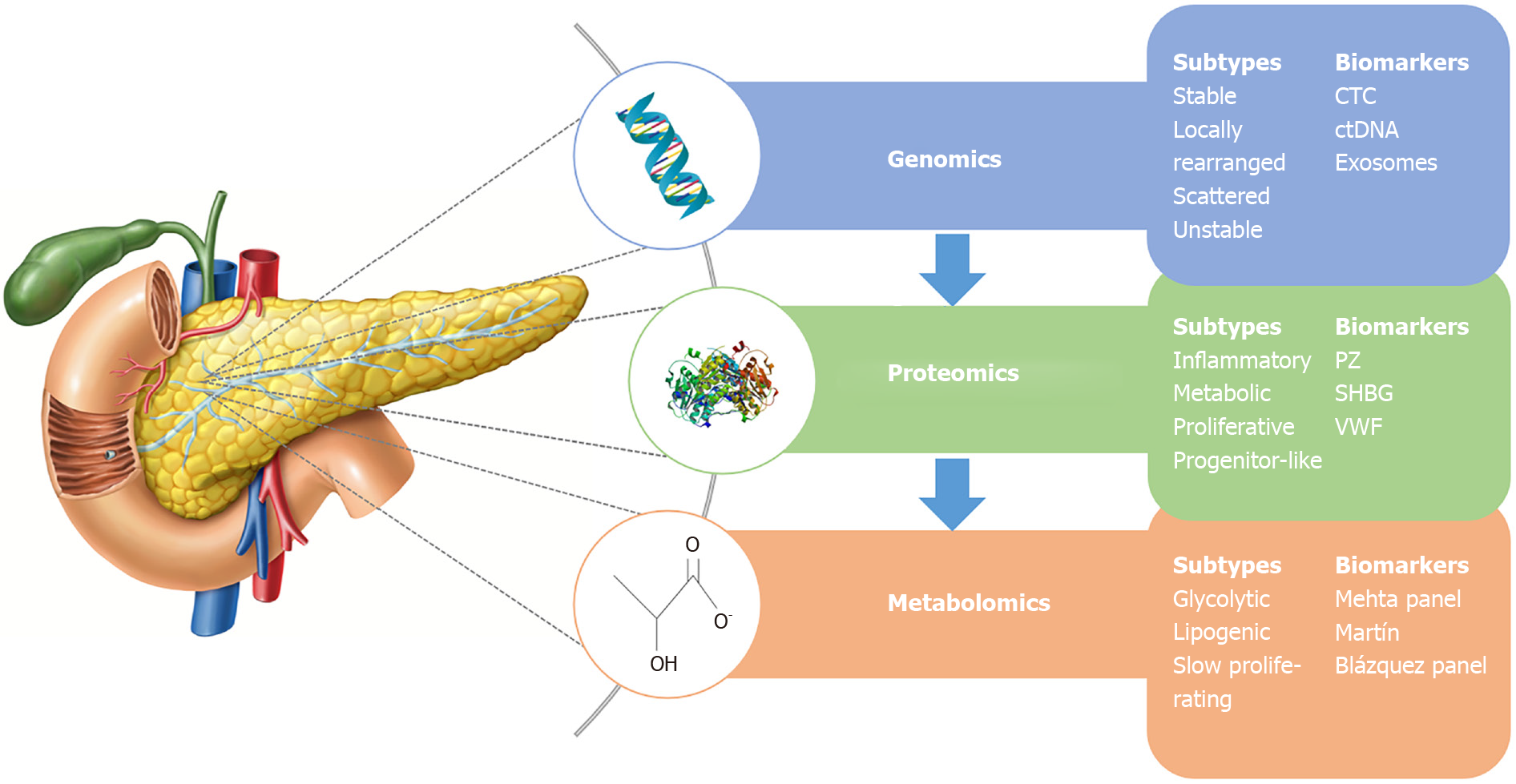
Waddell et al[11] utilised WGS to map the genome of 100 PDAC specimens. Their findings reinforced the known main drivers of PDAC (KRAS, TP53, CDKN2A and SMAD4) and also found multiple other mutations at much lower prevalence. The authors described four subtypes of PDAC based on the quantity of variations in chromosome structure: (1) Stable (20%; < 50 structural variations and widespread aneuploidy); (2) Locally rearranged (30%; significant events on 1-2 chromosomes); (3) Scattered (36%; < 200 structural variations and moderate chromosomal damage); and (4) Unstable (14%; large number of structural variation and defects in DNA damage repair). The unstable subtype was associated with a high BReast CAncer gene (BRCA) mutational signature, suggesting defects in DNA damage repair, possibly sensitising these to platinum based chemotherapy or poly (ADP-ribose) polymerase inhibitors (PARPi). Indeed, from their sample, 4 out of 5 of these patients treated with platinum based chemotherapy showed response to treatment[11].
Singhi et al[12] utilised real time targeted sequencing of exons and introns of 3594 PDAC specimens during the course of clinical care and reported that 17% of these may be susceptible to current therapies based on this genomic data. They found genetic alterations in receptor tyrosine kinase (RTK)/Ras/mitogen-activated protein kinase (MAPK) activation, DNA damage repair, cell cycle control, TGF-B signalling, histone modification, SWI/SNF protein complex, PI3L/mTOR signalling, WNT/B-catenin pathway, RNA splicing, NOTCH pathway, angiogenesis and Hedgehog signalling. Interestingly, the investigators also found that 14% of their sample exhibited mutations in DNA damage repair genes (BRCA-FANC family). Further to this, they identified genetic alterations in receptor tyrosine kinases as potential targets on a background of wild-type KRAS PDAC (12% of the sample)[12].
Aguirre et al[13] performed deep WES of PDAC primaries and metastases for 73 patients having clinically indicated biopsies. Average time for the results of WES to return to the clinicians was 39 d, longer than that of research only biopsies (28 d) due to the need for histological diagnosis prior to sequencing. Analysis of these findings resulted in three mutational signatures: SigA (homologous recombination deficiency), SigB (aging) and SigC (unknown aetiology). Around 40% of these patients had potentially targetable genomic findings, when excluding KRAS or CDKN2A, 48% were eligible for clinical trials or off-label use of other therapeutic agents and 24% of patients were indeed enrolled onto a clinical trial or treated with an off-label agent. This shows the feasibility of implementing WES clinically to guide treatment choice. Clinically relevant findings included DNA damage repair (DDR) mutations and BRAF mutations in KRAS wild-type PDAC which may confer sensitivity to platinum-based chemotherapy/PARPi and MAPK inhibition respectively. The authors also presented two such case studies with significant responses to treatment[13].
Using 56 PDAC liver metastases specimens, Law et al[14] performed liquid chromatography-mass spectrometry (LC-MS/MS) to identify 30811 peptides that mapped to 916 proteins comprising of at least 5 peptides in 80% of the sample. Functional analysis of these proteins showed that they play a role in, “extracellular matrix organization, protein processing and transport, translation, glycolytic processes, NADPH metabo
These proteins were analysed to categorise four PDAC subtypes and three protein clusters. The matched subtypes and clusters are: “Inflammatory” (cluster 3 - pentose phosphate pathway, adaptive immune response, complement activation, IL8 produc
Clinically, the “proliferative” subtype was associated with a history of alcohol use and the “metabolic” subtype was associated with tobacco use. The “metabolic” and “progenitor-like” subtypes had a decreased risk of death when treated with FOL
Another interesting finding from this study is the role of serine hydro- xymethyltransferase (SHMT1) (involved in the folic acid cycle) in gemcitabine resistance. Comparison of untreated samples and samples treated with only gemcitabine showed that SHMT1 was significantly down-regulated in the treatment group. The investigators further displayed increased EC50 of gemcitabine in cell lines with SHMT1 knockdown compared to the control group, showing that SHMT1 is a potential mediator of gemcitabine resistance. Expression of SHMT1 was higher in “metabolic” and “progenitor-like” subtypes compared to the other two subtypes regardless of gemcitabine treatment. Expression of this protein may potentially guide the choice of gemcitabine as treatment or monitor those on gemcitabine for resistance to treatment[14].
Humphrey et al[15] used liquid chromatography-mass spectrometry (LC-MS/MS) to stratify two cohorts of PDAC cell lines American Type Culture Collection (ATCC) and The Kinghorn Cancer Centre (TKCC), along tyrosine phosphorylation (pTyr) sites. The authors produced a list of 1622 pTyr sites from 797 proteins. Of these, 144 had significant subtype specificity. ATCC subtype 1 showed hypophosphorylation of 65 pTyr sites and the enriched proteins were involved in formation and regulation cell-cell adheren junctions and tight junctions. ATCC subtype 2 contained 54 up-regulated pTyr sites (specifically increased relative phosphorylation rather than increased pro
When this methodology was applied to the TKCC cohorts, 1220 pTyr sites were identified, of which 383 were subtype specific. TKCC subtypes 1 and 2 showed 101 and 73 down-regulated pTyr sites, while TKCC subtype 3 showed up-regulation of 209 pTyr sites and was enriched for Ephrin and EGFR signalling. Targeting RTK pTyr sites in the TKCC cohort showed increased phosphorylation of RTKs in subtype 3 including sites on EGFR, EPHA2, DDR1, FGFR1, INSR, MERTK, MET, and RON[15].
Of the subtype specific pTyr sites identified, 8 were identified as “common classifier sites”, able to predict the subtype in the ATCC cohort. Subtypes 1, 2 and 3 were identified to exhibit low medium and high phosphorylation of these sites respectively. As both cohorts had subtypes that were “RTK-enriched”, the investigators tested the cell lines in this cohort against erlotinib, an EGFR kinase inhibitor. Indeed, the cell lines in this subtype showed increased sensitivity to erlotinib. The authors recognised that the pTyr signature of these RTKs are what conferred sensitivity to RTK blockade rather than expression levels and so stratifying patients with such a signature can potentially allow targeted therapeutic regimes to be studied[15].
Metabolomics profiling by Daemen et al[16] using LC-MS/MS and gas chromatography-MS represents the only metabolomic study to stratify PDAC. The investigators examined 38 “PDAC-derived” cell lines to quantify 256 metabolites. Analysis of these metabolites revealed three subtypes of PDAC, described as: (1) Slow proliferating (34%); (2) Glycolytic (27%); and (3) Lipogenic (39%). The slow proliferating subtype was low in amino acids and carbohydrates, and the cells had a significantly longer doubling time than the other two subtypes. The glycolytic subtype showed increased levels of metabolites of the glycolytic and serine pathways (phosphoenolpyruvate, glyceraldehyde-3-phosphate, lactate, and serine) and decreased redox balance meta
Moreover, the authors predicted and demonstrated in vitro, sensitivity of the gly
| Ref. | Subtypes | Clinical significance |
| Genomic | ||
| Waddell et al[11] | Stable, locally re-arranged, scattered and unstable | High BRCA mutational signature in the unstable subtype, sensitizing to PARPi and PBC. |
| Singhi et al[12] | - | Real time genetic sequencing. 17% of specimens found to have sensitivities to available treatments. Potential therapeutic targets. |
| Aguirre et al[13] | SigA, SigB and SigC | Potential targets in 40% of patients. 48% eligible for trials/off-label use. Of 24% enrolled onto a clinical trial. |
| Proteomic | ||
| Law et al[14] | Inflammatory, proliferative, progenitor-like and metabolic | ↓Risk of death in metabolic and progenitor-like subtypes treated with FOLFIRINOX+Gemcitabine. |
| Humphrey et al[15] | TKCC subtypes 1, 2 and 3 | Subtypes 3 in both cohorts showed increased sensitivity to erlotinib, potentially mediated by tyrosine phosphorylation of RTK sites. |
| ATCC subtypes 1, 2 and 3 | ||
| Metabolomic | ||
| Daemen et al[16] | Slow proliferating, glycolytic and lipogenic | Glycolytic subtype sensitive to inhibitors of aerobic glycolysis, glutaminolysis, γ-glutamylcysteine and Xct. Lipogenic subtype sensitive to lipid synthesis inhibitors. |
Currently, the only biomarker for PDAC is carbohydrate antigen 19-9 (CA19-9), approved by the Food and Drug Administration for use in clinical practice[17]. Unfortunately, the median sensitivity and specificity of CA19-9 is 79% and 82% respectively, making it unsuitable for use as a diagnostic marker, with it being raised in other gastrointestinal pathology[18]. Further complicating this is the fact that roughly 10% of the population with a Lewis-negative genotype do not express CA19-9 (a sialyl-Lewis A tetrasaccheride) at all. Its use currently is limited to monitoring CA19-9 positive PDAC for progression or recurrence after resection[17]. There is an obvious need for further investigation for potential biomarkers for early diagnosis, prognosis and sensitivity/resistance to therapeutics. “Omics” techniques have the capability to produce large amounts of data which can be correlated with specific states of the biological system and so there is great potential for this data to be used for biomarker discovery.
As mentioned earlier, one of the main factors that influence the outcomes in PDAC is that the majority are diagnosed at an advanced stage. With surgical resection cur
An emerging area of research is the use of “liquid biopsy”, which is the sampling of tumour material which spills into the circulation[19]. The main components of such a biopsy include circulating tumour cells (CTC), cell-free DNA (cfDNA) and circulating exosomes. cfDNA is genetic material released into the circulation from benign and malignant cells during cell death, circulating tumour DNA (ctDNA) being the subset of this derived from malignant cells. Exosomes are extracellular vesicles released from cells into various bodily fluids and can contain proteins and genetic material for analysis. They have a longer half-life and are constantly being produced by cells ma
Zhu et al[20] conducted a systematic review and meta-analysis of 19 studies utilising ctDNA, CTCs and exosomes to diagnose “pancreatic adenocarcinoma”, “pancreatic ductal adenocarcinoma” or “pancreatic cancer”. They found that the overall sensiti
Overall sensitivity, specificity and AUC of the CTC studies was 0.74 (95%CI: 0.68-0.79), 0.83 (95%CI: 0.78-0.88), and 0.8166[20]. This was thought to be due to hepatic trapping of CTCs and due to reduced blood flow in pancreatic malignancies compared to normal tissue (decreasing the chances of cell shedding into the circulation)[20]. Indeed, it was found that for PC, the levels of CTC was the lowest compared to other types of cancer[21].
ctDNA’s overall sensitivity, specificity and AUC was 0.64 (95%CI: 0.58-0.70), 0.92 (95%CI: 0.88-0.95), and 0.9478[20]. All of the ctDNA studies utilised PCR of KRAS mutations to distinguish PC from either healthy controls, pancreatitis or benign lesions. As mentioned before, KRAS is the most common genetic driver of PDAC and so utilising this as the marker for detection may be the reason for the high specificity. The relatively low sensitivity however may have been due to the abundance of ctDNA in the circulation[20]. It has been found that the abundance of ctDNA has a positive correlation with tumour load, supported by the physiology of ctDNA mentioned above: It is released through cell death[22]. Due to this, ctDNA may not be an ideal candidate for use in early detection of PDAC but may have a role as a prognostic biomarker or a marker of response to treatment, especially in those that are CA19-9 negative.
In terms of protein biomarkers, many studies have been done to characterise the differences in the proteomes of PDAC and normal control (NC) specimens from various sources including tissue samples, cell lines, serum/plasma and pancreatic juice[8]. While quite a few studies have been done, only a few of the biomarkers described overlap between studies. Further to this, none of these biomarkers have been put to use clinically, mainly due to the lack of validation in clinical trials, and standardised, reproducible and cost-effective analytical methods.
MS data combined with the results of a literature review by Capello et al[23] yielded 17 plasma protein biomarkers to distinguish early stage PDAC from benign pancreatic disease and NC. They tested these biomarkers using ELISA, first in a triage set which narrowed these down to 7 biomarkers, followed by validation of these 7 biomarkers in three independent plasma sample sets. Statistical analysis of the performance of these 7 biomarkers led to the development of a 3 biomarker panel of metalloproteinase inhibitor 1, leucine rich alpha-2-glycoprotein 1 and CA 19-9 which was able to differentiate PDAC cases from healthy controls with an AUC of 0.887 (95%CI: 0.817-0.957) in a blinded test set, which was a statistically significant improvement compared to CA 19-9 alone. The sensitivities at a fixed 95% and 99% specificity were 0.667 and 0.410 respectively, compared to the sensitivities of 0.538 and 0.462 respectively for CA 19-9[23].
In terms of prognostic biomarkers, de Oliveira et al[24] recently conducted a meta-analysis of MS data from two systematic reviews of PDAC secretome and proteome. No protein was found to be present in all the studies and so the authors selected those that were presented in at least 2 studies, generating a list of 39 secreted proteins. Further gene expression analysis of 4747 tumours (of 10 types of cancers) and 2737 corresponding normal tissues for these proteins revealed that 31 of these were un
Peng et al[25] developed a protein signature to predict response to chemotherapy through LC-MS/MS of prepared serum samples from 16 stage IV PDAC patients. The three protein biomarker candidates vitamin-K dependent protein Z, sex hormone-binding globulin and von Willebrand factor, combined with CA 19-9 were used to make a biomarker panel, with biomarker positive patients having significantly shorter median survival in both stage III and stage IV patients [8.7 mo (95%CI: 6-11.7) for biomarker negtive vs 19.2 mo (95%CI: 11.4-22.1) for biomarker positive].
A meta-analysis of metabolomic biomarkers for PC by Mehta et al[26] yielded 21 deregulated blood based biomarkers that appeared in at least 2 studies. The authors developed a 10 metabolite diagnostic panel from these biomarkers which was tested on plasma samples of 192 patients from four diagnostic groups: PC (n = 59), NC (n = 48), colorectal cancer (CRC, n = 66) and type 2 diabetes mellitus (T2DM, n = 19). The AUC of PC, T2DM and CRC vs NC were 0.992 (95%CI: 0.977-1), 0.957 (95%CI: 0.868-1) and 0.986 (95%CI: 0.967-1) respectively. The AUC of PC vs CRC was only 0.653 (95%CI: 0.543-0.757), suggesting a lack of specificity of the panel between these two groups. An index of the 10 metabolite panel showed that higher index values correlated with increased risk of malignancy, with a value of ≥ 12.5 representing a 100% risk of PC[26]. While the authors used the term “pancreatic cancer”, in their study, the authors manually curated the literature to ensure the comparison groups were PDAC patients and controls.
More recently, Martín-Blázquez et al[27] analysed serum from unresectable PDAC patients and NC using reverse-phase liquid chromatography and high-resolution MS to identify 86 significant metabolites. With these, the researchers proposed a model of 9 markers that discriminated PDAC from healthy controls with an AUC of 0.992 (95%CI: 0.972-1.000). Table 2 summarises these molecular subtyping studies while Figure 1 demonstrates subtypes and biomarkers by “Omics” level[28].
| Ref. | Biomarkers | Sensitivity | Specificity | AUC |
| Zhu et al[20] | ctDNA | 0.64 | 0.92 | 0.9478 |
| CTC | 0.74 | 0.83 | 0.8166 | |
| Exosome | 0.93 | 0.92 | 0.9819 | |
| Capello et al[23] | TIMP1, LRG1 and CA19-9 | 0.667/0.410 | 0.95/0.99 | 0.887 |
| Mehta et al[26] | Panel of: Lactate, LysoPC (18:2), Alanine, Choline, Threonine, Asparagine, Tyrosine, Lysine, Palmitate and 3-hydroxybutyrate | - | - | 0.992 vs NC |
| 0.957 vs T2DM | ||||
| 0.653 vs CRC | ||||
| Biomarker + ve median survival | Biomarker - ve median survival | |||
| Peng et al[25] | Panel of: PZ, SHBG, VWF and CA19-9 | 19.2 mo | 8.7 mo | |
| AUC | ||||
| Martín-Blázquez et al[27] | Panel of: PS (12:0/15:1), TG (22:2/15:0/18:3), 4-oxo-Retinoic acid, Androsterone sulfate, LysoPE (18:2), Phenylalanylphenylalanine, all-trans-Decaprenyldiphosphate, LysoPC (18:2) and Dehydroepiandrosterone sulfate | 0.992 (CI: 0.972-1.000) |
“Omics” technologies have allowed mining of massive amounts of data, giving new insights into the complex, heterogeneous nature of PDAC. As described above, many studies have been done to describe molecular classifications and potential biomarkers, but none of these have yet been translated into clinical practice[28]. Several trials, as reviewed by Du et al[29], faced difficulties in sample procurement, low quality of samples and waiting time for sample analysis leading to patient deterioration or withdrawal. The authors suggest a multi-disciplinary approach with specialist input in sample acquisition in designated centres with high levels of experience. Issues re
Additionally, many of these studies are largely comparative, most of them de
Finally, with such high throughput technologies being used, cancer research is moving into the realm of “big data”. “Wide” data sets (where the number of variables exceed the number of subjects) such as those produced by “omics” technologies are better analysed through machine learning/artificial intelligence than traditional statistical analysis[34]. Over the past few years, many studies have been done using machine learning methods for molecular subtyping and biomarker discovery in other types of malignancies, simultaneously using data from multiple “omics” levels and there is great potential for machine learning and artificial intelligence applications in PDAC research[35].
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Singh I S-Editor: Liu M L-Editor: A P-Editor: Yuan YY
| 1. | Rawla P, Sunkara T, Gaduputi V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J Oncol. 2019;10:10-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 893] [Cited by in RCA: 1514] [Article Influence: 252.3] [Reference Citation Analysis (1)] |
| 2. | International Agency for Research in Cancer; World Health Organization. Globocan 2020 Pancreas Factsheet. [cited 10 November 2020]. Available from: https://gco.iarc.fr/today/data/factsheets/cancers/13-Pancreas-fact-sheet.pdf. |
| 3. | Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913-2921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5379] [Cited by in RCA: 5125] [Article Influence: 465.9] [Reference Citation Analysis (0)] |
| 4. | McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846-4861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1338] [Cited by in RCA: 1258] [Article Influence: 179.7] [Reference Citation Analysis (39)] |
| 5. | Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 900] [Cited by in RCA: 1397] [Article Influence: 174.6] [Reference Citation Analysis (0)] |
| 6. | Nakagawa H, Fujita M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018;109:513-522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 231] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 7. | Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, Romano A. Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification. Int J Mol Sci. 2017;18:308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 320] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 8. | Ansari D, Torén W, Zhou Q, Hu D, Andersson R. Proteomic and genomic profiling of pancreatic cancer. Cell Biol Toxicol. 2019;35:333-343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 9. | Horgan RP, Kenny LC. ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. The Obstetrician and Gynaecologist. Obstetrician Gynaecologist. 2011;13:189-195. [RCA] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 207] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 10. | Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1:a000588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 405] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 11. | Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grützmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA; Australian Pancreatic Cancer Genome Initiative; Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 1984] [Article Influence: 198.4] [Reference Citation Analysis (1)] |
| 12. | Singhi AD, George B, Greenbowe JR, Chung J, Suh J, Maitra A, Klempner SJ, Hendifar A, Milind JM, Golan T, Brand RE, Zureikat AH, Roy S, Schrock AB, Miller VA, Ross JS, Ali SM, Bahary N. Real-Time Targeted Genome Profile Analysis of Pancreatic Ductal Adenocarcinomas Identifies Genetic Alterations That Might Be Targeted With Existing Drugs or Used as Biomarkers. Gastroenterology. 2019;156:2242-2253.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 240] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 13. | Aguirre AJ, Nowak JA, Camarda ND, Moffitt RA, Ghazani AA, Hazar-Rethinam M, Raghavan S, Kim J, Brais LK, Ragon D, Welch MW, Reilly E, McCabe D, Marini L, Anderka K, Helvie K, Oliver N, Babic A, Da Silva A, Nadres B, Van Seventer EE, Shahzade HA, St Pierre JP, Burke KP, Clancy T, Cleary JM, Doyle LA, Jajoo K, McCleary NJ, Meyerhardt JA, Murphy JE, Ng K, Patel AK, Perez K, Rosenthal MH, Rubinson DA, Ryou M, Shapiro GI, Sicinska E, Silverman SG, Nagy RJ, Lanman RB, Knoerzer D, Welsch DJ, Yurgelun MB, Fuchs CS, Garraway LA, Getz G, Hornick JL, Johnson BE, Kulke MH, Mayer RJ, Miller JW, Shyn PB, Tuveson DA, Wagle N, Yeh JJ, Hahn WC, Corcoran RB, Carter SL, Wolpin BM. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov. 2018;8:1096-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 265] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 14. | Law HC, Lagundžin D, Clement EJ, Qiao F, Wagner ZS, Krieger KL, Costanzo-Garvey D, Caffrey TC, Grem JL, DiMaio DJ, Grandgenett PM, Cook LM, Fisher KW, Yu F, Hollingsworth MA, Woods NT. The Proteomic Landscape of Pancreatic Ductal Adenocarcinoma Liver Metastases Identifies Molecular Subtypes and Associations with Clinical Response. Clin Cancer Res. 2020;26:1065-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 15. | Humphrey ES, Su SP, Nagrial AM, Hochgräfe F, Pajic M, Lehrbach GM, Parton RG, Yap AS, Horvath LG, Chang DK, Biankin AV, Wu J, Daly RJ. Resolution of Novel Pancreatic Ductal Adenocarcinoma Subtypes by Global Phosphotyrosine Profiling. Mol Cell Proteomics. 2016;15:2671-2685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, Kowanetz K, Hong R, Moffat J, Gao M, Boudreau A, Mroue R, Corson L, O'Brien T, Qing J, Sampath D, Merchant M, Yauch R, Manning G, Settleman J, Hatzivassiliou G, Evangelista M. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci USA. 2015;112:E4410-E4417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 275] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 17. | Khomiak A, Brunner M, Kordes M, Lindblad S, Miksch RC, Öhlund D, Regel I. Recent Discoveries of Diagnostic, Prognostic and Predictive Biomarkers for Pancreatic Cancer. Cancers (Basel). 2020;12:3234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 18. | Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 611] [Article Influence: 32.2] [Reference Citation Analysis (1)] |
| 19. | Marrugo-Ramírez J, Mir M, Samitier J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int J Mol Sci. 2018;19:2877. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 281] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 20. | Zhu Y, Zhang H, Chen N, Hao J, Jin H, Ma X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: A systematic review and meta-analysis. Medicine (Baltimore). 2020;99:e18581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 21. | Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897-6904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1810] [Cited by in RCA: 1952] [Article Influence: 97.6] [Reference Citation Analysis (0)] |
| 22. | Grunvald MW, Jacobson RA, Kuzel TM, Pappas SG, Masood A. Current Status of Circulating Tumor DNA Liquid Biopsy in Pancreatic Cancer. Int J Mol Sci. 2020;21:7651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 23. | Capello M, Bantis LE, Scelo G, Zhao Y, Li P, Dhillon DS, Patel NJ, Kundnani DL, Wang H, Abbruzzese JL, Maitra A, Tempero MA, Brand R, Firpo MA, Mulvihill SJ, Katz MH, Brennan P, Feng Z, Taguchi A, Hanash SM. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J Natl Cancer Inst. 2017;109:djw266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 24. | de Oliveira G, Freire PP, Cury SS, de Moraes D, Oliveira JS, Dal-Pai-Silva M, Reis PP, Carvalho RF. An Integrated Meta-Analysis of Secretome and Proteome Identify Potential Biomarkers of Pancreatic Ductal Adenocarcinoma. Cancers (Basel). 2020;12:716. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 25. | Peng H, Chen R, Brentnall TA, Eng JK, Picozzi VJ, Pan S. Predictive proteomic signatures for response of pancreatic cancer patients receiving chemotherapy. Clin Proteomics. 2019;16:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Mehta KY, Wu HJ, Menon SS, Fallah Y, Zhong X, Rizk N, Unger K, Mapstone M, Fiandaca MS, Federoff HJ, Cheema AK. Metabolomic biomarkers of pancreatic cancer: a meta-analysis study. Oncotarget. 2017;8:68899-68915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Martín-Blázquez A, Jiménez-Luna C, Díaz C, Martínez-Galán J, Prados J, Vicente F, Melguizo C, Genilloud O, Pérez Del Palacio J, Caba O. Discovery of Pancreatic Adenocarcinoma Biomarkers by Untargeted Metabolomics. Cancers (Basel). 2020;12:1002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Watanabe. DNA double-strand B; 2018. [cited 10 November 2020]. DataBase: Center for Life Science (DBCLS). Available from: https://doi.org/10.7875/togopic.2018.23. |
| 29. | Du Y, Zhao B, Liu Z, Ren X, Zhao W, Li Z, You L, Zhao Y. Molecular Subtyping of Pancreatic Cancer: Translating Genomics and Transcriptomics into the Clinic. J Cancer. 2017;8:513-522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 30. | Chantrill LA, Nagrial AM, Watson C, Johns AL, Martyn-Smith M, Simpson S, Mead S, Jones MD, Samra JS, Gill AJ, Watson N, Chin VT, Humphris JL, Chou A, Brown B, Morey A, Pajic M, Grimmond SM, Chang DK, Thomas D, Sebastian L, Sjoquist K, Yip S, Pavlakis N, Asghari R, Harvey S, Grimison P, Simes J, Biankin AV; Australian Pancreatic Cancer Genome Initiative (APGI); Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) Trial Management Committee of the Australasian Gastrointestinal Trials Group (AGITG). Precision Medicine for Advanced Pancreas Cancer: The Individualized Molecular Pancreatic Cancer Therapy (IMPaCT) Trial. Clin Cancer Res. 2015;21:2029-2037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 185] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 31. | Dixon-Hughes J. Precision-Panc Master Protocol: Personalising Treatment For Pancreatic Cancer [accessed 2020 Dec 27]. In: ClinicalTrials.gov [Internet]. Bethesda (MD): U.S National Library of Medicine. Available from: https://clinicaltrials.gov/ct2/show/NCT04161417 ClinicalTrials.gov Identifier: NCT04161417. |
| 32. | Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1247] [Cited by in RCA: 1378] [Article Influence: 172.3] [Reference Citation Analysis (0)] |
| 33. | Follia L, Ferrero G, Mandili G, Beccuti M, Giordano D, Spadi R, Satolli MA, Evangelista A, Katayama H, Hong W, Momin AA, Capello M, Hanash SM, Novelli F, Cordero F. Integrative Analysis of Novel Metabolic Subtypes in Pancreatic Cancer Fosters New Prognostic Biomarkers. Front Oncol. 2019;9:115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 34. | Bzdok D, Altman N, Krzywinski M. Statistics vs machine learning. Nat Methods. 2018;15:233-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 658] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 35. | Nicora G, Vitali F, Dagliati A, Geifman N, Bellazzi R. Integrated Multi-Omics Analyses in Oncology: A Review of Machine Learning Methods and Tools. Front Oncol. 2020;10:1030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |