Published online Jul 28, 2021. doi: 10.3748/wjg.v27.i28.4555
Peer-review started: January 27, 2021
First decision: March 29, 2021
Revised: April 4, 2021
Accepted: July 9, 2021
Article in press: July 9, 2021
Published online: July 28, 2021
Processing time: 179 Days and 22 Hours
The hepatitis C virus (HCV), an obligatory intracellular pathogen, highly depends on its host cells to propagate successfully. The HCV life cycle can be simply divided into several stages including viral entry, protein translation, RNA replication, viral assembly and release. Hundreds of cellular factors involved in the HCV life cycle have been identified over more than thirty years of research. Characterization of these cellular factors has provided extensive insight into HCV replication strategies. Some of these cellular factors are targets for anti-HCV therapies. In this review, we summarize the well-characterized and recently identified cellular factors functioning at each stage of the HCV life cycle.
Core Tip: The hepatitis C virus (HCV) depends on its host cells to propagate successfully. Hundreds of cellular factors involved in the HCV life cycle have been identified. Some of these cellular factors are potential targets for anti-HCV therapies (e.g., scavenger receptor class B type 1, epidermal growth factor receptor, Niemann–Pick C1-like 1, microRNA-122, cyclophilin A). A successful vaccine for HCV is still a challenge in the near future. Investigating the cellular factors involved in viral entry should help vaccine development. HCV is also a unique model to study the interactions between viral infection and cellular lipid metabolisms.
- Citation: Li HC, Yang CH, Lo SY. Cellular factors involved in the hepatitis C virus life cycle. World J Gastroenterol 2021; 27(28): 4555-4581
- URL: https://www.wjgnet.com/1007-9327/full/v27/i28/4555.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i28.4555
Around 50%–80% of patients infected with the hepatitis C virus (HCV) will develop chronic infection. Approximately 71 million individuals are chronically infected by the HCV worldwide according to an estimation by the World Health Organization (https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/).
Chronic hepatitis C (CHC) patients are at high risk of developing liver cirrhosis and even hepatocellular carcinoma. Although CHC can now be cured using various direct-acting antivirals, the majority of CHC patients remain undiagnosed and, thus, untreated. Furthermore, a successful antiviral treatment does not prevent HCV reinfection. Therefore, HCV eradication remains a challenge and will probably require an effective vaccine[1] (for a review see References[2,3]).
HCV belongs to the family Flaviviridae and genus Hepacivirus. Its genome is a single-stranded RNA with positive polarity that is packaged by viral core protein and enveloped by a lipid membrane containing two viral glycoproteins (i.e., E1 and E2) to form the virion. HCV genomic RNA sequences are highly heterogeneous among different isolates. At present, HCV is classified into at least six major genotypes (1 to 6). HCV heterogeneity hinders the development of an effective vaccine to protect against infection from all HCV genotypes. Despite the sequence variations, all currently recognized HCV genotypes are pathogenic, hepatotropic and preserve the similarity of the life cycle in cells (for a review see References[4,5]).
The HCV life cycle begins with the binding of a virion to its specific entry factors (or receptors) on hepatocytes. After binding, the virion is internalized into the cytoplasm and its genomic RNA is released. Then, the HCV genomic RNA is used for both polyprotein translation and viral replication. HCV replication takes place within the membranous web (MW) in the endoplasmic reticulum (ER). At last, HCV uses the biosynthetic pathway of very-low-density lipoprotein (VLDL) to assemble and release the viral particles from cells (for a review see Reference[6]).
HCV relies significantly on its host cells to establish a successful infection due to the fact of its limited genetic content. Hundreds of cellular factors have been identified as being involved in the HCV life cycle over more than thirty years of research. In this review article, due to the limited space, we can only summarize the well-characterized and recently identified cellular factors involved in the HCV life cycle but not including immune responses against viral infections (for a review see Reference[7]).
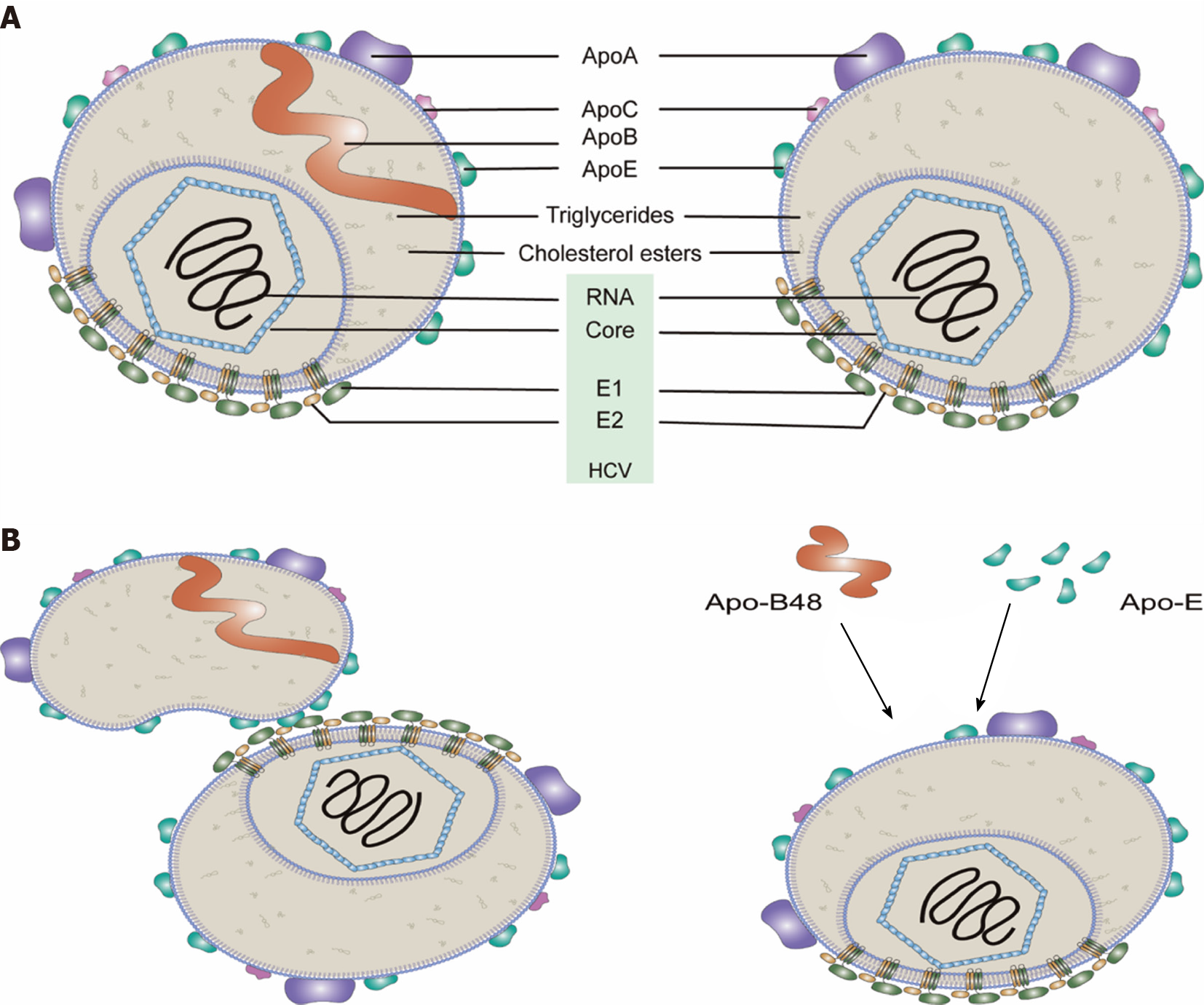
It has been demonstrated that purified HCV particles are spherical and heterogeneous in size (40–100 nm in diameter) with no obvious symmetry under cryo-EM[8]. The density of HCV particles in the plasma of CHC patients also varied (from 1.03 to 1.25 g/mL). Low-density HCV particles with 81–85 nm diameters are highly infectious[8,9]. The HCV particles with low density are associated with apolipoproteins (Apo such as Apo-E, Apo-AI, Apo-CI and Apo-B), phospholipids (such as phosphatidylcholine and sphingomyelin) and cholesterol (free and esterified), while they have almost no phosphatidylserine (PS) and very little amounts of phosphatidylethanolamine[8,10-12]. HCV particles derived from cultured cells show characteristics similar to those from CHC patients[9,10,13]. Thus, these circulating HCV particles are referred to as “lipo-viro particles” (LVPs)[14] and have very low buoyant densities due to the fact of their interactions with lipoproteins in the blood[15] (Figure 1) (for a review see Ref
Modifications of LVPs could be mediated by lipoprotein lipases (LPLs)[17,18]. LPL was shown to shift the HCV particles to higher densities with diminished HCV infectivity[19,20]. Indeed, LPL lipolytic activity in clinical samples has been reported to be inversely correlated with HCV viremia[21].
More than 40 cellular proteins were found in the HCV particles recently using proteomics analyses, including heat shock cognate protein 70 (HSC70) and nucleoporin 98 (Nup98); both proteins play a role in virus assembly/release[22,23].
Association of HCV particles with host lipoproteins has advantages for the virus: (1) Lipoproteins likely mask viral epitopes of viral E1 and E2 proteins thus helping the virus escape from the humoral immune response; and (2) Lipoproteins contribute to the hepatotropism of HCV, e.g., HCV attachment to hepatocytes involving Apo-E (and/or Apo-B) binding with the cellular factors [e.g., low-density lipoprotein receptor (LDLR), highly sulfated heparan sulfate proteoglycans (HSPGs) and scavenger receptor class B type 1 (SR-B1)] for viral entry[24].
HCV can infect hepatocytes and other cells (e.g., lymphocytes). In general, HCV infects hepatocytes through two distinct routes: Cell-free virus entry and cell-to-cell transmission. Viral entry requires interactions between the components of LPVs (particularly Apo-E and viral envelope proteins) and those of cells (particularly entry factors). Alternatively, HCV may infect cells through exosomes (for a review see Reference[25]).
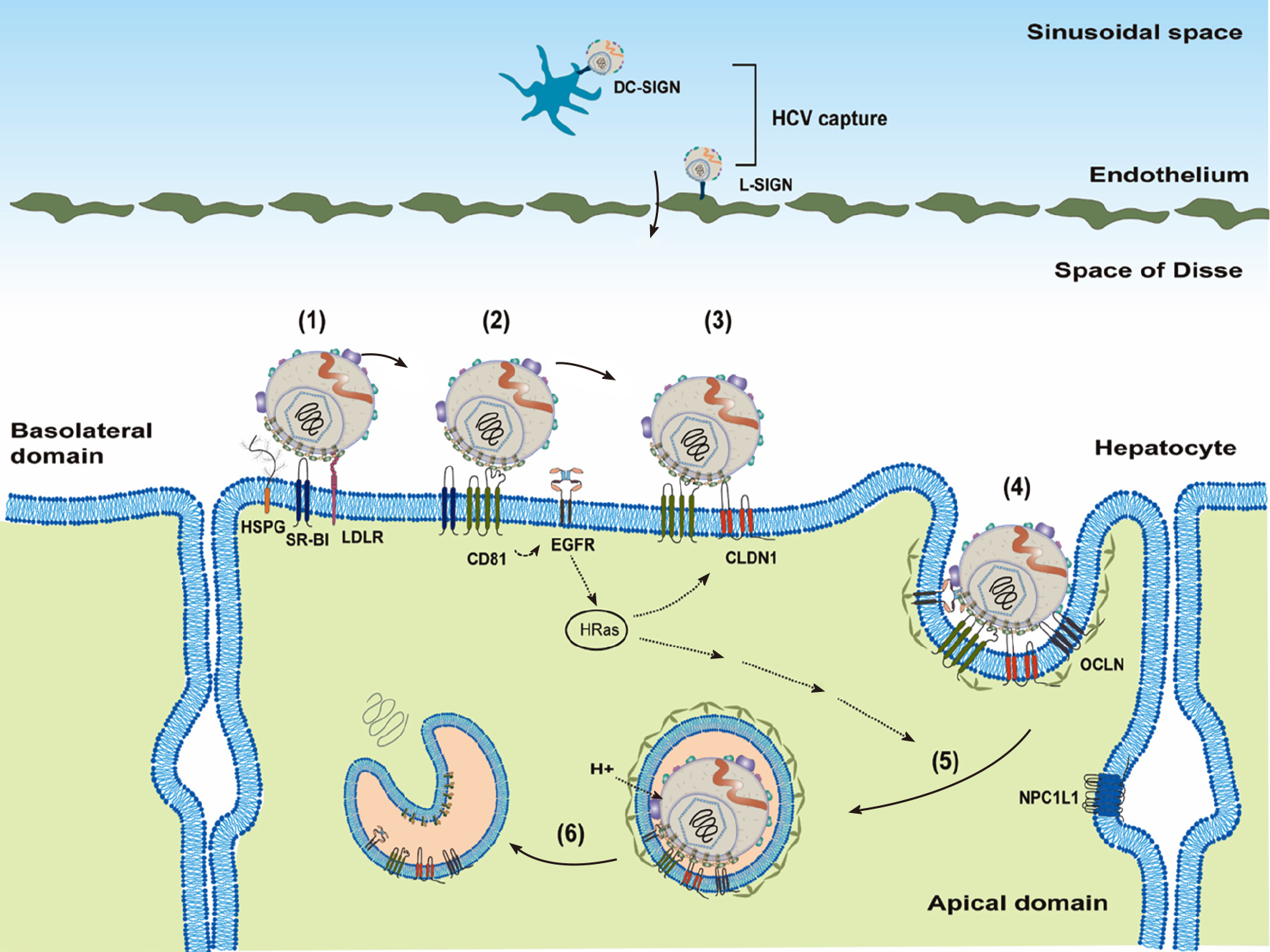
HCV transmission is through the parenteral route, and it needs to reach the liver by crossing the endothelium. Before attachment to the hepatocytes, HCV particles may be captured by DC-SIGN on dendritic cells or L-SIGN on endothelium[26,27] (Figure 2).
Cellular factors responsible for HCV entry into hepatocytes are divided into three groups based on their functions[25,28]: Attachment factors, entry factors (or receptors) and entry cofactors (or facilitators). Attachment factors help dock the virus on the cell’s surface, mostly through non-specific interactions. Entry factors mediate specific interactions with viral glycoproteins. Entry cofactors, although not interacting directly with the virus, play an important role in supporting viral entry.
To infect a new hepatocyte, the HCV needs to interact with the attachment factors on the basolateral side of hepatocytes first (Figure 2). The well-known attachment factors include HSPGs, (particularly syndecans: SDC-1 and SDC-2), LDLR and SR-B1[29-32]. Attachment of HCV LPVs to host cells is mediated mainly through the virus-associated lipoprotein components (particularly Apo-E[33]) and viral glycoproteins (E1 and E2, particularly E2)[32,34,35]. In addition to the binding of Apo-E with attachment factors, Apo-B100 could also interact with LDLR[36] and Apo-AI with SR-B1[37]. Recently, the redundant functions of Apo-C1 and Apo-E in the HCV infection have been demonstrated[38]. Attachment to SR-B1 may bring HCV particles to entry factor cluster of differentiation 81 (CD81)[39]. SR-B1, as both an entry factor and an attachment factor, has been shown to bind viral envelope proteins[29,40]. Interaction of HCV with CD81 activates epidermal growth factor receptor (EGFR) signaling and also facilitates CD81 diffusion and formation of the HCV–CD81–CLDN1 complex[39,41-43]. The HCV–CD81–CLDN1 complex then interacts with OCLN, which is believed to mediate the clathrin-dependent internalization through interacting with GTPase dynamin[44-46]. SR-B1, CD81, CLDN1 and OCLN are four well-characterized entry factors for HCV entry[28]. In addition, LRL-R could interact with both HCV and E2 proteins and, thus, function as an entry factor[47].
HCV particles are then internalized, mature in the acidic endosomes that promote low pH-dependent HCV fusion and, ultimately, release HCV genomic RNAs (un-coating) into cytosol[48-50] (Figure 2). Low endosomal pH and interactions of viral glycoproteins with CD81 are thought to induce conformational rearrangements of viral glycoproteins for HCV fusion[51], which is controlled by E1 protein[52]. Recently, cell death-inducing DFFA-like effector B (CIDEB) protein was identified as an entry cofactor to act at a late membrane fusion event[53].
Both HCV and coronavirus [e.g., severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)][54] are positive-strand RNA viruses. These two viruses enter their target cells through receptor-mediated endocytosis and release their genomic RNAs for translation in the cytosol. However, unlike HCV, coronavirus fusion for viral entry is unusual in that it is often biphasic and can occur at or near the cell’s surface or in late endosomes[55].
In addition to HSPGs, LDLR, and SR-B1, interferon receptor IFNAR2 was found as a novel attachment factor to facilitate HCV entry through interacting with Apo-E[56]. VLDL receptor (VLDLR), similar to LDLR, could also serve as an attachment factor for HCV entry[57]. Niemann–Pick C1-like 1 (NPC1L1) contributes to HCV entry possibly through its role as a cholesterol receptor, thus functioning as an HCV entry cofactor[58]. Similarly, PS receptor (TIM-1/human hepatitis A virus cellular receptor 1/CD365) has also been identified as an attachment factor through binding with PS on HCV LPVs[59,60]. It should be noticed that only tiny amounts of PS were detected in the HCV particles[12]. On the other hand, LPLs, hepatic triglyceride lipase and long-chain fatty acyl-coenzyme A can inhibit HCV attachment by targeting virus-associated lipoproteins[17,18,20,61].
In addition to CLDN1, other CLDNs (e.g., CLDN6, CLDN9 and CLDN12) have also been reported as HCV entry factors for some genotypes[62-64]. Recently, CD63 (binding directly with HCV) and CD36 (interacting directly with HCV E1 protein) were demonstrated to be entry factors[65,66]. Under hypoxic conditions, VLDLR could serve as an entry factor independent of canonical CD81-mediated HCV entry[67].
Most cellular factors, not directly interacting with the virus, involved in viral entry are served as the function of entry cofactors[25]. Some entry cofactors were identified to interact with SR-B1 [e.g., PDZK1 and UDP-glucose: Glycoprotein glucosyltransferase 1 (UGGT1)]. The scaffold protein PDZK1 may be involved in linking SR-B1 to the actin cytoskeleton and the endocytic network[68], while UGGT1, a key component of the calnexin cycle involved in protein glycosylation, may stabilize SR-B1[69].
In addition to EGFR signaling, the association of CD81–CLDN1 is also regulated by protein kinase A (PKA)[70], ephrin receptor A2[71] and phosphatidylinositol 4-kinase type III-alpha/beta[72]. These are also entry cofactors supporting viral entry. Several other entry cofactors were identified as CD81-associated factors including HRas[39], integrin 1 (ITGB1)[39], Ras-related protein Rap2B[39], calpain-5[73], ubiquitin ligase Casitas B-lineage lymphoma proto-oncogene B[73] and serum response factor binding protein 1[74]. The membrane lipid composition has been shown to affect CD81 expression on the cell’s surface and, in turn, modulate HCV entry[75]. Indeed, depleting cholesterol from the plasma membrane has been shown to decrease viral entry owing to reduced CD81 on the cell’s surface[76]. In agreement with these findings, lipid-free Apo-E was demonstrated to induce adenosine triphosphate-binding cassette subfamily G member 1 protein-dependent cholesterol efflux and inhibit HCV replication[77].
Several other entry cofactors affecting CLDN1 and/or OCLN localization are also important in the HCV entry. E-cadherin, tumor-associated calcium signal transducer 2 (TACSTD2) and possibly Rab13 could regulate the localization of CLDN1 and OCLN[78-80]. Cell surface localization of CLDN1 is also regulated by vesicular transport proteins (such as Sec24C)[81]. Serotonin 2A receptor (5-HT2AR) is shown to control CLDN1 localization through PKA-mediated phosphorylation[82].
Several cellular factors involved in the process of HCV internalization have been identified: the expression level of SR-B1[83,84], EGFR–MKNK1 signaling[85,86], Abl tyrosine kinase[87], signaling pathways of Rac/Rho/CDC42/mitogen-activated protein kinase[41], actin[50], transferrin receptor 1[88], adaptor proteins (AP)-2-associated protein kinase 1[89] and cyclin G-associated kinase[89].
Pim kinase[90], Ankyrin repeat domain 1[91] and solute carrier family 3 member 2[92] proteins are required for HCV entry, though their exact roles are not yet defined. In addition to the clathrin-dependent internalization pathway, HCV could also use clathrin-independent mechanisms to enter different cells[93].
In addition to cell-free virus entry into hepatocytes, HCV particles can also transmit directly from an infected hepatocyte to an adjacent hepatocyte[94-96]. A recent study demonstrated that the HCV core, E1, E2 and P7 genes were essential for not only cell-free viral transmission but also cell-to-cell transmission[97]. Several cellular factors were reported to contribute to HCV cell-to-cell transmission, including CD81[98,99], SR-B1[95], CLDN1[98], OCLN[98], LDLR[98], SDC-1[98], SDC-2[98], TIM-1[98], Apo-E in mature HCV particles[33,98,100], AP-1B and AP-4[101]. However, SR-B1-independent[102] or CD81-independent[103] HCV cell-to-cell transmission has also been reported. More studies are needed to clarify this issue.
Exosomes, small vesicles normally used for intercellular communication, have been reported containing HCV RNA[104-107]. HCV transmission through exosomes should be independent of any entry factor, and it is resistant to antibody neutralization. However, further investigation is required to understand the mechanisms of transmitting HCV genomic RNA through exosomes[108].
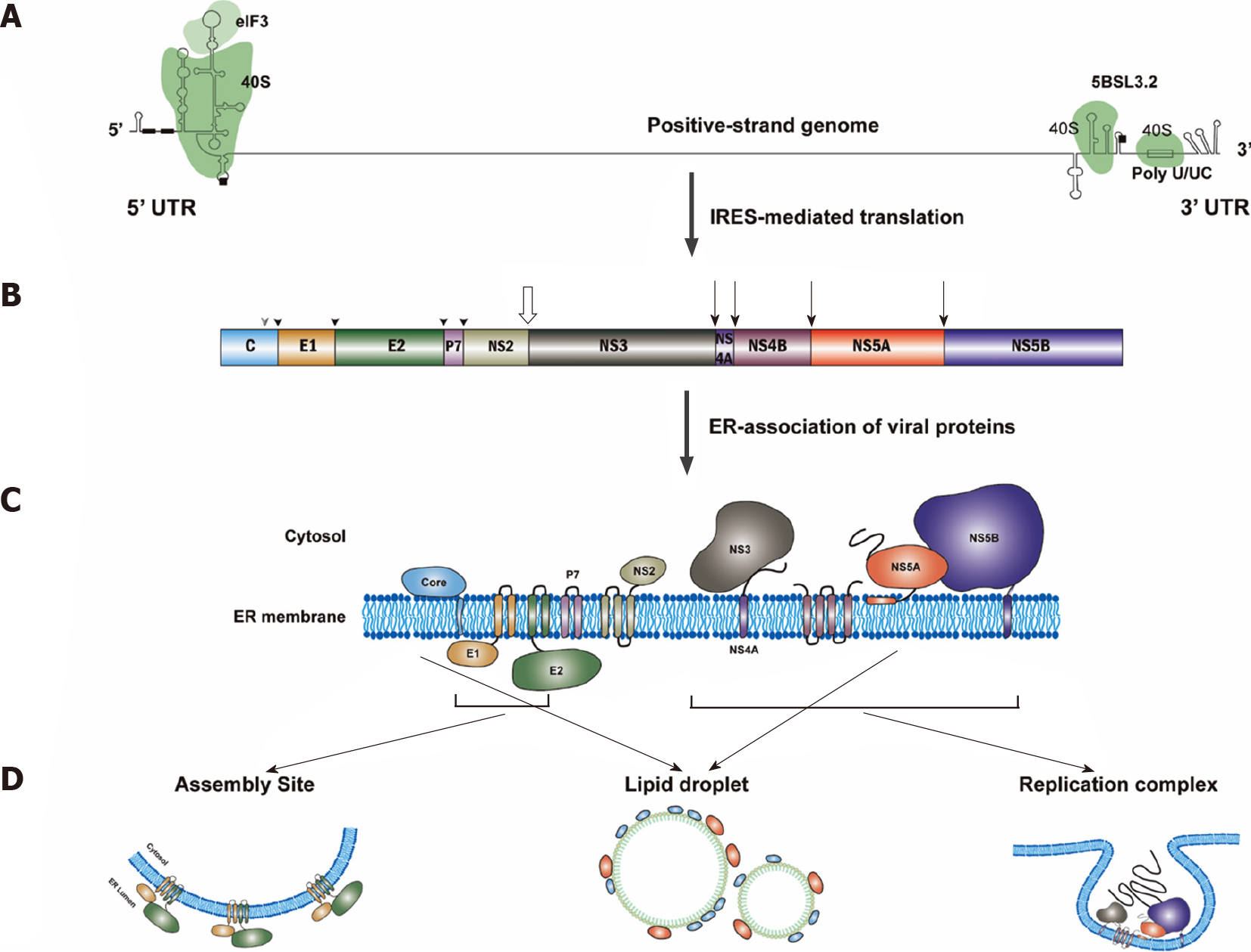
After the fusion between the viral envelope and the endosomal membrane, HCV positive-strand RNA will be released into the cytosol (Figure 2). Translation of HCV genomic RNA is modulated by the viral RNA structure, cellular translational machinery [e.g., 40S, translation initiation factors (eIFs)] and several cellular regulatory factors [e.g., microRNA-122 (miR122) and internal ribosome entry site (IRES) trans-acting factors (ITAFs)] (for a review see Reference[112]).
The most important structures of the HCV genomic RNA for translational regulation reside in the IRES of the 5’-untranslated region (5’UTR), a short segment of the core gene sequence, the cis-acting replication element (CRE) in the NS5B coding region[113] and the entire 3’UTR[114] (Figure 3). Many long-range RNA–RNA interactions among different regions of the HCV genomic RNA are involved in translational regulation[115]. The HCV IRES could bind to the ribosomal 40S subunit directly[116-118]. Interaction between the viral IRES and cellular 18S rRNA consisting of a three-nucleotide base pairing of these two molecules is crucial for HCV translation[119].
When eIF2–Met–tRNAi Met is available, canonical eIFs, including eIF3, eIF2, eIF1A, eIF5 and eIF5B, can initiate HCV translation[120]. On the other hand, when eIF2–Met–tRNAi Met is limited, eIF2A[121], eIF2D[122], eIF5B[123], a combination of eIF2A and eIF5B[124], or MCT-1 and a DENR protein complex[125] could substitute for eIF2 to initiate the translation (for a review see Reference[112]). A single RNA loop in domain II of the HCV IRES controls the translation from initiation to elongation[126].
The most important cellular factor involved in HCV translation is miR122. The liver-specific miR122 is crucial for HCV production (for a review see Reference[127]). miR122 can bind to two adjacent recognition sites on the HCV 5’ UTR with the help of human Argonaute protein 2 (hAgo2)[128-130]. The hAgo2/miR122 complex could alter the HCV 5’UTR structure and promote the IRES formation to enhance viral translation[131].
Several cellular proteins could bind to the HCV 5’UTR directly as ITAFs and enhance the viral protein translation such as La[132], heterogeneous nuclear RNA-interacting protein Q (hnRNP Q; NSAP1)[133], hnRNP L[134], hnRNP D[135], IGF2BP1 (insulin-like growth factor 2 mRNA binding protein 1; IMP-1)[136], poly(C)-binding protein 2 (PCBP2)[137], the LSm1-7 complex[138] and polypyrimidine tract binding protein (PTB)[139]. On the other hand, RNA binding protein 24[140] and Gemin5 would possibly suppress HCV translation[141].
Some proteins do not bind to HCV RNA directly but act through protein–protein interactions to help HCV translation, such as HuR (ELAVL1)[142], and the proteasome subunits[143].
Several other cellular proteins, identified as positive regulators of HCV translation through different mechanisms include MAP kinase interacting serine/threonine kinase 1 (MKNK1), phosphatidylinositol 4-kinase catalytical subunit beta (PI4K-beta)[144], ubiquitin-specific protease 15[145], NSAP1[146], RNA helicase DDX6 (RCK, p54)[147] and heat shock protein 70[148].
In addition to miRNAs and proteins, long non-coding RNA (lncRNA), such as HULC, could enhance HCV translation[149]. Moreover, the PI3K–Akt signal pathway could also upregulate HCV translation through the activation of SREBPs[150].
In addition to the synthesis of HCV polyprotein, another viral protein is produced through the core+1 reading frame (for a review see References[151,152]). Recently, the conserved RNA stem loops (SLs) SL47 and SL87 of the HCV core gene were identified to possess a novel cis-acting element and direct the internal translation initiation of the alternative reading frame (ARF)/core+1[153].
HCV genomic RNA replication, tightly linked to ER membrane alterations designated the MW[154,155], occurs in the replication organelles (ROs)[156], mainly consisting of double membrane vesicles (DMVs)[157]. The HCV RNA replication requires at least viral genomic RNA and viral RNA-dependent RNA polymerase (NS5B protein) in the ROs. Thus, HCV RNA replication could be modulated by the viral RNA structure (i.e., CREs), viral proteins (particularly, NS5B) and the biogenesis of ROs. Many cellular factors play important roles in the modulation of HCV RNA replication (for a review see References[158,159]).
The CREs of HCV genomic RNA were identified close to the 5’UTR and 3’UTR of the genome[160]. Several cellular proteins could bind to the 5’UTR and the 3’UTR of the viral RNA, facilitate the genome circularization and enhance RNA replication. These proteins include La[161], hnRNP L[162], the NFAR protein complex (NF90, NF45, and RHA)[163], PTB[164], PCBP2[137], and RNA binding protein 24[140]. High-mobility group box 1 interacting with SL 4 of 5’-UTR[165], Src-associated in mitosis 68 kDa protein binding with SL2 of 5’-UTR[166] and HSC70 with poly U/UC in the 3’-UTR[167] could also promote HCV replication.
In addition to cellular proteins, liver-specific miR122 could bind to the two adjacent sites of HCV 5’UTR[168], forming a ternary complex[169]. Through this interaction, in addition to stimulating translation[128], miR122 could protect the genome from cellular DUSP11 pyrophosphatase activity[170] and subsequent degradation by the exonucleases Xrn1[171,172] and Xrn2[172,173] to facilitate RNA replication. Cellular proteins involved in HCV RNA replication through the regulation of miR122 include DDB1-Cul4 associate factor 1[174], glycogen synthase kinase 3b[175], GTPase Rab27a[176], heterogeneous ribonucleoprotein K (hnRNP K1)[177] and DEAD-box RNA helicase (DDX6)[178]. Interestingly, various miRNAs were found to compensate for the role of miR122 on HCV replication in non-hepatic cells[179].
The HCV nonstructural proteins, NS3 to NS5B, are required for RNA replication, and NS5B constitutes the catalytic core of the HCV replication complex[180,181]. Cellular protein kinase C-related kinase 2 could phosphorylate NS5B to regulate HCV RNA replication[182]. Several cellular factors were identified to enhance HCV RNA replication through interacting with NS5B, including cellular chaperonin TRiC/CCT[183], ribonucleotide reductase M2[184], sphingomyelin[185], HuR[186], VAPB-MSP[187] and CYP4F12[188].
In addition to NS5B, NS5A is also essential to HCV RNA replication. DDX3[189], Y-box binding protein 1[189] and FKBP6[190] could interact with NS5A to facilitate HCV RNA replication. Cellular cyclophilin A (CypA)[191,192] and human replication protein A[193] could bind to NS5A and stimulate the binding of NS5A to NS5B and viral RNA to facilitate HCV RNA replication. Three domains in NS5A protein were identified using biochemical analyses: Domain I (a.a. 1–213), domain II (a.a. 250–342) and domain III (a.a. 356–448) in NS5A[194]. Domain I, with RNA-binding ability, is essential for RNA replication[195], while domain III is required for assembly[196]. A critical ratio between the different phospho-forms of NS5A protein must be maintained for productive HCV RNA replication[197]. Several cellular kinases could phosphorylate HCV NS5A, including casein kinase II (CKII)[198] and CK1a[199], lipid kinase PI4KIIIα[200] and c-Abl tyrosine kinase[201]. These kinases control the switch between virus replication and assembly by phosphorylating NS5A[202]. Vinexin b also regulates HCV replication via modulating the phosphorylation status of NS5A in a CK1a-dependent manner[203]. Notably, association of vesicle-associated membrane protein A (VAP-A) with NS5A depends on the phosphorylation status of NS5A[204]: VAP-A binds to hypophosphorylated NS5A and contributes to HCV RNA replication[202]. F-box/LRR-repeat protein 2, when geranylgeranylated, could also interact with NS5A and promote HCV replication[205]. On the other hand, proprotein convertase subtilisin/kexin type 9 interacts with NS5A and inhibits HCV replication, possibly through preventing the dimerization of NS5A[206]. SPRY domain- and SOSC box-containing protein 2 induces NS5A ubiquitination and degradation to suppress HCV replication[207].
Viral NS3 protein is also an important component of HCV replication complex. Rad51[208] and GBF1[209] could interact with NS3 and promote HCV RNA replication.
HCV NS3–NS5B proteins, in collaboration with cellular factors, could induce MW formation. Among these viral proteins, NS5A and NS4B play a major role in the induction of MW[210,211]. Cellular factors could modulate HCV replication through affecting MW biogenesis. For example, CypA plays a role in the formation of DMVs through interacting with NS5A, in addition to its role in facilitating HCV RNA replication[212]. Similarly, receptor for activated protein C kinase 1 and ATG14L were found to participate in the DMV formation through interacting NS5A[213].
Proline–serine–threonine phosphatase interacting protein 2, a protein with membrane-deforming activity, is critical for MW formation through directly interacting with NS4B and NS5A[214]. Cytosolic phospholipase A2 gamma group IVC (PLA2G4C) is also required in the biogenesis of the MW. PLA2G4C expression was upregulated after HCV infection, contributing to HCV replication and assembly through interacting with NS5A and NS4B[215].
The cellular protein Surf4, maintaining ER-Golgi intermediate compartments and the Golgi compartment, was recruited into ROs by NS4B and was involved in the formation of DMVs[216]. Another cellular protein prolactin regulatory element binding could promote HCV RNA replication by interacting with NS4B and participating in the formation of DMVs[217].
HCV replication also depends on the GBF1-Arf1-COPI complex[218,219] and phosphatidic acid phosphatase lipin1[220], possibly due to their involvement in the MW biogenesis. Sphingomyelin and ceramide transfer protein (CERT), which is in the sphingomyelin biosynthesis pathway, are also essential for the biosynthesis of DMVs[221].
Several Rabs (the Ras superfamily of small GTPases) are involved in the formation of HCV RNA replication machinery: (1) Rab5 can be recruited by NS4B and involved in HCV genome replication[222]; (2) Rab5 and 7 co-localize with NS4B and Rab2, 5 and 7 are required for HCV RNA replication[223]; (3) Rab1b and its negative regulator TBC1D20 are involved in the HCV replication[224,225]; and (4) Rab18, through associating with NS5A directly, is believed to promote the physical interaction between LDs and Ros[226].
Proteins in the nuclear transport machinery [including soluble nuclear transport factors, e.g., karyopherins (Kaps)] and nucleoporins (Nups) in the nuclear pore complexes are involved in the HCV life cycle[227,228]. Interaction of various HCV proteins with Nups and Kaps could potentially alter host cell nucleocytoplasmic transport to facilitate HCV replication[229].
Several studies have demonstrated that autophagy plays an early role in establishing HCV re
To shape an ER membrane into an RO requires not only viral and cellular proteins but also lipid synthesis (for a review see Refenence[236]). Many studies have shown that HCV could modulate lipid metabolism (e.g., cholesterol and fatty acid biosynthesis) to promote viral replication[237-239]. Furthermore, to achieve robust HCV replication, it is necessary to limit the oxidative degradation of lipids[240]. Sphingolipid is also required for HCV replication and might contribute to detergent resistance of HCV replication sites[241]. Modulation of the lipid environment of RO by HCV includes the recruitment and activation of the lipid kinase PI4KIIIα by NS5A and NS5B proteins to generate enhanced levels of phosphatidylinositol 4-phosphate (PI4P) at the RO[200]. PI4P could attract lipid transport proteins [oxysterolbinding protein (OSBP), four-phosphate adaptor protein 2 and CERT)] to deliver glycosphingolipids, cholesterol and ceramide respectively to the RO[242,243]. OSBP and CERT could interact with the human VAP residing in the ER[244]. Both VAP-A and VAP-B, enriched in purified DMVs[245], interact with NS5A and NS5B and assist in the formation of the replicase complex[204,245]. Two types of lncRNAs, lin-IGF2-AS and lnc-7SK, are involved in HCV replication through regulating PI4P[246]. Recently, HCV NS3/4A protease was reported to control the activity of 24-dehydrocholesterol reductase, catalyzing the conversion of desmosterol to cholesterol, to regulate the lipid environment for HCV RNA replication[247].
FUSE binding protein 1 is reported to be an essential cellular factor required for HCV replication through inhibiting the function of tumor suppressor p53[248]. Several other cellular factors were involved in the HCV RNA replication, mTORC1[249] and chloride channel[250], but their exact roles are not yet defined.
Assembly of HCV particles requires a viral genomic RNA, core proteins (for the capsid formation) and the viral envelope glycoproteins (E1 and E2). In addition to these viral factors, other viral nonstructural proteins and cellular factors, especially VLDL synthesis and secretion, are essential for the HCV assembly (for a review see Reference[251]).
Cleavage at the HCV core protein C-terminus by the intramembrane signal peptide peptidase is required for its maturation and targeting to LDs[252]. The mature core protein, forming the viral capsid, comprises two domains: The amino-terminal domain (D1; a.a. 1–118) and a central domain (D2; a.a. 119–177)[253]. D1 harbors basic aa residues that interact with viral RNA[254], while D2 is hydrophobic and associated with LDs[252]. LDs are important for the production of infectious HCV particles[255]. As expected, a reduction in the volume of LDs by the suppression of HSC70 expression[23] or disruption of LDs by the inhibitor of aryl hydrocarbon (AhR)[256] would inhibit HCV production. Thus, ADP-ribosylation factor-related protein 1 essential for LD growth is required for HCV propagation[257], while N-Myc Downstream-Regulated Gene 1 restricts HCV assembly by limiting LD formation[258].
Several cellular factors are involved in the association of the core with LDs. Diacylglycerol acyltransferase-1 (DGAT1) interacts with both the core and NS5A proteins and is required for the trafficking of these two proteins to LDs[259]. PLA2G4A also plays a role in recruiting core to LDs, and its specific cleavage of lipids containing arachidonic acid is essential for the production of infectious viral particles[260]. Interaction of core and Nup98 in LDs is important for HCV propagation[22], while heterogeneous nuclear ribonucleoprotein K is recruited to sites in close proximity to LDs and suppress HCV production[261].
HCV genomic RNAs synthesized by the HCV replication complex (NS3–NS5B proteins) in the DMVs will be transferred by NS5A and NS3-4A proteins and encapsidated by the viral capsid to form the nucleocapsid. The HCV RNA structure[262] responsible for its encapsidation by core proteins has been suggested to be (1) a highly conserved secondary structure within the core D2 region[263]; (2) the conserved apical motifs of the 3’X region[264]; or (3) multiple RNA motifs with a secondary structure[265].
Lipid mobilization from cytoplasmic LDs favors the morphogenesis and secretion of HCV particles[266,267]. HCV infection suppresses the cellular lysophosphati
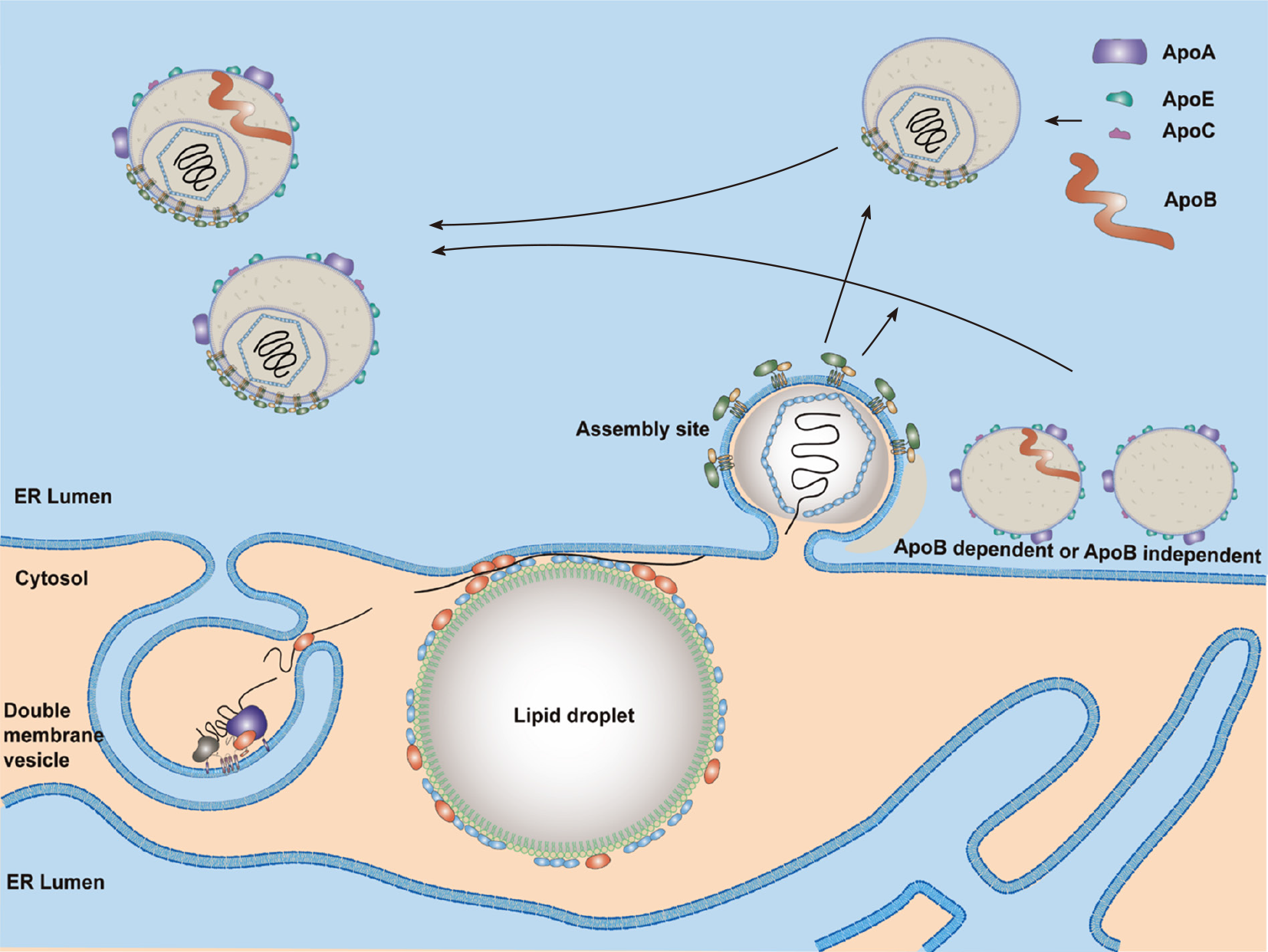
PLA2G4C[215] and AAM-B[269] recruit the NS4B protein to LDs. Thus, these two proteins may bridge the steps of HCV RNA replication and assembly by translocation of RCs to LDs. In addition to DGAT1, CD2AP also participates in the transfer of NS5A to LDs[270]. Interactions between NS5A and core proteins are crucial for productive HCV infection[271]. Protein kinase C and CK substrate in neurons protein 2[272] and cortactin[273] promote interactions between HCV core and NS5A in the LDs. HCV NS5A protein domain I interacts with the D1 region of core protein[274]. Indeed, core and NS5A proteins are found associated with LDs at 12 h post-infection[275]. The LDs associated with core and NS5A proteins are close to the DMVs and the assembly sites on the ER membrane (Figure 4). Several studies suggested that the NS5A protein might link DMVs with assembly sites. Two LD-associated proteins, Rab18[226] and TIP47[276,277], were found to interact with NS5A and might help the juxtaposition of replication and assembly sites.
Formation of HCV nucleocapsid may occur in the LDs and/or assembly sites (Figure 4). Then, HCV nucleocapsid will move to the assembly sites and interact with viral E1/E2 proteins (envelopment) and bud into the ER lumen (egress) (Figure 4). All viral proteins are involved in HCV assembly[278,279]. The core and E1/E2 proteins are the integral protein components of an HCV particle. The other viral proteins do help viral assembly and egress, especially NS5A, p7, and NS2[278-281]. NS2, ubiquitinated by MARCH8[282], is a key regulator of viral assembly by bringing together the structural and nonstructural proteins required for particle formation. The cellular signal peptidase complex subunit 1 interacts with both NS2 and E2 proteins and mediates membrane association of the NS2–E2 complex to control HCV assembly[283]. Then, PLA1A plays a role in bridging NS2–E2 complex and NS5A-associated replication complex through its interaction with E2, NS2 and NS5A[284,285]. It is likely that NS2 protein brings E1, E2, NS3, NS5A and core proteins together to form a complex within the detergent-resistant membranes in the ER as an assembly platform to initiate HCV assembly[286]. Meanwhile, the clathrin Adaptor Related Protein Complex 2 Subunit Mu 1[287] and a small GTPase, Rab32[288], may transfer nucleocapsids to the sites of envelopment. HRS (hepatocyte growth factor-regulated tyrosine kinase substrate), an endosomal-sorting complex required for transport (ESCRT)-0 complex component, is involved in the viral envelopment[289].
HCV assembly and envelopment are linked to the VLDL synthesis and secretion[290]. Indeed, CIDEB, an important regulator of the VLDL pathway, contributes to the HCV assembly through interacting with NS5A[291]. However, inhibitors of microsomal triglyceride transfer protein (MTTP) affect secretion of HCV more severely than that of VLDL[292]. HCV is also reported to modify VLDL secretion[293,294]. These results suggest that HCV assembly occurs possibly through modification of the VLDL secretion. Indeed, colocalization of the core with Apo-E but not with Apo-B was demonstrated[295]. Therefore, it is more likely that HCV suppresses VLDL secretion and then uses the excess lipid to produce lipid-rich viral particles. Components of VLDL synthesis, such as MTTP[290], Apo-B[296] and especially Apo-E[10], have been implicated in HCV assembly. HCV production in HuH7 cells with double knockout of Apo-B/Apo-E was reduced significantly compared to that of single knockout cells, and ectopic expression of Apo-E in cells with double knockout of Apo-B/Apo-E restored production of infectious viruses. Furthermore, ectopic expression of Apo-E or MTTP in cells with double knockout of Apo-B/MTTP could restore infectious virus production[297]. These studies suggested that there are Apo-B-dependent and -independent virus assembly pathways (Figure 4). Similar to the effect of ectopic expression of Apo-E in Apo-B/Apo-E double knockout cells, expression of exchangeable apolipoproteins (e.g., Apo-A1, A2, C1, C2 and C3), the peptides of amphipathic α-helices containing the amino-terminal domain of Apo-E[297] or even human cathelicidin antimicrobial peptide[298] also restored infectious virus production. These results suggest that infectious virus production is regulated redundantly by exchangeable apolipoproteins expressed in the liver. Annexin A3 (ANXA3) through facilitating the incorporation of Apo-E[299] and Golgi protein 73, a resident Golgi membrane protein, through facilitating the interaction of HCV NS5A with Apo-E[300], promote HCV virion maturation. Recently, Apo-M, interacting with E2, was reported to be a novel virus particle-associated protein[301].
After envelopment, HCV particles then traffic to Golgi likely within COPII secretory vesicles[295,302]. Secretion of infectious HCV particles relies in part on components of the ESCRT pathway[303]. HCV egress but not VLDL secretion is blocked by silencing Rabs and the transGolgi network (TGN)-associating adaptors[304]. Moreover, inhibition of Apo-E secretion using monensin does not impair HCV release. These results suggested that HCV and VLDL use distinct secretion pathways[305]. Altogether, these results suggest that the release of HCV particles occur via a TGN-endosomal secretion pathway that is different from that of VLDL. The lipid-associated TM6SF2 (transmembrane 6 superfamily 2) has been demonstrated to promote lipidation and secretion of HCV particles[306]. The secreted HCV particle is likely a single particle fusion of viral structural proteins with various apolipoproteins and lipids (Figure 1).
Autophagy triggered by HCV-induced oxidative stress favors the release of HCV particles[307]. Thus, autophagy may play a role not only in the induction of DMVs but also in the secretion of HCV particles (for a review see Reference[235]).
HCV particles from the sera of HCV-infected patients harbor higher amounts of Apo-E than those derived from cell culture[308]. The interaction of Apo-E and HCV enhances specific infectivity and may aid HCV in evading neutralizing antibodies[309]. HCV particles from the blood of HCV-infected patients contain Apo-B100 or Apo-B48, indicating that a significant fraction of HCV particles in blood is also associated with Apo-B48-containing lipoproteins[310]. These results suggested that the interactions between HCV particles and lipoproteins (e.g., Apo-E and Apo-B48) in the blood of HCV patients (Figure 1B). Besides lipoproteins, specific serum factors, including albumin, also promote extracellular maturation of HCV particles[311].
Several cellular factors were involved in the assembly and secretion of HCV particles, such as ANXA2[312], sorcin (soluble resistance-related calcium-binding protein)[313], AP-1A, AP-1B, AP-4[101] and O-linked N-acetylglucosamine transferase[314], but their exact roles are not yet defined.
Study on the life cycle of the HCV has progressed tremendously after the development of in vitro HCV culture systems[315]. Understanding the HCV replication cycle led to the huge success of direct-acting anti-virals (DAAs) targeting NS3, NS5A, and NS5B. Hundreds of cellular factors involved in various stages of the HCV’s life cycle have also been identified after more than 30 years of research on HCV–host cell interactions. Some of these cellular factors have been selected as targets for anti-HCV therapy (e.g., SR-B1, EGFR, NPC1L1, miR122, CypA)[7]. Inhibitors against these cellular factors may complement existing DAAs. A successful vaccine for HCV is still a challenge in the near future. Understanding the mechanisms of viral entry, especially E2–CD81 interactions, should help in the development of a vaccine.
HCV particle, a hybrid lipo-viro-particle, does not look like a canonical enveloped virus. Thus, HCV has become a unique model for studying virus–host interactions, e.g., between HCV and cellular lipid metabolisms. Furthermore, all positive-strand RNA viruses, including coronaviruses and picornaviruses, induce the reorganization of cellular membranes to replicate their genomes, similar to HCV[316]. Using HCV as a paradigm to study how HCV induces cellular membrane re-organization may lead to identification of broad-spectrum antivirals targeting cellular factors commonly used by these viruses.
Despite the impressive advances, many issues are still far from being clarified regarding HCV–host cell interactions. More studies are needed to understand the detailed mechanisms.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Taiwan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Pham TTT S-Editor: Fan JR L-Editor: A P-Editor: Liu JH
| 1. | Roingeard P, Beaumont E. Hepatitis C Vaccine: 10 Good Reasons for Continuing. Hepatology. 2020;71:1845-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | Ansaldi F, Orsi A, Sticchi L, Bruzzone B, Icardi G. Hepatitis C virus in the new era: perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J Gastroenterol. 2014;20:9633-9652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 113] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 3. | Li HC, Lo SY. Hepatitis C virus: Virology, diagnosis and treatment. World J Hepatol. 2015;7:1377-1389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (3)] |
| 4. | Jacka B, Lamoury F, Simmonds P, Dore GJ, Grebely J, Applegate T. Sequencing of the Hepatitis C Virus: A Systematic Review. PLoS One. 2013;8:e67073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (2)] |
| 5. | Houghton M. Hepatitis C Virus: 30 Years after Its Discovery. Cold Spring Harb Perspect Med. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (1)] |
| 6. | Alazard-Dany N, Denolly S, Boson B, Cosset FL. Overview of HCV Life Cycle with a Special Focus on Current and Possible Future Antiviral Targets. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 7. | Shulla A, Randall G. Hepatitis C Virus-Host Interactions. Springer, 2016: 197-233. [DOI] [Full Text] |
| 8. | Catanese MT, Uryu K, Kopp M, Edwards TJ, Andrus L, Rice WJ, Silvestry M, Kuhn RJ, Rice CM. Ultrastructural analysis of hepatitis C virus particles. Proc Natl Acad Sci USA. 2013;110:9505-9510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 210] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 9. | Bradley D, McCaustland K, Krawczynski K, Spelbring J, Humphrey C, Cook EH. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J Med Virol. 1991;34:206-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 118] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 10. | Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783-13793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 342] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 11. | Dreux M, Boson B, Ricard-Blum S, Molle J, Lavillette D, Bartosch B, Pécheur EI, Cosset FL. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J Biol Chem. 2007;282:32357-32369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018-3032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 285] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 13. | Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci USA. 2006;103:3805-3809. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 344] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 14. | André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919-6928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 517] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 15. | Hijikata M, Shimizu YK, Kato H, Iwamoto A, Shih JW, Alter HJ, Purcell RH, Yoshikura H. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953-1958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 254] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 16. | Marcotrigiano J, Catanese MT. The Structure of HCV. Springer, 2016: 31-64. |
| 17. | Andréo U, Maillard P, Kalinina O, Walic M, Meurs E, Martinot M, Marcellin P, Budkowska A. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell Microbiol. 2007;9:2445-2456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Shimizu Y, Hishiki T, Sugiyama K, Ogawa K, Funami K, Kato A, Ohsaki Y, Fujimoto T, Takaku H, Shimotohno K. Lipoprotein lipase and hepatic triglyceride lipase reduce the infectivity of hepatitis C virus (HCV) through their catalytic activities on HCV-associated lipoproteins. Virology. 2010;407:152-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Thomssen R, Bonk S. Virolytic action of lipoprotein lipase on hepatitis C virus in human sera. Med Microbiol Immunol. 2002;191:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 20. | Maillard P, Walic M, Meuleman P, Roohvand F, Huby T, Le Goff W, Leroux-Roels G, Pécheur EI, Budkowska A. Lipoprotein lipase inhibits hepatitis C virus (HCV) infection by blocking virus cell entry. PLoS One. 2011;6:e26637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | Sun HY, Lin CC, Lee JC, Wang SW, Cheng PN, Wu IC, Chang TT, Lai MD, Shieh DB, Young KC. Very low-density lipoprotein/Lipo-viro particles reverse lipoprotein lipase-mediated inhibition of hepatitis C virus infection via apolipoprotein C-III. Gut. 2013;62:1193-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Lussignol M, Kopp M, Molloy K, Vizcay-Barrena G, Fleck RA, Dorner M, Bell KL, Chait BT, Rice CM, Catanese MT. Proteomics of HCV virions reveals an essential role for the nucleoporin Nup98 in virus morphogenesis. Proc Natl Acad Sci USA. 2016;113:2484-2489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 23. | Parent R, Qu X, Petit MA, Beretta L. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology. 2009;49:1798-1809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 24. | Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr Top Microbiol Immunol. 2013;369:87-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 25. | Gerold G, Moeller R, Pietschmann T. Hepatitis C Virus Entry: Protein Interactions and Fusion Determinants Governing Productive Hepatocyte Invasion. Cold Spring Harb Perspect Med. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 26. | Pöhlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, Granelli-Piperno A, Doms RW, Rice CM, McKeating JA. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070-4080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 301] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 27. | Gardner JP, Durso RJ, Arrigale RR, Donovan GP, Maddon PJ, Dragic T, Olson WC. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc Natl Acad Sci USA. 2003;100:4498-4503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 215] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 28. | Tawar RG, Schuster C, Baumert TF. HCV Receptors and Virus Entry. Springer, 2016; 81-103. [DOI] [Full Text] |
| 29. | Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017-5025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 888] [Cited by in RCA: 891] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 30. | Owen DM, Huang H, Ye J, Gale M Jr. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 174] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 31. | Shi Q, Jiang J, Luo G. Syndecan-1 serves as the major receptor for attachment of hepatitis C virus to the surfaces of hepatocytes. J Virol. 2013;87:6866-6875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 32. | Lefèvre M, Felmlee DJ, Parnot M, Baumert TF, Schuster C. Syndecan 4 is involved in mediating HCV entry through interaction with lipoviral particle-associated apolipoprotein E. PLoS One. 2014;9:e95550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 33. | Qiao L, Luo GG. Functional Characterization of Apolipoproteins in the HCV Life Cycle. Methods Mol Biol. 2019;1911:235-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 34. | Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278:41003-41012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 350] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 35. | Jiang J, Wu X, Tang H, Luo G. Apolipoprotein E mediates attachment of clinical hepatitis C virus to hepatocytes by binding to cell surface heparan sulfate proteoglycan receptors. PLoS One. 2013;8:e67982. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 36. | Wattis JA, O'Malley B, Blackburn H, Pickersgill L, Panovska J, Byrne HM, Jackson KG. Mathematical model for low density lipoprotein (LDL) endocytosis by hepatocytes. Bull Math Biol. 2008;70:2303-2333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 37. | Li Y, Kakinami C, Li Q, Yang B, Li H. Human apolipoprotein A-I is associated with dengue virus and enhances virus infection through SR-BI. PLoS One. 2013;8:e70390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 38. | Zhang H, Qiao L, Luo G. Characterization of apolipoprotein C1 in hepatitis C virus infection and morphogenesis. Virology. 2018;524:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 39. | Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M, Durand SC, Duong FH, Heim MH, Cosset FL, Hirsch I, Samuel D, Brino L, Zeisel MB, Le Naour F, McKeating JA, Baumert TF. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 40. | Douam F, Dao Thi VL, Maurin G, Fresquet J, Mompelat D, Zeisel MB, Baumert TF, Cosset FL, Lavillette D. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology. 2014;59:776-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 41. | Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol. 2008;82:8316-8329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 144] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 42. | Diao J, Pantua H, Ngu H, Komuves L, Diehl L, Schaefer G, Kapadia SB. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol. 2012;86:10935-10949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 43. | Davis C, Harris HJ, Hu K, Drummer HE, McKeating JA, Mullins JG, Balfe P. In silico directed mutagenesis identifies the CD81/claudin-1 hepatitis C virus receptor interface. Cell Microbiol. 2012;14:1892-1903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Farquhar MJ, Hu K, Harris HJ, Davis C, Brimacombe CL, Fletcher SJ, Baumert TF, Rappoport JZ, Balfe P, McKeating JA. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J Virol. 2012;86:4305-4316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 45. | Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, Narbus CM, Parra Martín A, Evans MJ. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog. 2013;9:e1003244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 46. | Liu S, Kuo W, Yang W, Liu W, Gibson GA, Dorko K, Watkins SC, Strom SC, Wang T. The second extracellular loop dictates Occludin-mediated HCV entry. Virology. 2010;407:160-170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 47. | Wünschmann S, Medh JD, Klinzmann D, Schmidt WN, Stapleton JT. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74:10055-10062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 163] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 48. | Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80:1734-1741. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 308] [Cited by in RCA: 308] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 49. | Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271-7276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 644] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 50. | Coller KE, Berger KL, Heaton NS, Cooper JD, Yoon R, Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009;5:e1000702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 51. | Sharma NR, Mateu G, Dreux M, Grakoui A, Cosset FL, Melikyan GB. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J Biol Chem. 2011;286:30361-30376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 52. | Banda DH, Perin PM, Brown RJP, Todt D, Solodenko W, Hoffmeyer P, Kumar Sahu K, Houghton M, Meuleman P, Müller R, Kirschning A, Pietschmann T. A central hydrophobic E1 region controls the pH range of hepatitis C virus membrane fusion and susceptibility to fusion inhibitors. J Hepatol. 2019;70:1082-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 53. | Wu X, Lee EM, Hammack C, Robotham JM, Basu M, Lang J, Brinton MA, Tang H. Cell death-inducing DFFA-like effector b is required for hepatitis C virus entry into hepatocytes. J Virol. 2014;88:8433-8444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 54. | V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155-170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2164] [Cited by in RCA: 2020] [Article Influence: 505.0] [Reference Citation Analysis (0)] |
| 55. | Millet JK, Whittaker GR. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3-8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 200] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 56. | Qing J, Wu M, Luo R, Chen J, Cao L, Zeng D, Shang L, Nong J, Wu Q, Ding BS, Chen X, Rao Z, Liu L, Lou Z. Identification of Interferon Receptor IFNAR2 As a Novel HCV Entry Factor by Using Chemical Probes. ACS Chem Biol. 2020;15:1232-1241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 57. | Yamamoto S, Fukuhara T, Ono C, Uemura K, Kawachi Y, Shiokawa M, Mori H, Wada M, Shima R, Okamoto T, Hiraga N, Suzuki R, Chayama K, Wakita T, Matsuura Y. Lipoprotein Receptors Redundantly Participate in Entry of Hepatitis C Virus. PLoS Pathog. 2016;12:e1005610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 58. | Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med. 2012;18:281-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 353] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 59. | Wang J, Qiao L, Hou Z, Luo G. TIM-1 Promotes Hepatitis C Virus Cell Attachment and Infection. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 60. | Kachko A, Costafreda MI, Zubkova I, Jacques J, Takeda K, Wells F, Kaplan G, Major ME. Determinants in the Ig Variable Domain of Human HAVCR1 (TIM-1) Are Required To Enhance Hepatitis C Virus Entry. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 61. | Li X, Li J, Feng Y, Cai H, Li YP, Peng T. Long-chain fatty acyl-coenzyme A suppresses hepatitis C virus infection by targeting virion-bound lipoproteins. Antiviral Res. 2020;177:104734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 62. | Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol. 2007;81:12465-12471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 176] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 63. | Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J Virol. 2008;82:3555-3560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 64. | Huang J, Yin P, Zhang L. COPII cargo claudin-12 promotes hepatitis C virus entry. J Viral Hepat. 2019;26:308-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 65. | Park JH, Park S, Yang JS, Kwon OS, Kim S, Jang SK. Discovery of cellular proteins required for the early steps of HCV infection using integrative genomics. PLoS One. 2013;8:e60333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 66. | Cheng JJ, Li JR, Huang MH, Ma LL, Wu ZY, Jiang CC, Li WJ, Li YH, Han YX, Li H, Chen JH, Wang YX, Song DQ, Peng ZG, Jiang JD. CD36 is a co-receptor for hepatitis C virus E1 protein attachment. Sci Rep. 2016;6:21808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 67. | Ujino S, Nishitsuji H, Hishiki T, Sugiyama K, Takaku H, Shimotohno K. Hepatitis C virus utilizes VLDLR as a novel entry pathway. Proc Natl Acad Sci USA. 2016;113:188-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 68. | Eyre NS, Drummer HE, Beard MR. The SR-BI partner PDZK1 facilitates hepatitis C virus entry. PLoS Pathog. 2010;6:e1001130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 69. | Huang J, Yin H, Yin P, Jian X, Song S, Luan J, Zhang L. SR-BI Interactome Analysis Reveals a Proviral Role for UGGT1 in Hepatitis C Virus Entry. Front Microbiol. 2019;10:2043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 70. | Farquhar MJ, Harris HJ, Diskar M, Jones S, Mee CJ, Nielsen SU, Brimacombe CL, Molina S, Toms GL, Maurel P, Howl J, Herberg FW, van Ijzendoorn SC, Balfe P, McKeating JA. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J Virol. 2008;82:8797-8811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 71. | Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoël M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011;17:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 72. | Trotard M, Lepère-Douard C, Régeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780-3789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 73. | Bruening J, Lasswitz L, Banse P, Kahl S, Marinach C, Vondran FW, Kaderali L, Silvie O, Pietschmann T, Meissner F, Gerold G. Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLoS Pathog. 2018;14:e1007111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 74. | Gerold G, Meissner F, Bruening J, Welsch K, Perin PM, Baumert TF, Vondran FW, Kaderali L, Marcotrigiano J, Khan AG, Mann M, Rice CM, Pietschmann T. Quantitative Proteomics Identifies Serum Response Factor Binding Protein 1 as a Host Factor for Hepatitis C Virus Entry. Cell Rep. 2015;12:864-878. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 75. | Voisset C, Lavie M, Helle F, Op De Beeck A, Bilheu A, Bertrand-Michel J, Tercé F, Cocquerel L, Wychowski C, Vu-Dac N, Dubuisson J. Ceramide enrichment of the plasma membrane induces CD81 internalization and inhibits hepatitis C virus entry. Cell Microbiol. 2008;10:606-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 76. | Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol. 2007;81:374-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 205] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 77. | Crouchet E, Lefèvre M, Verrier ER, Oudot MA, Baumert TF, Schuster C. Extracellular lipid-free apolipoprotein E inhibits HCV replication and induces ABCG1-dependent cholesterol efflux. Gut. 2017;66:896-907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 78. | Sekhar V, Pollicino T, Diaz G, Engle RE, Alayli F, Melis M, Kabat J, Tice A, Pomerenke A, Altan-Bonnet N, Zamboni F, Lusso P, Emerson SU, Farci P. Infection with hepatitis C virus depends on TACSTD2, a regulator of claudin-1 and occludin highly downregulated in hepatocellular carcinoma. PLoS Pathog. 2018;14:e1006916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 79. | Li Q, Sodroski C, Lowey B, Schweitzer CJ, Cha H, Zhang F, Liang TJ. Hepatitis C virus depends on E-cadherin as an entry factor and regulates its expression in epithelial-to-mesenchymal transition. Proc Natl Acad Sci USA. 2016;113:7620-7625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 80. | Takeda M, Ikeda M, Satoh S, Dansako H, Wakita T, Kato N. Rab13 Is Involved in the Entry Step of Hepatitis C Virus Infection. Acta Med Okayama. 2016;70:111-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 81. | Yin P, Li Y, Zhang L. Sec24C-Dependent Transport of Claudin-1 Regulates Hepatitis C Virus Entry. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 82. | Cao L, Chen J, Wang Y, Yang Y, Qing J, Rao Z, Chen X, Lou Z. Identification of serotonin 2A receptor as a novel HCV entry factor by a chemical biology strategy. Protein Cell. 2019;10:178-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 83. | Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, Schwarz A, Moreau M, Owen JS, Leroux-Roels G, Balfe P, McKeating JA. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J Virol. 2007;81:3162-3169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 116] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 84. | Schwarz AK, Grove J, Hu K, Mee CJ, Balfe P, McKeating JA. Hepatoma cell density promotes claudin-1 and scavenger receptor BI expression and hepatitis C virus internalization. J Virol. 2009;83:12407-12414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 85. | Baktash Y, Madhav A, Coller KE, Randall G. Single Particle Imaging of Polarized Hepatoma Organoids upon Hepatitis C Virus Infection Reveals an Ordered and Sequential Entry Process. Cell Host Microbe. 2018;23:382-394.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 83] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 86. | Kim S, Ishida H, Yamane D, Yi M, Swinney DC, Foung S, Lemon SM. Contrasting roles of mitogen-activated protein kinases in cellular entry and replication of hepatitis C virus: MKNK1 facilitates cell entry. J Virol. 2013;87:4214-4224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 87. | Min S, Lim YS, Shin D, Park C, Park JB, Kim S, Windisch MP, Hwang SB. Abl Tyrosine Kinase Regulates Hepatitis C Virus Entry. Front Microbiol. 2017;8:1129. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 88. | Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci USA. 2013;110:10777-10782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 89. | Neveu G, Ziv-Av A, Barouch-Bentov R, Berkerman E, Mulholland J, Einav S. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J Virol. 2015;89:4387-4404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 106] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 90. | Park C, Min S, Park EM, Lim YS, Kang S, Suzuki T, Shin EC, Hwang SB. Pim Kinase Interacts with Nonstructural 5A Protein and Regulates Hepatitis C Virus Entry. J Virol. 2015;89:10073-10086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 91. | Than TT, Tran GV, Son K, Park EM, Kim S, Lim YS, Hwang SB. Ankyrin Repeat Domain 1 is Up-regulated During Hepatitis C Virus Infection and Regulates Hepatitis C Virus Entry. Sci Rep. 2016;6:20819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 92. | Nguyen NNT, Lim YS, Nguyen LP, Tran SC, Luong TTD, Nguyen TTT, Pham HT, Mai HN, Choi JW, Han SS, Hwang SB. Hepatitis C Virus Modulates Solute carrier family 3 member 2 for Viral Propagation. Sci Rep. 2018;8:15486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 93. | Matsuda M, Suzuki R, Kataoka C, Watashi K, Aizaki H, Kato N, Matsuura Y, Suzuki T, Wakita T. Alternative endocytosis pathway for productive entry of hepatitis C virus. J Gen Virol. 2014;95:2658-2667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 94. | Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, Desombere I, Roels GL, Balfe P, McKeating JA. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 272] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 95. | Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL, Wong-Staal F, Balfe P, McKeating JA. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol. 2011;85:596-605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 198] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 96. | Xiao F, Fofana I, Heydmann L, Barth H, Soulier E, Habersetzer F, Doffoël M, Bukh J, Patel AH, Zeisel MB, Baumert TF. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog. 2014;10:e1004128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 97. | Zhao F, Zhao T, Deng L, Lv D, Zhang X, Pan X, Xu J, Long G. Visualizing the Essential Role of Complete Virion Assembly Machinery in Efficient Hepatitis C Virus Cell-to-Cell Transmission by a Viral Infection-Activated Split-Intein-Mediated Reporter System. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 98. | Fan H, Qiao L, Kang KD, Fan J, Wei W, Luo G. Attachment and Postattachment Receptors Important for Hepatitis C Virus Infection and Cell-to-Cell Transmission. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 99. | Fofana I, Xiao F, Thumann C, Turek M, Zona L, Tawar RG, Grunert F, Thompson J, Zeisel MB, Baumert TF. A novel monoclonal anti-CD81 antibody produced by genetic immunization efficiently inhibits Hepatitis C virus cell-cell transmission. PLoS One. 2013;8:e64221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 100. | Gondar V, Molina-Jiménez F, Hishiki T, García-Buey L, Koutsoudakis G, Shimotohno K, Benedicto I, Majano PL. Apolipoprotein E, but Not Apolipoprotein B, Is Essential for Efficient Cell-to-Cell Transmission of Hepatitis C Virus. J Virol. 2015;89:9962-9973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 101. | Xiao F, Wang S, Barouch-Bentov R, Neveu G, Pu S, Beer M, Schor S, Kumar S, Nicolaescu V, Lindenbach BD, Randall G, Einav S. Interactions between the Hepatitis C Virus Nonstructural 2 Protein and Host Adaptor Proteins 1 and 4 Orchestrate Virus Release. mBio. 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 102. | Catanese MT, Loureiro J, Jones CT, Dorner M, von Hahn T, Rice CM. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free vs cell-to-cell transmission. J Virol. 2013;87:8282-8293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 103. | Witteveldt J, Evans MJ, Bitzegeio J, Koutsoudakis G, Owsianka AM, Angus AG, Keck ZY, Foung SK, Pietschmann T, Rice CM, Patel AH. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol. 2009;90:48-58. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 146] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 104. | Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJ. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci USA. 2013;110:13109-13113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 414] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 105. | Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014;10:e1004424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 342] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 106. | Longatti A, Boyd B, Chisari FV. Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol. 2015;89:2956-2961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 107. | Shrivastava S, Devhare P, Sujijantarat N, Steele R, Kwon YC, Ray R, Ray RB. Knockdown of Autophagy Inhibits Infectious Hepatitis C Virus Release by the Exosomal Pathway. J Virol. 2016;90:1387-1396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 108. | Medvedev R, Hildt E, Ploen D. Look who's talking-the crosstalk between oxidative stress and autophagy supports exosomal-dependent release of HCV particles. Cell Biol Toxicol. 2017;33:211-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 109. | Chen CL, Huang JY, Wang CH, Tahara SM, Zhou L, Kondo Y, Schechter J, Su L, Lai MM, Wakita T, Cosset FL, Jung JU, Machida K. Hepatitis C virus has a genetically determined lymphotropism through co-receptor B7.2. Nat Commun. 2017;8:13882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 110. | Sarhan MA, Pham TN, Chen AY, Michalak TI. Hepatitis C virus infection of human T lymphocytes is mediated by CD5. J Virol. 2012;86:3723-3735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 111. | Liu Y, Wang W, Zou Z, Hu Z, Fan Q, Xiong J. Hepatitis C Virus Entry into Macrophages/Monocytes Mainly Depends on the Phagocytosis of Macrophages. Dig Dis Sci. 2019;64:1226-1237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 112. | Niepmann M, Gerresheim GK. Hepatitis C Virus Translation Regulation. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 113. | Romero-López C, Ríos-Marco P, Berzal-Herranz B, Berzal-Herranz A. The HCV genome domains 5BSL3.1 and 5BSL3.3 act as managers of translation. Sci Rep. 2018;8:16101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 114. | Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 700] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 115. | Fricke M, Dünnes N, Zayas M, Bartenschlager R, Niepmann M, Marz M. Conserved RNA secondary structures and long-range interactions in hepatitis C viruses. RNA. 2015;21:1219-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 116. | Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959-1962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 383] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 117. | Lytle JR, Wu L, Robertson HD. The ribosome binding site of hepatitis C virus mRNA. J Virol. 2001;75:7629-7636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 118. | Yamamoto H, Collier M, Loerke J, Ismer J, Schmidt A, Hilal T, Sprink T, Yamamoto K, Mielke T, Bürger J, Shaikh TR, Dabrowski M, Hildebrand PW, Scheerer P, Spahn CM. Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA. EMBO J. 2015;34:3042-3058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 119. | Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 155] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 120. | Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci USA. 2004;101:16990-16995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 141] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 121. | Kim JH, Park SM, Park JH, Keum SJ, Jang SK. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 2011;30:2454-2464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 97] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 122. | Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779-26787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 123. | Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol. 2008;15:836-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 124. | Kim E, Kim JH, Seo K, Hong KY, An SWA, Kwon J, Lee SV, Jang SK. eIF2A, an initiator tRNA carrier refractory to eIF2α kinases, functions synergistically with eIF5B. Cell Mol Life Sci. 2018;75:4287-4300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 125. | Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787-1801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 126. | Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat Struct Mol Biol. 2013;20:150-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 127. | Kunden RD, Khan JQ, Ghezelbash S, Wilson JA. The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 128. | Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300-3310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 527] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 129. | Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716-7729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 177] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 130. | Nieder-Röhrmann A, Dünnes N, Gerresheim GK, Shalamova LA, Herchenröther A, Niepmann M. Cooperative enhancement of translation by two adjacent microRNA-122/Argonaute 2 complexes binding to the 5' untranslated region of hepatitis C virus RNA. J Gen Virol. 2017;98:212-224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 131. | Chahal J, Gebert LFR, Gan HH, Camacho E, Gunsalus KC, MacRae IJ, Sagan SM. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5' terminus. Nucleic Acids Res. 2019;47:5307-5324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 132. | Ali N, Siddiqui A. The La antigen binds 5' noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249-2254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 215] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 133. | Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol Cell Biol. 2004;24:7878-7890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 134. | Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. Heterogeneous nuclear ribonucleoprotein L interacts with the 3' border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782-8788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 124] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 135. | Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol. 2008;82:12082-12093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 136. | Weinlich S, Hüttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3'UTR. RNA. 2009;15:1528-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 137. | Wang L, Jeng KS, Lai MM. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5' untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol. 2011;85:7954-7964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 138. | Scheller N, Mina LB, Galão RP, Chari A, Giménez-Barcons M, Noueiry A, Fischer U, Meyerhans A, Díez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci USA. 2009;106:13517-13522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 139. | Gosert R, Chang KH, Rijnbrand R, Yi M, Sangar DV, Lemon SM. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol Cell Biol. 2000;20:1583-1595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 140. | Cao H, Zhao K, Yao Y, Guo J, Gao X, Yang Q, Guo M, Zhu W, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Lu M, Chen X, Pei R. RNA binding protein 24 regulates the translation and replication of hepatitis C virus. Protein Cell. 2018;9:930-944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 141. | Francisco-Velilla R, Azman EB, Martinez-Salas E. Impact of RNA-Protein Interaction Modes on Translation Control: The Versatile Multidomain Protein Gemin5. Bioessays. 2019;41:e1800241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 142. | Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverría F, Dormoy-Raclet V, Rodríguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, López-Lastra M. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 143. | Fedorova OA, Moiseeva TN, Nikiforov AA, Tsimokha AS, Livinskaya VA, Hodson M, Bottrill A, Evteeva IN, Ermolayeva JB, Kuznetzova IM, Turoverov KK, Eperon I, Barlev NA. Proteomic analysis of the 20S proteasome (PSMA3)-interacting proteins reveals a functional link between the proteasome and mRNA metabolism. Biochem Biophys Res Commun. 2011;416:258-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 144. | Lupberger J, Casanova C, Fischer B, Weiss A, Fofana I, Fontaine N, Fujiwara T, Renaud M, Kopp A, Schuster C, Brino L, Baumert TF, Thoma C. PI4K-beta and MKNK1 are regulators of hepatitis C virus IRES-dependent translation. Sci Rep. 2015;5:13344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 145. | Kusakabe S, Suzuki T, Sugiyama Y, Haga S, Horike K, Tokunaga M, Hirano J, Zhang H, Chen DV, Ishiga H, Komoda Y, Ono C, Fukuhara T, Yamamoto M, Ikawa M, Satoh T, Akira S, Tanaka T, Moriishi K, Fukai M, Taketomi A, Yoshio S, Kanto T, Okamoto T, Matsuura Y. USP15 Participates in Hepatitis C Virus Propagation through Regulation of Viral RNA Translation and Lipid Droplet Formation. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 146. | Park SM, Paek KY, Hong KY, Jang CJ, Cho S, Park JH, Kim JH, Jan E, Jang SK. Translation-competent 48S complex formation on HCV IRES requires the RNA-binding protein NSAP1. Nucleic Acids Res. 2011;39:7791-7802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 147. | Huys A, Thibault PA, Wilson JA. Modulation of hepatitis C virus RNA accumulation and translation by DDX6 and miR-122 are mediated by separate mechanisms. PLoS One. 2013;8:e67437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 148. | Khachatoorian R, Riahi R, Ganapathy E, Shao H, Wheatley NM, Sundberg C, Jung CL, Ruchala P, Dasgupta A, Arumugaswami V, Gestwicki JE, French SW. Allosteric heat shock protein 70 inhibitors block hepatitis C virus assembly. Int J Antimicrob Agents. 2016;47:289-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 149. | Kitabayashi J, Shirasaki T, Shimakami T, Nishiyama T, Welsch C, Funaki M, Murai K, Sumiyadorj A, Takatori H, Kitamura K, Kawaguchi K, Arai K, Yamashita T, Sakai Y, Mizukoshi E, Honda M, Kaneko S; Hokuriku Liver Study Group. Upregulation of the Long Non-Coding RNA HULC by Hepatitis C Virus and its Regulation of Viral Replication. J Infect Dis. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 150. | Shi Q, Hoffman B, Liu Q. PI3K-Akt signaling pathway upregulates hepatitis C virus RNA translation through the activation of SREBPs. Virology. 2016;490:99-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 151. | Vassilaki N, Mavromara P. The HCV ARFP/F/core+1 protein: production and functional analysis of an unconventional viral product. IUBMB Life. 2009;61:739-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 152. | Li HC, Ma HC, Yang CH, Lo SY. Production and pathogenicity of hepatitis C virus core gene products. World J Gastroenterol. 2014;20:7104-7122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 24] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 153. | Vassilaki N, Frakolaki E, Kalliampakou KI, Sakellariou P, Kotta-Loizou I, Bartenschlager R, Mavromara P. A Novel Cis-Acting RNA Structural Element Embedded in the Core Coding Region of the Hepatitis C Virus Genome Directs Internal Translation Initiation of the Overlapping Core+1 ORF. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 154. | Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487-5492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 470] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 155. | Moradpour D, Gosert R, Egger D, Penin F, Blum HE, Bienz K. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antiviral Res. 2003;60:103-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 113] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 156. | den Boon JA, Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 351] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 157. | Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol. 2003;77:4160-4168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 215] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 158. | Tabata K, Neufeldt CJ, Bartenschlager R. Hepatitis C Virus Replication. Cold Spring Harb Perspect Med. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 159. | Romero-Brey I, Lohmann V. The HCV Replicase Complex and Viral RNA Synthesis. Springer, 2016: 149-196. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 160. | Romero-López C, Barroso-Deljesus A, García-Sacristán A, Briones C, Berzal-Herranz A. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3'X-tail region. Nucleic Acids Res. 2014;42:567-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 161. | Kumar A, Ray U, Das S. Human La protein interaction with GCAC near the initiator AUG enhances hepatitis C Virus RNA replication by promoting linkage between 5' and 3' untranslated regions. J Virol. 2013;87:6713-6726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 162. | Li Y, Masaki T, Shimakami T, Lemon SM. hnRNP L and NF90 interact with hepatitis C virus 5'-terminal untranslated RNA and promote efficient replication. J Virol. 2014;88:7199-7209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 163. | Isken O, Baroth M, Grassmann CW, Weinlich S, Ostareck DH, Ostareck-Lederer A, Behrens SE. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13:1675-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 133] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 164. | Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3'-terminal structure of the hepatitis C virus genome, the 3'X. J Virol. 1997;71:6720-6726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 88] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 165. | Yu R, Yang D, Lei S, Wang X, Meng X, Xue B, Zhu H. HMGB1 Promotes Hepatitis C Virus Replication by Interaction with Stem-Loop 4 in the Viral 5' Untranslated Region. J Virol. 2015;90:2332-2344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 166. | Qin Y, Xun Z, Guo Y, Chen S, Zhu H. Sam68 Promotes Hepatitis C Virus Replication by Interaction with Stem-Loop 2 of Viral 5' Untranslated Region. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 167. | Wang Y, Lee S, Ha Y, Lam W, Chen SR, Dutschman GE, Gullen EA, Grill SP, Cheng Y, Fürstner A, Francis S, Baker DC, Yang X, Lee KH, Cheng YC. Tylophorine Analogs Allosterically Regulates Heat Shock Cognate Protein 70 And Inhibits Hepatitis C Virus Replication. Sci Rep. 2017;7:10037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 168. | Machlin ES, Sarnow P, Sagan SM. Masking the 5' terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci USA. 2011;108:3193-3198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 229] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 169. | Mortimer SA, Doudna JA. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013;41:4230-4240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 170. | Kincaid RP, Lam VL, Chirayil RP, Randall G, Sullivan CS. RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus. Proc Natl Acad Sci USA. 2018;115:8197-8202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 171. | Li Y, Masaki T, Yamane D, McGivern DR, Lemon SM. Competing and noncompeting activities of miR-122 and the 5' exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc Natl Acad Sci USA. 2013;110:1881-1886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 186] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 172. | Li Y, Yamane D, Lemon SM. Dissecting the roles of the 5' exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J Virol. 2015;89:4857-4865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 173. | Sedano CD, Sarnow P. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014;16:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 174. | Yan Y, Li C, Sun B, Yang R. DCAF1 is involved in HCV replication through regulation of miR-122. Arch Virol. 2018;163:977-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 175. | Saleh M, Rüschenbaum S, Welsch C, Zeuzem S, Moradpour D, Gouttenoire J, Lange CM. Glycogen Synthase Kinase 3β Enhances Hepatitis C Virus Replication by Supporting miR-122. Front Microbiol. 2018;9:2949. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 176. | Chen TC, Hsieh CH, Sarnow P. Supporting Role for GTPase Rab27a in Hepatitis C Virus RNA Replication through a Novel miR-122-Mediated Effect. PLoS Pathog. 2015;11:e1005116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 177. | Fan B, Sutandy FX, Syu GD, Middleton S, Yi G, Lu KY, Chen CS, Kao CC. Heterogeneous Ribonucleoprotein K (hnRNP K) Binds miR-122, a Mature Liver-Specific MicroRNA Required for Hepatitis C Virus Replication. Mol Cell Proteomics. 2015;14:2878-2886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 178. | Biegel JM, Henderson E, Cox EM, Bonenfant G, Netzband R, Kahn S, Eager R, Pager CT. Cellular DEAD-box RNA helicase DDX6 modulates interaction of miR-122 with the 5' untranslated region of hepatitis C virus RNA. Virology. 2017;507:231-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 179. | Ono C, Fukuhara T, Li S, Wang J, Sato A, Izumi T, Fauzyah Y, Yamamoto T, Morioka Y, Dokholyan NV, Standley DM, Matsuura Y. Various miRNAs compensate the role of miR-122 on HCV replication. PLoS Pathog. 2020;16:e1008308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 180. | Lohmann V, Overton H, Bartenschlager R. Selective stimulation of hepatitis C virus and pestivirus NS5B RNA polymerase activity by GTP. J Biol Chem. 1999;274:10807-10815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 181. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1154] [Cited by in RCA: 1150] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 182. | Kim SJ, Kim JH, Kim YG, Lim HS, Oh JW. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J Biol Chem. 2004;279:50031-50041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 183. | Inoue Y, Aizaki H, Hara H, Matsuda M, Ando T, Shimoji T, Murakami K, Masaki T, Shoji I, Homma S, Matsuura Y, Miyamura T, Wakita T, Suzuki T. Chaperonin TRiC/CCT participates in replication of hepatitis C virus genome via interaction with the viral NS5B protein. Virology. 2011;410:38-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 184. | Kitab B, Satoh M, Ohmori Y, Munakata T, Sudoh M, Kohara M, Tsukiyama-Kohara K. Ribonucleotide reductase M2 promotes RNA replication of hepatitis C virus by protecting NS5B protein from hPLIC1-dependent proteasomal degradation. J Biol Chem. 2019;294:5759-5773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 185. | Weng L, Hirata Y, Arai M, Kohara M, Wakita T, Watashi K, Shimotohno K, He Y, Zhong J, Toyoda T. Sphingomyelin activates hepatitis C virus RNA polymerase in a genotype-specific manner. J Virol. 2010;84:11761-11770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 186. | Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, Das S. HuR Displaces Polypyrimidine Tract Binding Protein To Facilitate La Binding to the 3' Untranslated Region and Enhances Hepatitis C Virus Replication. J Virol. 2015;89:11356-11371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 187. | Gupta G, Song J. C-Terminal Auto-Regulatory Motif of Hepatitis C Virus NS5B Interacts with Human VAPB-MSP to Form a Dynamic Replication Complex. PLoS One. 2016;11:e0147278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 188. | Zhu SL, Wang L, Cao ZY, Wang J, Jing MZ, Xia ZC, Ao F, Ye LB, Liu S, Zhu Y. Inducible CYP4F12 enhances Hepatitis C virus infection via association with viral nonstructural protein 5B. Biochem Biophys Res Commun. 2016;471:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 189. | Wang WT, Tsai TY, Chao CH, Lai BY, Wu Lee YH. Y-Box Binding Protein 1 Stabilizes Hepatitis C Virus NS5A via Phosphorylation-Mediated Interaction with NS5A To Regulate Viral Propagation. J Virol. 2015;89:11584-11602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 190. | Kasai H, Kawakami K, Yokoe H, Yoshimura K, Matsuda M, Yasumoto J, Maekawa S, Yamashita A, Tanaka T, Ikeda M, Kato N, Okamoto T, Matsuura Y, Sakamoto N, Enomoto N, Takeda S, Fujii H, Tsubuki M, Kusunoki M, Moriishi K. Involvement of FKBP6 in hepatitis C virus replication. Sci Rep. 2015;5:16699. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 191. | Liu Z, Yang F, Robotham JM, Tang H. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J Virol. 2009;83:6554-6565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 192. | Foster TL, Gallay P, Stonehouse NJ, Harris M. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J Virol. 2011;85:7460-7464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 193. | Mani N, Yuzhakov A, Yuzhakov O, Coll JT, Black J, Saxena K, Fulghum JR, Lippke JA, Rao BG, Rijnbrand R, Kwong AD. Nonstructural protein 5A (NS5A) and human replication protein A increase the processivity of hepatitis C virus NS5B polymerase activity in vitro. J Virol. 2015;89:165-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 194. | Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J Biol Chem. 2004;279:48576-48587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 271] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 195. | Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem. 2005;280:36417-36428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 204] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 196. | Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J Virol. 2008;82:1073-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 167] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 197. | Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, Bartholomew L, De Francesco R. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J Virol. 2004;78:13306-13314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 198. | Kim J, Lee D, Choe J. Hepatitis C virus NS5A protein is phosphorylated by casein kinase II. Biochem Biophys Res Commun. 1999;257:777-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 63] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 199. | Quintavalle M, Sambucini S, Summa V, Orsatti L, Talamo F, De Francesco R, Neddermann P. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J Biol Chem. 2007;282:5536-5544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 200. | Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 2013;9:e1003359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 201. | Yamauchi S, Takeuchi K, Chihara K, Sun X, Honjoh C, Yoshiki H, Hotta H, Sada K. Hepatitis C Virus Particle Assembly Involves Phosphorylation of NS5A by the c-Abl Tyrosine Kinase. J Biol Chem. 2015;290:21857-21864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 202. | Tellinghuisen TL, Foss KL, Treadaway J. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 2008;4:e1000032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 321] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 203. | Xiong W, Yang J, Wang M, Wang H, Rao Z, Zhong C, Xin X, Mo L, Yu S, Shen C, Zheng C. Vinexin β Interacts with Hepatitis C Virus NS5A, Modulating Its Hyperphosphorylation To Regulate Viral Propagation. J Virol. 2015;89:7385-7400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 204. | Evans MJ, Rice CM, Goff SP. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci USA. 2004;101:13038-13043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 255] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 205. | Wang C, Gale M Jr, Keller BC, Huang H, Brown MS, Goldstein JL, Ye J. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol Cell. 2005;18:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 233] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 206. | Li Z, Liu Q. Proprotein convertase subtilisin/kexin type 9 inhibits hepatitis C virus replication through interacting with NS5A. J Gen Virol. 2018;99:44-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 207. | Wang M, Wang Y, Liu Y, Wang H, Xin X, Li J, Hao Y, Han L, Yu F, Zheng C, Shen C. SPSB2 inhibits hepatitis C virus replication by targeting NS5A for ubiquitination and degradation. PLoS One. 2019;14:e0219989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 208. | Son K, Nguyen TTT, Choi JW, Pham LV, Luong TTD, Lim YS, Hwang SB. Rad51 Interacts with Non-structural 3 Protein of Hepatitis C Virus and Regulates Viral Production. Front Microbiol. 2017;8:1249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 209. | Lebsir N, Goueslain L, Farhat R, Callens N, Dubuisson J, Jackson CL, Rouillé Y. Functional and Physical Interaction between the Arf Activator GBF1 and Hepatitis C Virus NS3 Protein. J Virol. 2019;93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 210. | Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 402] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 211. | Egger D, Wölk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974-5984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 639] [Cited by in RCA: 633] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 212. | Madan V, Paul D, Lohmann V, Bartenschlager R. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology. 2014;146:1361-1372.e1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 213. | Lee JS, Tabata K, Twu WI, Rahman MS, Kim HS, Yu JB, Jee MH, Bartenschlager R, Jang SK. RACK1 mediates rewiring of intracellular networks induced by hepatitis C virus infection. PLoS Pathog. 2019;15:e1008021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 214. | Chao TC, Su WC, Huang JY, Chen YC, Jeng KS, Wang HD, Lai MM. Proline-serine-threonine phosphatase-interacting protein 2 (PSTPIP2), a host membrane-deforming protein, is critical for membranous web formation in hepatitis C virus replication. J Virol. 2012;86:1739-1749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 215. | Xu S, Pei R, Guo M, Han Q, Lai J, Wang Y, Wu C, Zhou Y, Lu M, Chen X. Cytosolic phospholipase A2 gamma is involved in hepatitis C virus replication and assembly. J Virol. 2012;86:13025-13037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 216. | Kong L, Aoyagi H, Yang Z, Ouyang T, Matsuda M, Fujimoto A, Watashi K, Suzuki R, Arita M, Yamagoe S, Dohmae N, Suzuki T, Muramatsu M, Wakita T, Aizaki H. Surfeit 4 Contributes to the Replication of Hepatitis C Virus Using Double-Membrane Vesicles. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 217. | Kong L, Fujimoto A, Nakamura M, Aoyagi H, Matsuda M, Watashi K, Suzuki R, Arita M, Yamagoe S, Dohmae N, Suzuki T, Sakamaki Y, Ichinose S, Wakita T, Aizaki H. Prolactin Regulatory Element Binding Protein Is Involved in Hepatitis C Virus Replication by Interaction with NS4B. J Virol. 2016;90:3093-3111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 218. | Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298-307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 354] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 219. | Zhang L, Hong Z, Lin W, Shao RX, Goto K, Hsu VW, Chung RT. ARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replication. PLoS One. 2012;7:e32135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 220. | Mingorance L, Castro V, Ávila-Pérez G, Calvo G, Rodriguez MJ, Carrascosa JL, Pérez-Del-Pulgar S, Forns X, Gastaminza P. Host phosphatidic acid phosphatase lipin1 is rate limiting for functional hepatitis C virus replicase complex formation. PLoS Pathog. 2018;14:e1007284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 221. | Gewaid H, Aoyagi H, Arita M, Watashi K, Suzuki R, Sakai S, Kumagai K, Yamaji T, Fukasawa M, Kato F, Hishiki T, Mimata A, Sakamaki Y, Ichinose S, Hanada K, Muramatsu M, Wakita T, Aizaki H. Sphingomyelin Is Essential for the Structure and Function of the Double-Membrane Vesicles in Hepatitis C Virus RNA Replication Factories. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 222. | Stone M, Jia S, Heo WD, Meyer T, Konan KV. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J Virol. 2007;81:4551-4563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 223. | Manna D, Aligo J, Xu C, Park WS, Koc H, Heo WD, Konan KV. Endocytic Rab proteins are required for hepatitis C virus replication complex formation. Virology. 2010;398:21-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 224. | Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282:36354-36361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 225. | Nevo-Yassaf I, Yaffe Y, Asher M, Ravid O, Eizenberg S, Henis YI, Nahmias Y, Hirschberg K, Sklan EH. Role for TBC1D20 and Rab1 in hepatitis C virus replication via interaction with lipid droplet-bound nonstructural protein 5A. J Virol. 2012;86:6491-6502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 226. | Salloum S, Wang H, Ferguson C, Parton RG, Tai AW. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 2013;9:e1003513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 227. | Ide Y, Zhang L, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene. 1996;182:203-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 79] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 228. | Isoyama T, Kuge S, Nomoto A. The core protein of hepatitis C virus is imported into the nucleus by transport receptor Kap123p but inhibits Kap121p-dependent nuclear import of yeast AP1-like transcription factor in yeast cells. J Biol Chem. 2002;277:39634-39641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 229. | Neufeldt CJ, Joyce MA, Levin A, Steenbergen RH, Pang D, Shields J, Tyrrell DL, Wozniak RW. Hepatitis C virus-induced cytoplasmic organelles use the nuclear transport machinery to establish an environment conducive to virus replication. PLoS Pathog. 2013;9:e1003744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 230. | Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046-14051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 388] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 231. | Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405:1-7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 232. | Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230-2237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 124] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 233. | Mohl BP, Bartlett C, Mankouri J, Harris M. Early events in the generation of autophagosomes are required for the formation of membrane structures involved in hepatitis C virus genome replication. J Gen Virol. 2016;97:680-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 234. | Paul D, Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol. 2013;2:32-48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 209] [Cited by in RCA: 222] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 235. | Chan ST, Ou JJ. Hepatitis C Virus-Induced Autophagy and Host Innate Immune Response. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 236. | Alvisi G, Madan V, Bartenschlager R. Hepatitis C virus and host cell lipids: an intimate connection. RNA Biol. 2011;8:258-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 237. | Kapadia SB, Chisari FV. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci USA. 2005;102:2561-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 406] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 238. | Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122-8130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 212] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 239. | Diamond DL, Syder AJ, Jacobs JM, Sorensen CM, Walters KA, Proll SC, McDermott JE, Gritsenko MA, Zhang Q, Zhao R, Metz TO, Camp DG 2nd, Waters KM, Smith RD, Rice CM, Katze MG. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6:e1000719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 331] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 240. | Yamane D, McGivern DR, Wauthier E, Yi M, Madden VJ, Welsch C, Antes I, Wen Y, Chugh PE, McGee CE, Widman DG, Misumi I, Bandyopadhyay S, Kim S, Shimakami T, Oikawa T, Whitmire JK, Heise MT, Dittmer DP, Kao CC, Pitson SM, Merrill AH Jr, Reid LM, Lemon SM. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat Med. 2014;20:927-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 241. | Hirata Y, Ikeda K, Sudoh M, Tokunaga Y, Suzuki A, Weng L, Ohta M, Tobita Y, Okano K, Ozeki K, Kawasaki K, Tsukuda T, Katsume A, Aoki Y, Umehara T, Sekiguchi S, Toyoda T, Shimotohno K, Soga T, Nishijima M, Taguchi R, Kohara M. Self-enhancement of hepatitis C virus replication by promotion of specific sphingolipid biosynthesis. PLoS Pathog. 2012;8:e1002860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 242. | Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol. 2014;88:12276-12295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 243. | Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J Virol. 2013;87:10612-10627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 209] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 244. | Amako Y, Sarkeshik A, Hotta H, Yates J 3rd, Siddiqui A. Role of oxysterol binding protein in hepatitis C virus infection. J Virol. 2009;83:9237-9246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 245. | Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 187] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 246. | Xiong Y, Jia M, Yuan J, Zhang C, Zhu Y, Kuang X, Lan L, Wang X. STAT3regulated long noncoding RNAs lnc7SK and lncIGF2AS promote hepatitis C virus replication. Mol Med Rep. 2015;12:6738-6744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 247. | Tallorin L, Villareal VA, Hsia CY, Rodgers MA, Burri DJ, Pfeil MP, Llopis PM, Lindenbach BD, Yang PL. Hepatitis C virus NS3-4A protease regulates the lipid environment for RNA replication by cleaving host enzyme 24-dehydrocholesterol reductase. J Biol Chem. 2020;295:12426-12436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 248. | Dixit U, Pandey AK, Liu Z, Kumar S, Neiditch MB, Klein KM, Pandey VN. FUSE Binding Protein 1 Facilitates Persistent Hepatitis C Virus Replication in Hepatoma Cells by Regulating Tumor Suppressor p53. J Virol. 2015;89:7905-7921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 249. | Stöhr S, Costa R, Sandmann L, Westhaus S, Pfaender S, Anggakusuma, Dazert E, Meuleman P, Vondran FW, Manns MP, Steinmann E, von Hahn T, Ciesek S. Host cell mTORC1 is required for HCV RNA replication. Gut. 2016;65:2017-2028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 250. | Igloi Z, Mohl BP, Lippiat JD, Harris M, Mankouri J. Requirement for chloride channel function during the hepatitis C virus life cycle. J Virol. 2015;89:4023-4029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 251. | Shimotohno K. HCV Assembly and Egress via Modifications in Host Lipid Metabolic Systems. Cold Spring Harb Perspect Med. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (1)] |
| 252. | Targett-Adams P, Hope G, Boulant S, McLauchlan J. Maturation of hepatitis C virus core protein by signal peptide peptidase is required for virus production. J Biol Chem. 2008;283:16850-16859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 253. | Hope RG, McLauchlan J. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J Gen Virol. 2000;81:1913-1925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 178] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 254. | Ivanyi-Nagy R, Kanevsky I, Gabus C, Lavergne JP, Ficheux D, Penin F, Fossé P, Darlix JL. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34:2618-2633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 255. | Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 937] [Cited by in RCA: 981] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 256. | Ohashi H, Nishioka K, Nakajima S, Kim S, Suzuki R, Aizaki H, Fukasawa M, Kamisuki S, Sugawara F, Ohtani N, Muramatsu M, Wakita T, Watashi K. The aryl hydrocarbon receptor-cytochrome P450 1A1 pathway controls lipid accumulation and enhances the permissiveness for hepatitis C virus assembly. J Biol Chem. 2018;293:19559-19571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 257. | Lim YS, Ngo HT, Lee J, Son K, Park EM, Hwang SB. ADP-ribosylation Factor-related Protein 1 Interacts with NS5A and Regulates Hepatitis C Virus Propagation. Sci Rep. 2016;6:31211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 258. | Schweitzer CJ, Zhang F, Boyer A, Valdez K, Cam M, Liang TJ. N-Myc Downstream-Regulated Gene 1 Restricts Hepatitis C Virus Propagation by Regulating Lipid Droplet Biogenesis and Viral Assembly. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 259. | Herker E, Harris C, Hernandez C, Carpentier A, Kaehlcke K, Rosenberg AR, Farese RV Jr, Ott M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat Med. 2010;16:1295-1298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 272] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 260. | Menzel N, Fischl W, Hueging K, Bankwitz D, Frentzen A, Haid S, Gentzsch J, Kaderali L, Bartenschlager R, Pietschmann T. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 2012;8:e1002829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 261. | Poenisch M, Metz P, Blankenburg H, Ruggieri A, Lee JY, Rupp D, Rebhan I, Diederich K, Kaderali L, Domingues FS, Albrecht M, Lohmann V, Erfle H, Bartenschlager R. Identification of HNRNPK as regulator of hepatitis C virus particle production. PLoS Pathog. 2015;11:e1004573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 262. | Shi G, Suzuki T. Molecular Basis of Encapsidation of Hepatitis C Virus Genome. Front Microbiol. 2018;9:396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 263. | Li Q, Tong Y, Xu Y, Niu J, Zhong J. Genetic Analysis of Serum-Derived Defective Hepatitis C Virus Genomes Revealed Novel Viral cis Elements for Virus Replication and Assembly. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 264. | Shi G, Ando T, Suzuki R, Matsuda M, Nakashima K, Ito M, Omatsu T, Oba M, Ochiai H, Kato T, Mizutani T, Sawasaki T, Wakita T, Suzuki T. Involvement of the 3' Untranslated Region in Encapsidation of the Hepatitis C Virus. PLoS Pathog. 2016;12:e1005441. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 265. | Stewart H, Bingham RJ, White SJ, Dykeman EC, Zothner C, Tuplin AK, Stockley PG, Twarock R, Harris M. Identification of novel RNA secondary structures within the hepatitis C virus genome reveals a cooperative involvement in genome packaging. Sci Rep. 2016;6:22952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 266. | Vieyres G, Welsch K, Gerold G, Gentzsch J, Kahl S, Vondran FW, Kaderali L, Pietschmann T. ABHD5/CGI-58, the Chanarin-Dorfman Syndrome Protein, Mobilises Lipid Stores for Hepatitis C Virus Production. PLoS Pathog. 2016;12:e1005568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 267. | Beilstein F, Lemasson M, Pène V, Rainteau D, Demignot S, Rosenberg AR. Lysophosphatidylcholine acyltransferase 1 is downregulated by hepatitis C virus: impact on production of lipo-viro-particles. Gut. 2017;66:2160-2169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 268. | Vieyres G, Reichert I, Carpentier A, Vondran FWR, Pietschmann T. The ATGL lipase cooperates with ABHD5 to mobilize lipids for hepatitis C virus assembly. PLoS Pathog. 2020;16:e1008554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 269. | Park EM, Lim YS, Ahn BY, Hwang SB. AAM-B Interacts with Nonstructural 4B and Regulates Hepatitis C Virus Propagation. PLoS One. 2015;10:e0132839. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 270. | Zhang H, Zhang C, Tang H, Gao S, Sun F, Yang Y, Zhou W, Hu Y, Ke C, Wu Y, Ding Z, Guo L, Pei R, Chen X, Sy MS, Zhang B, Li C. CD2-Associated Protein Contributes to Hepatitis C, Virus Propagation and Steatosis by Disrupting Insulin Signaling. Hepatology. 2018;68:1710-1725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 271. | Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J Virol. 2008;82:7964-7976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 295] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 272. | Nguyen LP, Tran SC, Suetsugu S, Lim YS, Hwang SB. PACSIN2 Interacts with Nonstructural Protein 5A and Regulates Hepatitis C Virus Assembly. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 273. | Nguyen LP, Nguyen TTT, Nguyen HC, Pham HT, Han KM, Choi DH, Park EM, Kang SM, Tark D, Lim YS, Hwang SB. Cortactin Interacts with Hepatitis C Virus Core and NS5A Proteins: Implications for Virion Assembly. J Virol. 2020;94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 274. | Gawlik K, Baugh J, Chatterji U, Lim PJ, Bobardt MD, Gallay PA. HCV core residues critical for infectivity are also involved in core-NS5A complex formation. PLoS One. 2014;9:e88866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 275. | Galli A, Scheel TKH, Prentoe JC, Mikkelsen LS, Gottwein JM, Bukh J. Analysis of hepatitis C virus core/NS5A protein co-localization using novel cell culture systems expressing core-NS2 and NS5A of genotypes 1-7. J Gen Virol. 2013;94:2221-2235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 276. | Ploen D, Hafirassou ML, Himmelsbach K, Sauter D, Biniossek ML, Weiss TS, Baumert TF, Schuster C, Hildt E. TIP47 plays a crucial role in the life cycle of hepatitis C virus. J Hepatol. 2013;58:1081-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 277. | Ploen D, Hafirassou ML, Himmelsbach K, Schille SA, Biniossek ML, Baumert TF, Schuster C, Hildt E. TIP47 is associated with the hepatitis C virus and its interaction with Rab9 is required for release of viral particles. Eur J Cell Biol. 2013;92:374-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 278. | Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J Virol. 2007;81:8374-8383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 346] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 279. | Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Héliot L, Rouillé Y, Dubuisson J. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 2011;7:e1001278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 280. | Ma Y, Yates J, Liang Y, Lemon SM, Yi M. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J Virol. 2008;82:7624-7639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 167] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 281. | Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 2010;6:e1001233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 282. | Kumar S, Barouch-Bentov R, Xiao F, Schor S, Pu S, Biquand E, Lu A, Lindenbach BD, Jacob Y, Demeret C, Einav S. MARCH8 Ubiquitinates the Hepatitis C Virus Nonstructural 2 Protein and Mediates Viral Envelopment. Cell Rep. 2019;26:1800-1814.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 283. | Suzuki R, Matsuda M, Watashi K, Aizaki H, Matsuura Y, Wakita T, Suzuki T. Signal peptidase complex subunit 1 participates in the assembly of hepatitis C virus through an interaction with E2 and NS2. PLoS Pathog. 2013;9:e1003589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 284. | Yang Q, Guo M, Zhou Y, Hu X, Wang Y, Wu C, Yang M, Pei R, Chen X, Chen J. Phosphatidylserine-Specific Phospholipase A1 is the Critical Bridge for Hepatitis C Virus Assembly. Virol Sin. 2019;34:521-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 285. | Guo M, Pei R, Yang Q, Cao H, Wang Y, Wu C, Chen J, Zhou Y, Hu X, Lu M, Chen X. Phosphatidylserine-specific phospholipase A1 involved in hepatitis C virus assembly through NS2 complex formation. J Virol. 2015;89:2367-2377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 286. | Boyer A, Dreneau J, Dumans A, Burlaud-Gaillard J, Bull-Maurer A, Roingeard P, Meunier JC. Endoplasmic Reticulum Detergent-Resistant Membranes Accommodate Hepatitis C Virus Proteins for Viral Assembly. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 287. | Neveu G, Barouch-Bentov R, Ziv-Av A, Gerber D, Jacob Y, Einav S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012;8:e1002845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 126] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 288. | Pham TM, Tran SC, Lim YS, Hwang SB. Hepatitis C Virus-Induced Rab32 Aggregation and Its Implications for Virion Assembly. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 289. | Barouch-Bentov R, Neveu G, Xiao F, Beer M, Bekerman E, Schor S, Campbell J, Boonyaratanakornkit J, Lindenbach B, Lu A, Jacob Y, Einav S. Hepatitis C Virus Proteins Interact with the Endosomal Sorting Complex Required for Transport (ESCRT) Machinery via Ubiquitination To Facilitate Viral Envelopment. mBio. 2016;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 290. | Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M Jr, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci USA. 2007;104:5848-5853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 429] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 291. | Cai H, Yao W, Li L, Li X, Hu L, Mai R, Peng T. Cell-death-inducing DFFA-like Effector B Contributes to the Assembly of Hepatitis C Virus (HCV) Particles and Interacts with HCV NS5A. Sci Rep. 2016;6:27778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 292. | Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J Virol. 2009;83:12680-12691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 293. | Perlemuter G, Sabile A, Letteron P, Vona G, Topilco A, Chrétien Y, Koike K, Pessayre D, Chapman J, Barba G, Bréchot C. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 2002;16:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 428] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 294. | Mancone C, Montaldo C, Santangelo L, Di Giacomo C, Costa V, Amicone L, Ippolito G, Pucillo LP, Alonzi T, Tripodi M. Ferritin heavy chain is the host factor responsible for HCV-induced inhibition of apoB-100 production and is required for efficient viral infection. J Proteome Res. 2012;11:2786-2797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 295. | Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012;8:e1002466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 140] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 296. | Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Ramière C, Bartenschlager R, Penin F, Lotteau V, André P. Secretion of hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS One. 2009;4:e4233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 297. | Fukuhara T, Wada M, Nakamura S, Ono C, Shiokawa M, Yamamoto S, Motomura T, Okamoto T, Okuzaki D, Yamamoto M, Saito I, Wakita T, Koike K, Matsuura Y. Amphipathic α-helices in apolipoproteins are crucial to the formation of infectious hepatitis C virus particles. PLoS Pathog. 2014;10:e1004534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 298. | Puig-Basagoiti F, Fukuhara T, Tamura T, Ono C, Uemura K, Kawachi Y, Yamamoto S, Mori H, Kurihara T, Okamoto T, Aizaki H, Matsuura Y. Human Cathelicidin Compensates for the Role of Apolipoproteins in Hepatitis C Virus Infectious Particle Formation. J Virol. 2016;90:8464-8477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 299. | Rösch K, Kwiatkowski M, Hofmann S, Schöbel A, Grüttner C, Wurlitzer M, Schlüter H, Herker E. Quantitative Lipid Droplet Proteome Analysis Identifies Annexin A3 as a Cofactor for HCV Particle Production. Cell Rep. 2016;16:3219-3231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 300. | Zhang X, Wang T, Dai X, Zhang Y, Jiang H, Zhang Q, Liu F, Wu K, Liu Y, Zhou H, Wu J. Golgi protein 73 facilitates the interaction of hepatitis C virus NS5A with apolipoprotein E to promote viral particle secretion. Biochem Biophys Res Commun. 2016;479:683-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 301. | Cai H, Yao W, Huang J, Xiao J, Chen W, Hu L, Mai R, Liang M, Chen D, Jiang N, Zhou L, Peng T. Apolipoprotein M, identified as a novel hepatitis C virus (HCV) particle associated protein, contributes to HCV assembly and interacts with E2 protein. Antiviral Res. 2020;177:104756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 302. | Syed GH, Khan M, Yang S, Siddiqui A. Hepatitis C Virus Lipoviroparticles Assemble in the Endoplasmic Reticulum (ER) and Bud off from the ER to the Golgi Compartment in COPII Vesicles. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 303. | Corless L, Crump CM, Griffin SD, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. 2010;91:362-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 304. | Mankouri J, Walter C, Stewart H, Bentham M, Park WS, Heo WD, Fukuda M, Griffin S, Harris M. Release of Infectious Hepatitis C Virus from Huh7 Cells Occurs via a trans-Golgi Network-to-Endosome Pathway Independent of Very-Low-Density Lipoprotein Secretion. J Virol. 2016;90:7159-7170. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 305. | Bayer K, Banning C, Bruss V, Wiltzer-Bach L, Schindler M. Hepatitis C Virus Is Released via a Noncanonical Secretory Route. J Virol. 2016;90:10558-10573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 306. | Boyer A, Park SB, de Boer YS, Li Q, Liang TJ. TM6SF2 Promotes Lipidation and Secretion of Hepatitis C Virus in Infected Hepatocytes. Gastroenterology 2018; 155: 1923-1935. e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 307. | Medvedev R, Ploen D, Spengler C, Elgner F, Ren H, Bunten S, Hildt E. HCV-induced oxidative stress by inhibition of Nrf2 triggers autophagy and favors release of viral particles. Free Radic Biol Med. 2017;110:300-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 308. | Bankwitz D, Doepke M, Hueging K, Weller R, Bruening J, Behrendt P, Lee JY, Vondran FWR, Manns MP, Bartenschlager R, Pietschmann T. Maturation of secreted HCV particles by incorporation of secreted ApoE protects from antibodies by enhancing infectivity. J Hepatol. 2017;67:480-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 309. | Li Z, Li Y, Bi Y, Zhang H, Yao Y, Li Q, Cun W, Dong S. Extracellular Interactions between Hepatitis C Virus and Secreted Apolipoprotein E. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 310. | Diaz O, Delers F, Maynard M, Demignot S, Zoulim F, Chambaz J, Trépo C, Lotteau V, André P. Preferential association of Hepatitis C virus with apolipoprotein B48-containing lipoproteins. J Gen Virol. 2006;87:2983-2991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 78] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 311. | Denolly S, Granier C, Fontaine N, Pozzetto B, Bourlet T, Guérin M, Cosset FL. A serum protein factor mediates maturation and apoB-association of HCV particles in the extracellular milieu. J Hepatol. 2019;70:626-638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 312. | Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. Role of annexin A2 in the production of infectious hepatitis C virus particles. J Virol. 2010;84:5775-5789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 313. | Tran GV, Luong TT, Park EM, Kim JW, Choi JW, Park C, Lim YS, Hwang SB. Nonstructural 5A Protein of Hepatitis C Virus Regulates Soluble Resistance-Related Calcium-Binding Protein Activity for Viral Propagation. J Virol. 2015;90:2794-2805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 314. | Herzog K, Bandiera S, Pernot S, Fauvelle C, Jühling F, Weiss A, Bull A, Durand SC, Chane-Woon-Ming B, Pfeffer S, Mercey M, Lerat H, Meunier JC, Raffelsberger W, Brino L, Baumert TF, Zeisel MB. Functional microRNA screen uncovers O-linked N-acetylglucosamine transferase as a host factor modulating hepatitis C virus morphogenesis and infectivity. Gut. 2020;69:380-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 315. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1843] [Cited by in RCA: 1850] [Article Influence: 92.5] [Reference Citation Analysis (0)] |
| 316. | Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol. 2018;16:125-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 291] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 317. | McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 371] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 318. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583-10587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 283] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 319. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832-2843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 420] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 320. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 655] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 321. | Lee JY, Cortese M, Haselmann U, Tabata K, Romero-Brey I, Funaya C, Schieber NL, Qiang Y, Bartenschlager M, Kallis S, Ritter C, Rohr K, Schwab Y, Ruggieri A, Bartenschlager R. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell Rep. 2019;27:3602-3617.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |