Published online Jun 7, 2021. doi: 10.3748/wjg.v27.i21.2850
Peer-review started: February 6, 2021
First decision: March 14, 2021
Revised: March 30, 2021
Accepted: May 10, 2021
Article in press: May 10, 2021
Published online: June 7, 2021
Processing time: 109 Days and 12.1 Hours
The coronavirus disease 2019 (COVID-19), a pandemic contributing to more than 105 million cases and more than 2.3 million deaths worldwide, was described to be frequently accompanied by extrapulmonary manifestations, including liver dysfunction. Liver dysfunction and elevated liver enzymes were observed in about 53% of COVID-19 patients.
To gain insight into transcriptional abnormalities in liver tissue of severe COVID-19 patients that may result in liver dysfunction.
The transcriptome of liver autopsy samples from severe COVID-19 patients against those of non-COVID donors was analyzed. Differentially expressed genes were identified from normalized RNA-seq data and analyzed for the enrichment of functional clusters and pathways. The differentially expressed genes were then compared against the genetic signatures of liver diseases including cirrhosis, fibrosis, non-alcoholic fatty liver disease (NAFLD), and hepatitis A/B/C. Gene expression of some differentially expressed genes was assessed in the blood samples of severe COVID-19 patients with liver dysfunction using qRT-PCR.
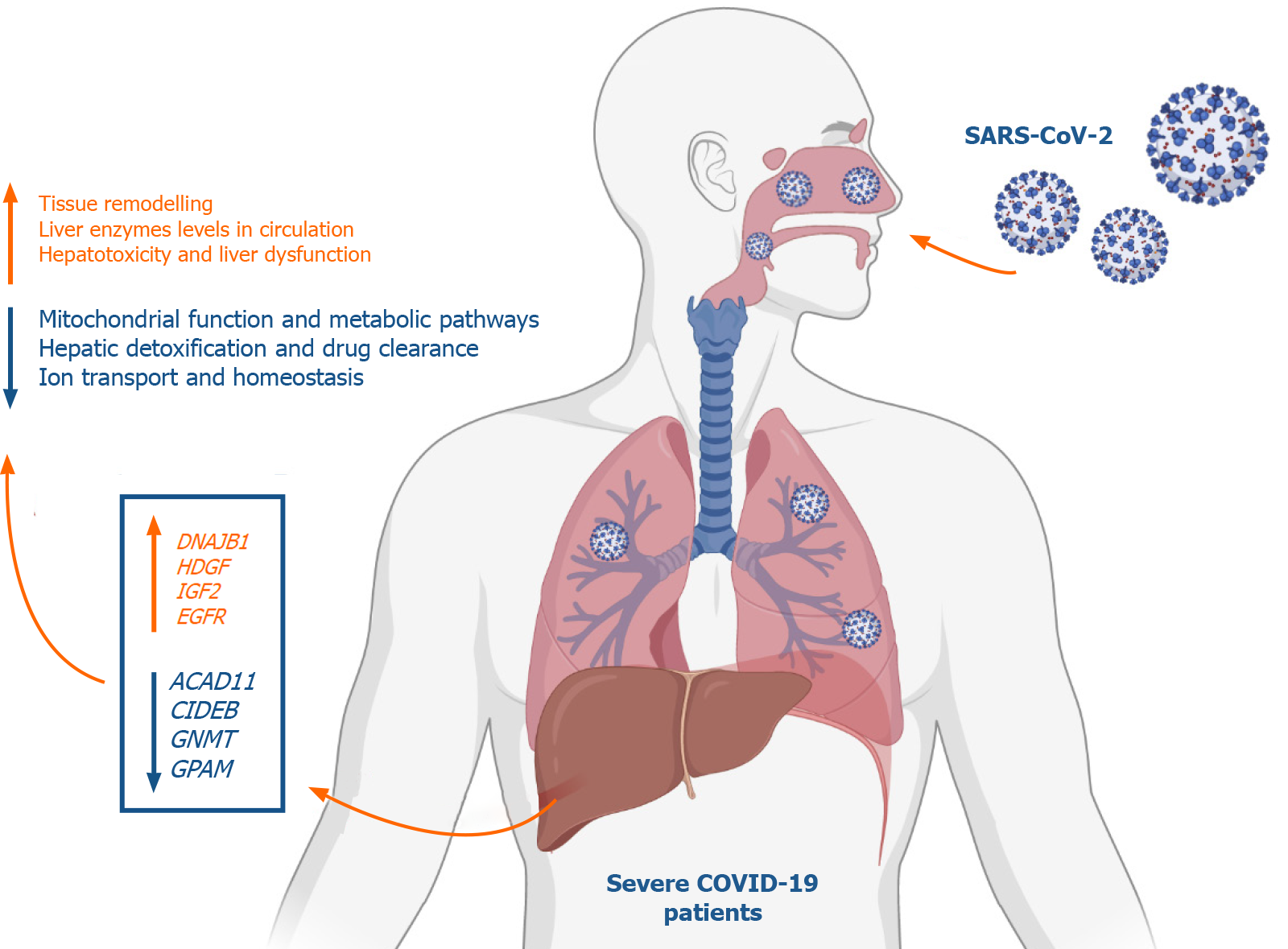
Analysis of the differential transcriptome of the liver tissue of severe COVID-19 patients revealed a significant upregulation of transcripts implicated in tissue remodeling including G-coupled protein receptors family genes, DNAJB1, IGF2, EGFR, and HDGF. Concordantly, the differential transcriptome of severe COVID-19 liver tissues substantially overlapped with the disease signature of liver diseases characterized with pathological tissue remodeling (liver cirrhosis, Fibrosis, NAFLD, and hepatitis A/B/C). Moreover, we observed a significant suppression of transcripts implicated in metabolic pathways as well as mitoch
Severe COVID-19 patients appear to experience significant transcriptional shift that may ensue tissue remodeling, mitochondrial dysfunction and lower hepatic detoxification resulting in the clinically observed liver dysfunction.
Core Tip: Liver dysfunction was frequently observed in severe coronavirus disease 2019 (COVID-19) patients. However, the mechanism through which severe acute respiratory syndrome coronavirus 2 potentially elicits liver function abnormality is not fully understood. We report a thorough analysis of changes occurring at the gene expression level in liver tissue of severe COVID-19 patients. Our findings suggest that severe COVID-19 patients may have a lower hepatic detoxification capacity and may experience liver tissue remodeling resulting in liver dysfunction.
- Citation: Hammoudeh SM, Hammoudeh AM, Bhamidimarri PM, Mahboub B, Halwani R, Hamid Q, Rahmani M, Hamoudi R. Insight into molecular mechanisms underlying hepatic dysfunction in severe COVID-19 patients using systems biology. World J Gastroenterol 2021; 27(21): 2850-2870
- URL: https://www.wjgnet.com/1007-9327/full/v27/i21/2850.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i21.2850
The coronavirus disease 2019 (COVID-19) pandemic resulted in 105 million corona
The incidence of liver dysfunction in COVID-19 patients was found to correlate with the severity of the disease, with a higher prevalence in severe COVID-19 cases than non-severe COVID-19 cases and requiring the administration of liver protective drugs[6]. Moreover, therapeutic agents administered to COVID-19 patients were shown to independently correlate with increased elevation of aminotransferases levels indicating dysregulated hepatic drug metabolism and consequent hepatotoxicity[7]. Therefore, understanding the mechanism through which SARS-CoV-2 infection results in hepatic dysfunction can contribute to patient management and prognosis.
The observed hepatic dysfunction was speculated to be either a result of liver injury caused by direct effect of the virus, organ injury induced by drugs used to treat COVID-19 patients, cytokine storm consequence, or sequelae of multi organ damage caused by severe disease[8]. However, the mechanism through which SARS-CoV-2 potentially elicits liver function abnormality is yet to be clearly understood. Therefore, the main aim of this study is to gain some insight into the molecular mechanisms employed by SARS-CoV-2 infection to induce liver dysfunction. In this study, we analyzed the transcriptome of liver autopsy samples from severe COVID-19 patients to identify significantly affected cellular pathways and processes.
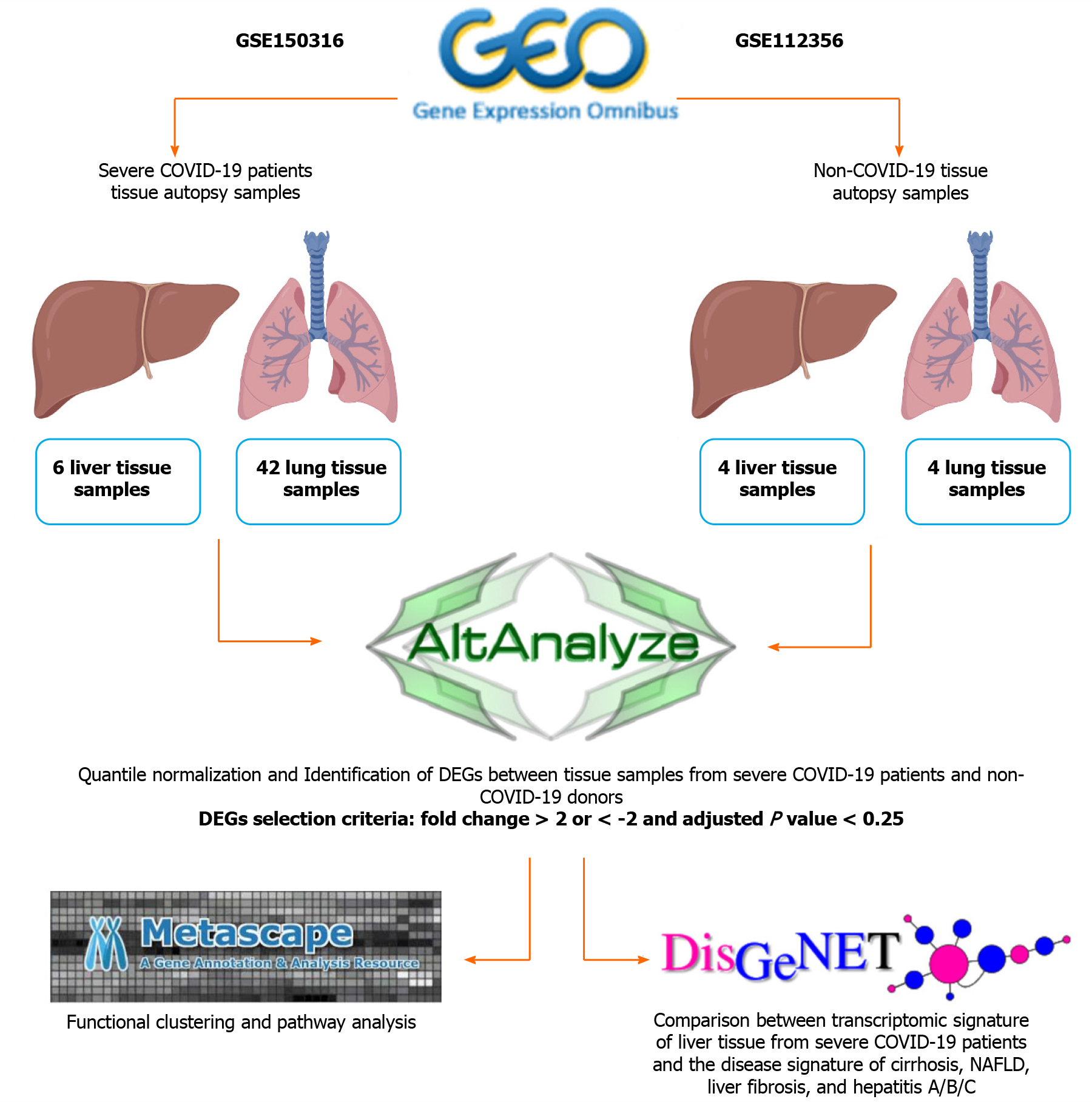
Datasets were retrieved from Gene Expression Omnibus for lung and liver autopsy samples from 12 patients severely infected with SARS-CoV-2 (GSE150316) (42 lung samples and 6 liver samples; Table 1), and lung and liver autopsy samples from 4 non-infected Caucasian male donors (GSE112356) (4 lung samples and 4 liver samples; Table 1). The COVID-19 autopsy samples were reported by the authors depositing the dataset[9] to be from donors with severe COVID-19 symptoms. The patients were hospitalized, required mechanical ventilation (9 out of 12 cases), and presented with elevated levels of D-Dimer (Cases 1-10 and 12), CRP (Cases 1-10 and 12), and AST (Cases 2-10 and 12).
| Tissue source | Non-COVID-19 autopsy samples | COVID-19 autopsy samples | Total number of analyzed samples |
| Lung | H1Lu; H2Lu; H3Lu; H4Lu | Case1: Samples 1, 2, 3, 4; Case2: Samples 1, 2, 3; Case3: Samples 1, 2; Case4: Samples 1, 2; Case5: Samples 1, 2, 3, 4, 5; Case6: Samples 1, 2, 3, 4, 5; Case7: Samples 1, 2, 3, 4, 5; Case8: Samples 1, 2, 3, 4, 5; Case9: Samples 1, 2, 3, 4, 5; Case10: Samples 1, 2, 3; Case11: Samples 1, 2, 3 | 4 healthy samples and 42 COVID-19 samples |
| Liver | H1Li; H2Li; H3Li; H4Li | Case3: Sample 1; Case4: Sample 1; Case5: Sample 1; Case8: Sample 1; Case10: Sample 1; Case12: Sample1 | 4 healthy samples and 6 COVID-19 samples |
The general workflow is summarized in Figure 1. Initially, raw gene counts data were retrieved from the deposited datasets and the successfully mapped genes overlapping between the samples from both datasets (28131 genes) were filtered and normalized using the quantile normalization function in AltAnalyze software[10]. Differentially expressed genes (DEGs) were identified by comparing each SARS-CoV-2 infected tissue against a healthy control tissue according to cutoff values of > 2 or < -2 for the fold change and 0.25 for the adjusted P value (q-value), based on the P value cut-off described by Li et al[11] and Subramanian et al[12]. Commonly upregulated and downregulated DEGs between infected lung and liver tissues were intersected and identified using InteractiVenn[13].
Functional clustering and pathways enrichment analysis of DEGs was performed using Metascape[14]. We further validated the significance of the differential expression of some DEGs by conducting two-tailed Student’s t test statistical analysis using GraphPad Prism.
The transcriptomic signature that map to liver cirrhosis (C0023890, C0023893, C0156189, C0400943), liver fibrosis (C0239946), non-alcoholic fatty liver disease (NAFLD) (C0400966), hepatitis A (C0019159 and C0276434), hepatitis B (C0019163, C0524909, C4728019, and C0276609), and hepatitis C (C0019196, C0524910, C3872662, and C0400914) were retrieved from DisGeNET[15]. The signatures were then compared against the upregulated transcriptome of SARS-CoV-2 infected liver tissue samples to identify overlapping transcripts.
Blood Serum was isolated from fresh blood samples collected from 4 healthy donors, 4 severe COVID-19 patients without liver dysfunction, and 9 severe COVID-19 patients with liver dysfunction (elevated alanine aminotransferase levels: > 41 U/L). The samples were collected following the approval of the ethical committee by Dubai Scientific Research Ethics Committee (DSREC-04/2020_09). Histopaque gradient separation (Sigma) was used to extract blood plasma and the RNA content was extracted from 300 µl of plasma using QIAamp Viral RNA Mini Kit (Qiagen). cDNA synthesis was carried out using the High-Capacity cDNA Reverse Transcription Kit for RT-PCR (applied Biosystems) and qRT-PCR was performed using about 50 ng of cDNA in triplicates with the Maxima SYBR Green/ROX qPCR Master Mix (Thermoscientific) using QuantStudio3 real-time PCR instrument (applied biosystems). qRT-PCR was performed using primers for 18SrRNA, DNAJB1, HSP90A1 and CYP39A1 as per the sequences in Table 2.
| Gene symbol | Forward primer sequence | Reverse primer sequence | Amplicon size (bp) |
| 18SrRNA | TGACTCAACACGGGAAACC | TCGCTCCACCAACTAAGAAC | 114 |
| DNAJB1 | GTTTTAAAGGACAAGCCCCACA | TCCAGAGTGGGGACGTTCA | 119 |
| HSP90AB1 | CTCTGTCAGAGTATGTTTCTCGC | GTTTCCGCACTCGCTCCACAAA | 114 |
| CYP39A1 | GTTCCAGTGTCCTGCAAGGT | TGGGTAATGGGTCCAGAAGAC | 101 |
Unpaired, two-tailed t test statistical analysis was performed using GraphPad Prism (version 5.01) to analyze the statistical significance of the gene expression. The significance was taken to be P < 0.05. Heatmap representations were generated using R (version 3.6.0); boxplot representations were generated using GraphPad Prism (version 5.01).
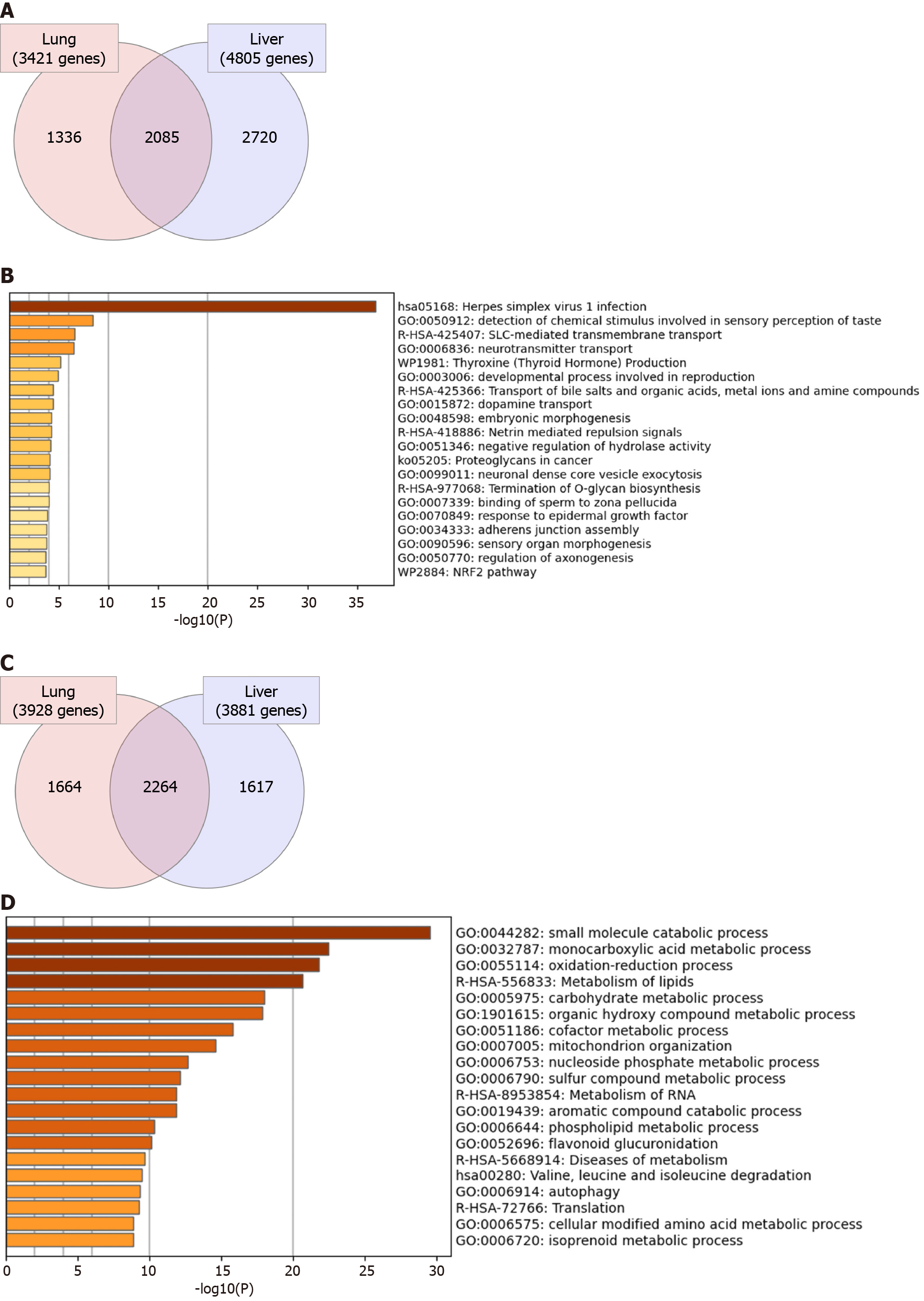
Initially, we aimed at investigating the general transcriptomic changes in the liver tissues of severe COVID-19 patients in comparison to those of lung tissues. Our analysis of the differential transcriptome of the two tissue types revealed the dysregulation of a larger number of genes in liver tissue (8686 genes) in comparison to lung tissue (7349 genes) (Figure 2). 2085 genes were commonly differentially expressed in the two types of tissues (Figure 2A). Functional clustering and pathway analysis of the commonly upregulated transcripts revealed their contribution to transmembrane transport and vesicle formation, cell adhesions, tissue morphogenesis, development, and intracellular signaling pathways [e.g. G-coupled protein receptor (GPCR) and Hippo signaling] (Figure 2B). On the other hand, the commonly downregulated genes (2264 genes) were significantly implicated in cellular metabolic pathways and mitochondrial function (Figure 2C and D). The enrichment of these pathways suggests that SARS-CoV-2 elicits similar molecular effects in lung and liver tissue that may translate into similar phenotypic presentations (e.g. tissue remodeling and dysfun
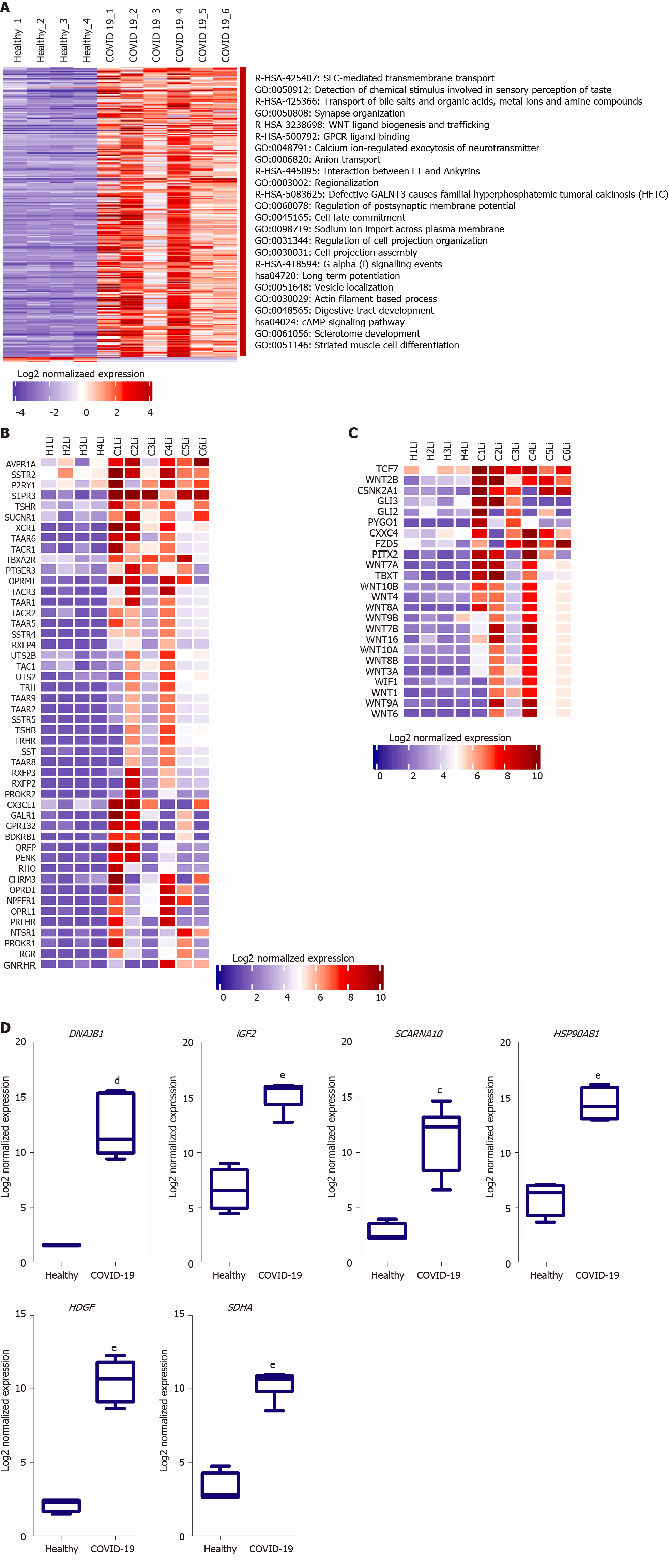
We next aimed at analyzing the effect of SARS-CoV-2 infection on the dysregulation of liver function at advanced stages in more detail. Functional clustering and pathway analysis of the upregulated genes in the liver tissues of severe COVID-19 patients revealed the enrichment of transcripts implicated in DNA and RNA transcription mechanisms, GPCRs signaling, and transmembrane transport (Figure 3A).
Multiple GPCR classes are dysregulated in liver tissue collected from severe COVID-19 patient autopsies including class A-rhodopsin-like receptors (R-HSA-373076: class A/1 rhodopsin-like receptors) and class F-frizzled and smoothened receptors (R-HSA-3238698: WNT ligand biogenesis and trafficking; hsa04310: Wnt signaling pathway) (Figure 3B and C). Consequently to the dysregulation of these GPCRs, numerous downstream signaling pathways, including WNT signaling pathway, sonic hedgehog signaling and hippo signaling pathways, are dysregulated affecting thereby cell proliferation, cell survival, host response to infection, immune response, chemotactic pathways, and tissue and local microenvironment remodeling [16-19]. Moreover, Wnt signaling pathway through frizzled binding has been shown to be hijacked by viral infections (e.g., influenza virus) to increase viral production[20]. GPCRs were found as well to be exploited by viruses (e.g., Filoviruses) to facilitate viral cellular entry, they can be explored as potential facilitators of SARS-CoV-2 entry into hepatocytes in the absence of ACE2 and TMPRSS2[21]. Therefore, the significant dysregulation of GPCRs and Wnt signaling, coupled with the significant upregulation of transmembrane and intracellular transport, in liver tissue of severe COVID-19 patients may indicate that SARS-CoV-2 may hijack these mechanisms to facilitate and enhance viral infectivity and production.
One of the top 10 upregulated genes in the liver tissue of severe COVID-19 patients was the DnaJ heat shock protein family (Hsp40) member B1, DNAJB1 (Figure 3D), which was found to contribute to liver inflammation, activation of uncontrolled fibrosis, and development of fibrolamellar carcinoma as a part of the DNAJB1-PRKACA fusion[22]. Another top upregulated gene was insulin like growth factor 2 (IGF2), a pro-steatosis and tissue remodeling factor[23]. Hepatoma-derived growth factor, HDGF, a pro-fibrogenic protein implicated in liver fibrosis through TGF-beta pathway, was as well amongst the top 10 upregulated genes[24]. Another significantly upregulated gene was the epidermal growth factor receptor (EGFR), a transmembrane receptor implicated in the regulation of various downstream intracellular signaling pathways including tissue remodeling pathways[25,26]. The upregulation of these genes suggests that SARS-CoV-2 infection may elicit tissue remodeling and fibrosis-like changes in liver tissue at advanced stages.
Based on the enrichment of transcripts implicated in pathways central to the activation and regulation of tissue remodeling, we next compared the differential transcriptome of the liver tissues from COVID-19 patients with the disease signature of liver cirrhosis, liver fibrosis, and NAFLD, which are commonly associated with pathological tissue remodeling.
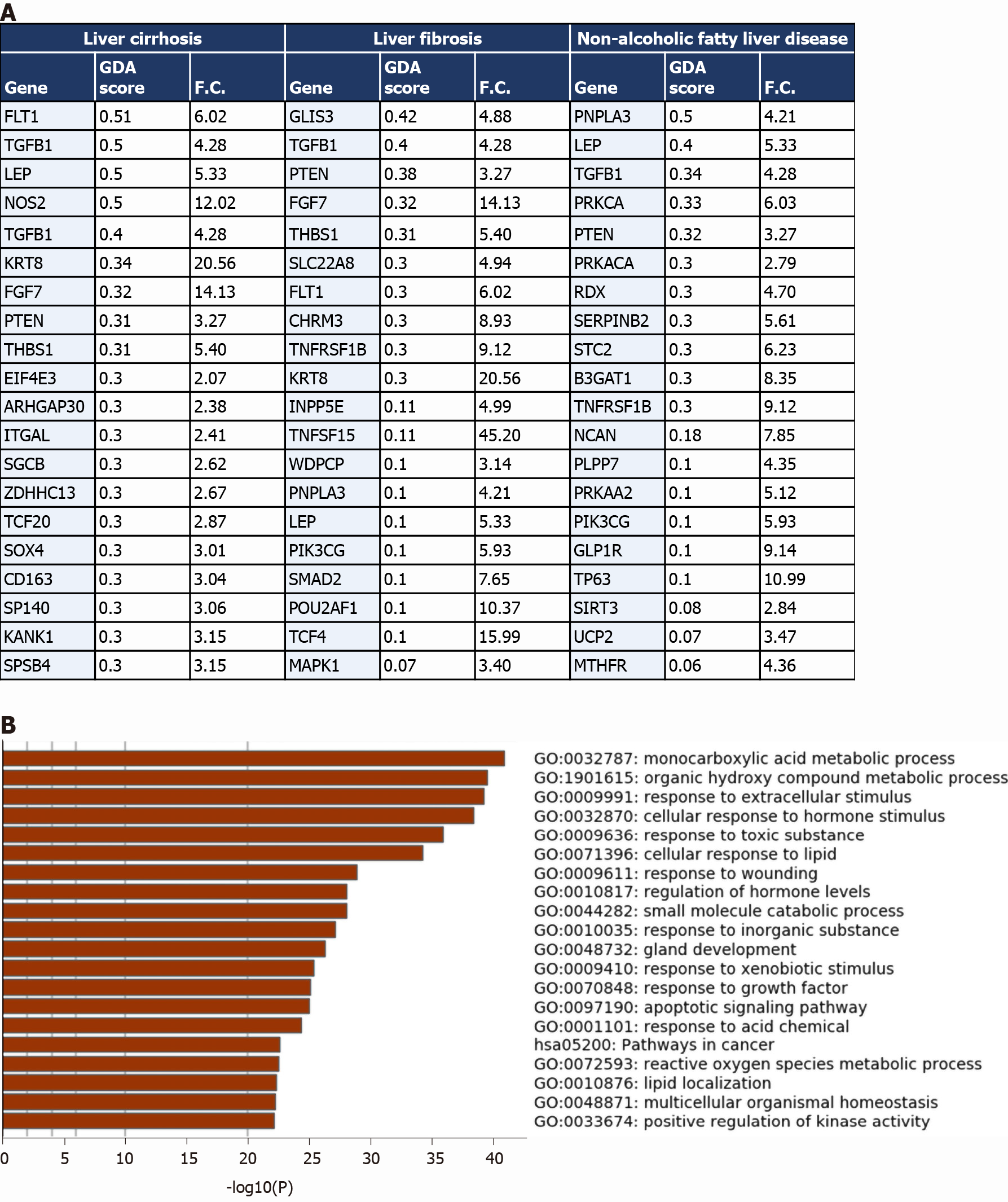
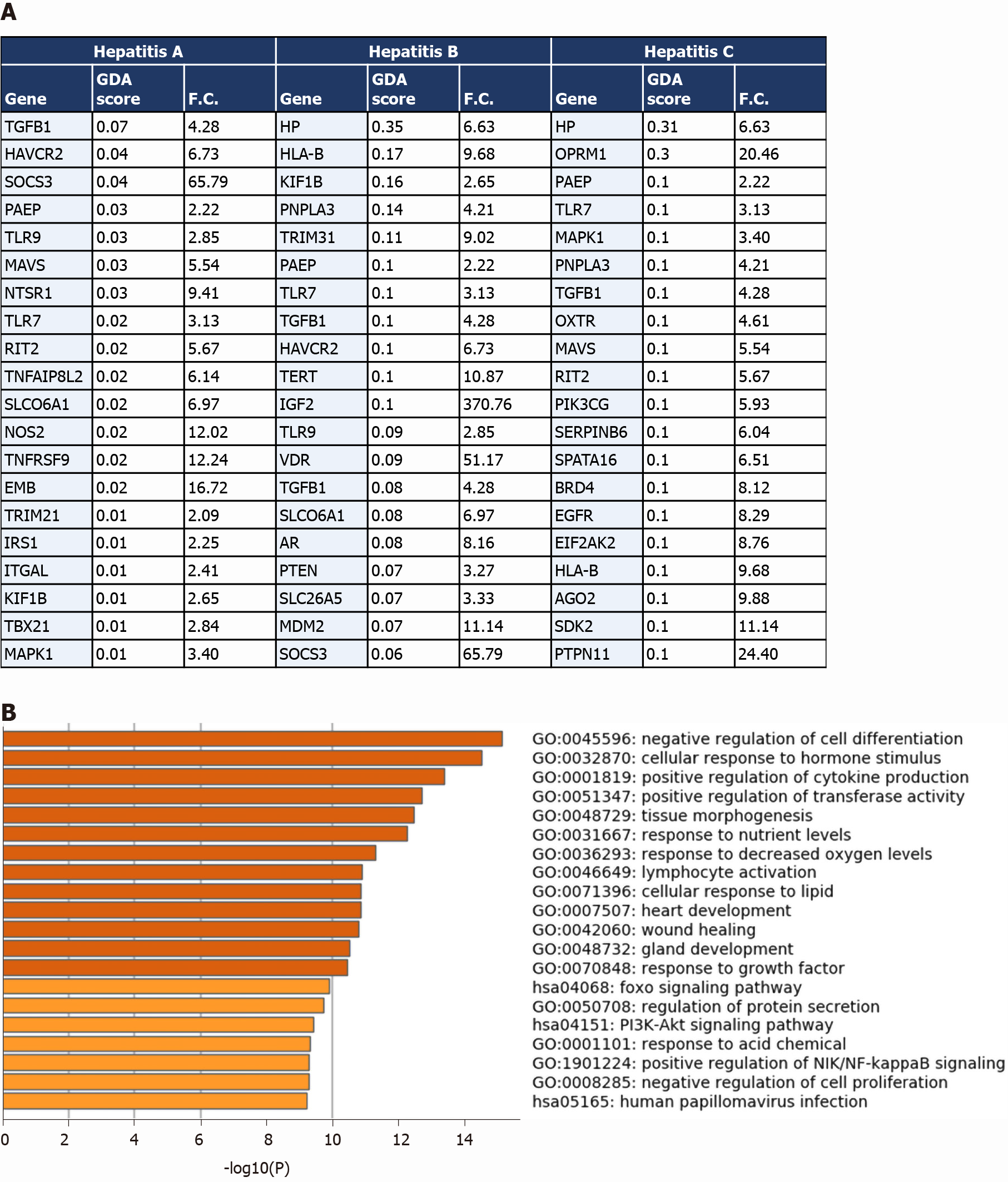
The comparison revealed that 1027 genes (318 upregulated and 709 downregulated) from the upregulated transcriptome of COVID-19 patients’ liver samples overlap with liver cirrhosis disease signature; 630 genes (206 upregulated and 424 downregulated) overlap with the signature of liver fibrosis; and 583 genes (190 upregulated and 393 downregulated) overlap with the signature of NAFLD (top 20 genes overlapping with every disease signature are presented in Figure 4A).
The transcriptome shared with the liver cirrhosis, liver fibrosis and NAFLD signature was significantly enriched in transcripts implicated in response to wounding and wound healing, cellular response to external stimulus (e.g., peptides, inorganic substance, growth factors, and hormone stimulus), response to pathogens from bacterial and viral origins (e.g., human papillomavirus infection, response to lipopolysaccharides), metabolic pathways, hormone and insulin mediated signaling pathways, cytokine-mediated signaling, MAPK signaling cascade, regulation of cell adhesions, regulation of cell differentiation, tissue/organ development and morphogenesis, blood vessels morphogenesis and development, regulation of programmed cell death, cell chemotaxis (e.g., leukocytes, endothelial cells, and epithelial cells), and immune response activation (Figure 4B).
One of the identified overlapping genes with the signature of liver cirrhosis and liver fibrosis was the vascular endothelial growth factor receptor 1 (FLT1), shown to be significantly upregulated during the pathogenesis of liver cirrhosis in which it centrally contributes to the tissue remodeling and increased vascularization of liver tissue[27]. Another overlapping gene with the signatures of the three diseases was leptin (LEP), a central mediator of hepatic fibrosis and tissue remodeling in response to chronic liver injury[28]. PNPLA3, overexpressed in the liver tissue of COVID-19 patients, has been previously shown to be implicated in the pathogenesis of non-alcoholic fatty liver disease, hepatic steatosis, liver cirrhosis and fibrosis[29].
Moreover, we observed the upregulation of a number of metalloproteinases (e.g., MMP3, MMP16, MMP17, TIMP1, TIMP2, and TIMP4), collagens (COL6A3, COL18A1-AS1, COL20A1, COL24A1, COLEC12, COL13A1, COL22A1, COLGALT2), and VCAM1 which were proposed to contribute to pathological tissue remodeling in liver diseases (e.g. non-alcoholic fatty liver disease and liver fibrosis)[30]. Furthermore, we observed a significant upregulation in the expression of TREM2 which was shown to be a protective mechanism activated in response to liver damage (e.g., immune-mediated hepatocellular damage and hepatotoxic injuries) (acts as a natural brake on inflammation during hepatocellular injury)[31]; which supports the potential occurrence of liver inflammation and injury in response to SARS-CoV-2 infection. Altogether, these results suggest the potential activation of tissue remodeling pathways (e.g., fibrosis, increased vascularization, and steatosis) in the liver tissue of severe COVID-19 patients at advanced stages in response to liver injury and inflammation that may result in cirrhosis/fibrosis-like phenotype.
We then compared the differential transcriptome of liver samples from severe COVID-19 patients with the disease signature of hepatitis A, hepatitis B and hepatitis C infection. We observed the overlap of 84 genes from the upregulated transcriptome of COVID-19 liver samples overlap with hepatitis A infection signature; 278 genes overlap with the signature of hepatitis B infection; and 336 genes overlap with the signature of hepatitis C infection (top overlapping genes are presented in Figure 5A). The overlapping signatures between the COVID-19 samples and the three types of hepatitis infections were enriched in transcripts contributing to response to infection, activation of immune response mechanisms, cytokines-mediated signaling, tissue remodeling and homeostasis (Figure 5B).
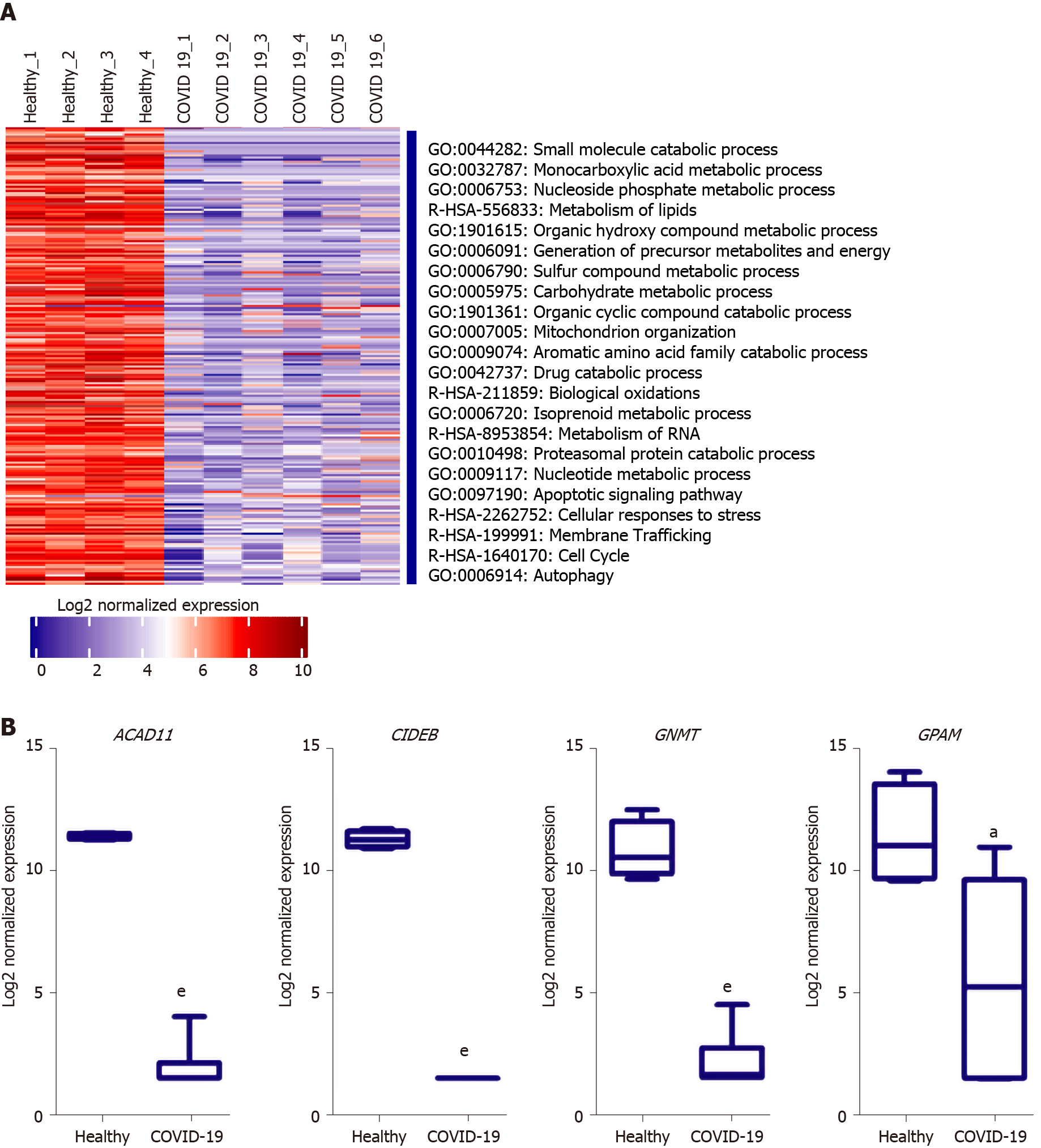
Analysis of the downregulated genes in the liver tissues of severe COVID-19 patients revealed a significant suppression of various metabolic and biosynthetic pathways including the metabolism of amines, aromatic compounds, amino acids, steroids, glycoproteins, lipids, and flavones (Figure 6A). The suppression of these metabolic pathways is concordant with the dysregulation of mitochondrial function reflected by the suppression of oxidation-reduction process, electron transport and respiratory chain, and the assembly of mitochondrial structures. These findings are consistent with previously published data on the accumulation of amino acids and steroids in the sera of COVID-19 patients in consequence to the suppression of lipid and amino acid metabolism[32]. These data show that metabolic pathways are potentially hijacked in severe COVID-19 patients’ liver tissue through the dysregulating of mitochondrial function. Analysis of key transcriptional regulators of the dysregulated gene implicated in mitochondrial function using TRRUST database suggested SP1, GATA6, SP3, HNF4A, and SMAD3 as candidates. SP1 has been previously implicated in pathogenesis of diseases characterized by mitochondrial dysfunction such as schizophrenia[33].
One of the top downregulated genes was acyl-CoA dehydrogenases 11 (ACAD11) (Figure 6B), a mitochondrial flavoenzyme involved in mitochondrial β-oxidation and metabolism of long chain fatty acyl-CoAs[34]. Another top downregulated gene was the cell death-inducing DNA fragmentation factor alpha-like effector B (CIDEB), a liver-specific regulator of lipid droplet dynamics and lipids metabolism[35]. Glycine N-methyltransferase (GNMT), another top downregulated gene in SARS-CoV-2 infected liver samples, was found to contribute to the development of liver steatosis and fibrosis when suppressed due to the metabolic dysregulation and accumulation of S-adenosylmethionine[36]. Glycerol-3-phosphate acyltransferase and metabolism (GPAM), another of the top 10 downregulated genes, is implicated triglyceride biosynthesis[37]. Moreover, 17β-hydroxysteroid dehydrogenase type 4 (HSD17B4), a regulator of fatty acid β-oxidation and steroid metabolism[38], was downregulated. Altogether, the downregulation of these genes suggests metabolic and biosynthetic pathways are suppressed in the liver tissue of severe COVID-19 patients.
Moreover, our analysis showed a significant enrichment of downregulated transcripts implicated in iron, copper, sodium, potassium, zinc, and calcium binding, transport, homeostasis and signaling. Imbalance of some of these ions (e.g. calcium and potassium ions) has been shown to induce mitochondrial swelling through shifting osmotic pressure[39].
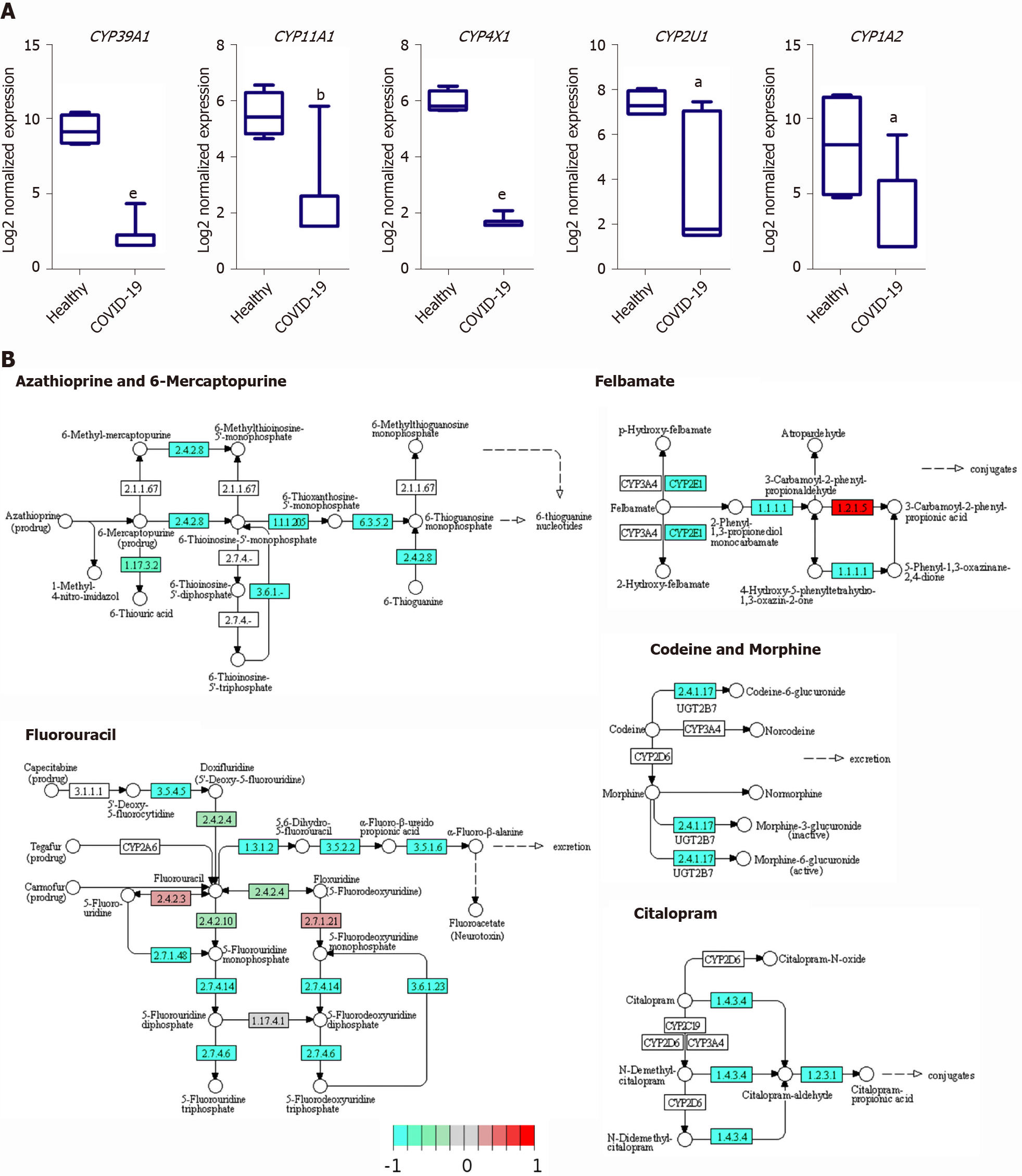
The downregulated transcriptome of COVID-19 patients’ liver tissue samples was enriched in members of the cytochromes P450 family enzymes (CYPs), including CYP39A1, CYP11A1, CYP4X1, CYP2U1, and CYP1A2, which play central roles in various metabolic pathways including the metabolism of drugs and xenobiotics (Figure 7A). Moreover, we observed the downregulation of the CYPs, CYP2C8 (fold change -4.18) and CYP3A4/5 (fold change -2.44), which have been shown to be centrally implicated in the metabolism of chloroquine and hydroxychloroquine; therapeutic agents administered to COVID-19 patients[40,41]. Functional clustering analysis revealed the enrichment of transcripts implicated in xenobiotic and drug metabolism processes (GO:0006805 and GO:0017144) in the downregulated transcriptome of SARS-CoV-2 infected liver samples (adjusted P values 0.01215 and 0.06111, respectively for xenobiotic and drug metabolism processes). KEGG pathway enrichment analysis confirmed the suppression of the metabolism of various types of drugs through CYPs as well as other enzymes (Figure 7B).
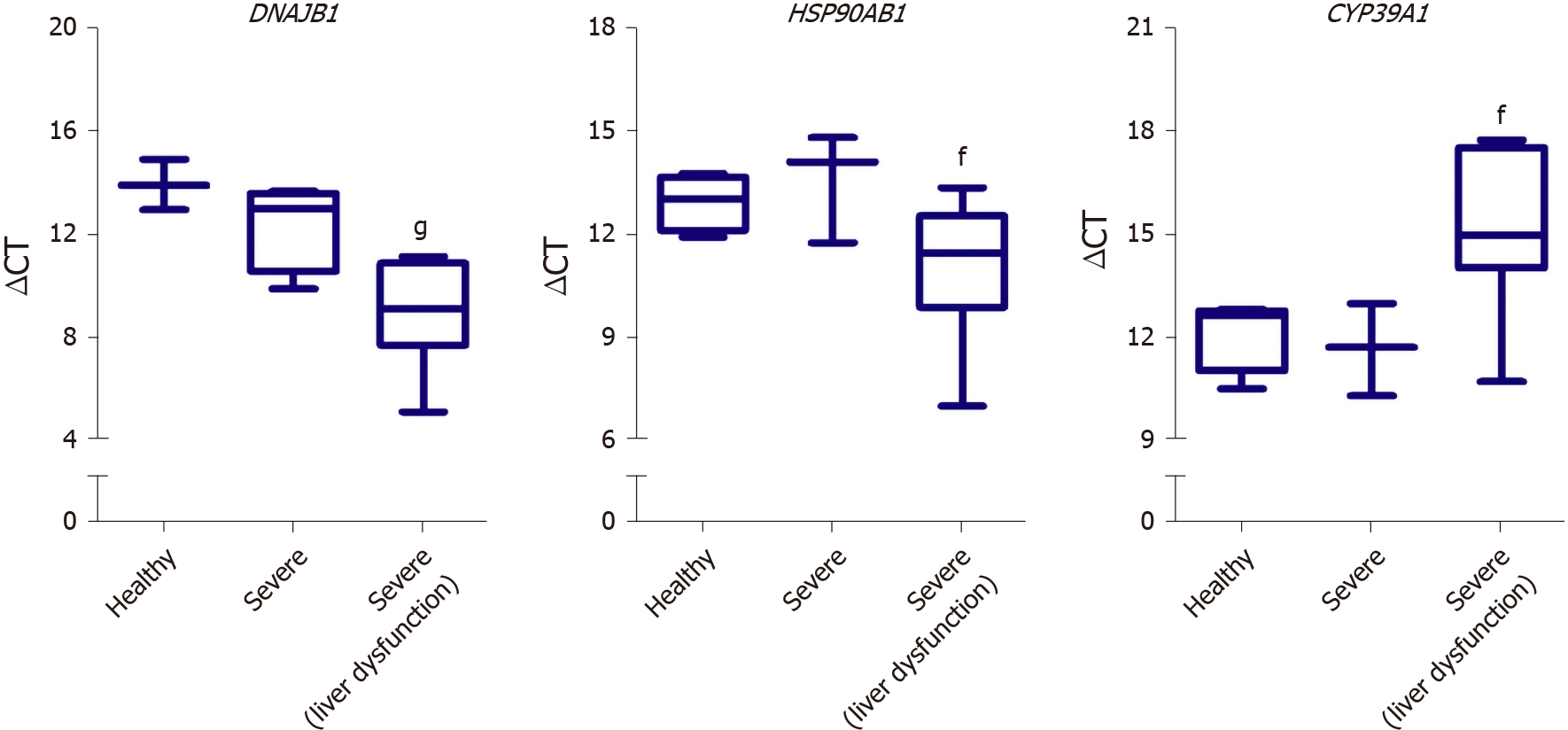
We next aimed at investigating the potential of utilizing some of the identified DEGs as putative biomarkers for liver dysfunction in COVID-19 patients. Therefore, we assessed the gene expression of some of the differentially expressed genes in the liver tissue of severe COVID-19 patients using blood plasma samples of severe COVID-19 patients with liver dysfunction. In correlation with the RNA-seq data analysis, qRT-PCR analysis confirmed the significant elevation in the gene expression of DNAJB1 and HSP90AB1 and the significant downregulation of CYP39A1 in response to liver dysfunction in severe COVID-19 patients (Figure 8).
Manifestations of SARS-CoV-2 infection were observed systemically and included indications of liver dysfunction. However, the molecular changes underlying the phenotypic systemic observations linking to liver dysfunction at advanced stages of COVID-19 are yet to be thoroughly explored. Therefore, in this study, we explored an RNA-seq dataset of severe COVID-19 patients’ liver tissues to get some insight on the dysregulated molecular mechanisms resulting in the observed phenotypic manifestations (summary of findings in Figure 9).
Our analysis of the RNA-seq data revealed that severe COVID-19 patients experience transcriptional shifts in their liver tissue that supersede those observed in lung tissue at advanced stages. These transcriptional shifts include the significant dysregulation of signaling through GPCRs [class A-rhodopsin-like receptors (R-HSA-373076: class A/1 Rhodopsin-like receptors] and class F-frizzled and smoothened receptors). Subsequently, various intracellular signaling pathways (e.g., WNT, hippo and SHH signaling pathways) are dysregulated resulting in the transcriptional dysregulation of viral reproduction, cell proliferation, cells survival, cellular chemotaxis as well as local environment, inflammation, and tissue remodeling[16,19,20].
A number of the top upregulated transcripts, including DNAJB1, IGF2, and HDGF, were implicated in the activation of tissue remodeling pathways[22-24]. The upregulation of these genes suggests that severe COVID-19 patients may experience tissue remodeling of the liver tissue. IGF2 overexpression was proposed to induce fatty liver and hepatic steatosis by increasing lipid accumulation in liver tissue[23]. In our analysis of the SARS-CoV-2 infected liver samples, we observed an increase in the pro-steatosis factor, IGF2, and a concordant decrease of the steatosis and tissue remodeling preventive factors, IGF1 and GHR (growth hormone receptor)[42]. In addition to IGF2, numerous genes proposed to contribute to the pathogenesis of liver diseases characterized by tissue remodeling (e.g. liver steatosis, liver fibrosis, and liver cirrhosis) were upregulated in the liver tissue of severe COVID-19 patients (e.g. FLT1, LEP, PNPLA3, VCAM1, TREM2, metalloproteinases-expressing genes, and collagen-expressing genes)[27-31].
Furthermore, consistently with the upregulation of these genes implicated in liver tissue remodeling, we observed the significant upregulation of EGFR which was proposed to contribute to the development of pulmonary fibrosis[25]. The enrichment of these transcripts in the transcriptome of SARS-CoV-2 infected liver samples suggests the potential activation of tissue remodeling and fat accumulation in liver tissue. Consistently with these findings, immunohistochemical analysis of liver tissue from COVID-19 patients revealed signs of hepatic steatosis and hepatic siderosis [43,44]. These morphologic changes might be attributed to the activation of the tissue remodeling mechanisms in response to inflammation; to the dysregulation of intracellular signaling pathways (e.g., EFGR, WNT, and SHH signaling pathways) in response to direct viral infection; or to the dysregulation of metabolic and biosynthetic pathways resulting the accumulation of lipids and other metabolic substrates in liver tissue and subsequently the development of liver steatosis.
Altogether, the dysregulation of these transcripts and pathways in the liver tissue of severe COVID-19 patients may indicate that the patients may experience pathological tissue remodeling similar to that observed in liver steatosis/cirrhosis/fibrosis that potentially contributes to the phenotypically observed liver dysfunction. Moreover, despite the observed recovery of liver function in some patients in response to treatment, the activation of tissue remodeling mechanisms may have long-term consequences in patients experiencing advanced stages of COVID-19 and would require long term follow-up.
On the other hand, the downregulated transcriptome of the SARS-CoV-2 infected liver tissue samples was enriched in transcripts implicated in metabolic and biosynthetic pathways regulation, including ACAD11, CIDEB, GNMT, GPAM, and HSD17B4. Previous investigations in the metabolomic profile of the SARS-CoV-2 infected patients’ sera similarly revealed the suppressive effect of SARS-CoV-2 infection on lipid and amino acid metabolism[32]. Moreover, accumulating evidence suggested the localization of SARS-CoV-2 viral RNA in the mitochondrial matrix and the direct viral interaction with mitochondrial proteins and metabolic regulators (e.g. MRPS2, MRPS5, MRPS25, MRPS27, NDUFAF1, NDUFB9, NDUFAF2, ATP1B1, ATP6V1A, ACADM, AASS, PMPCB, PITRM1, COQ8B, and PMPCA)[45-47]. By localizing to the mitochon
Further investigations on the potential secondary mechanisms resulting in mitochondrial dysfunction divulged the imbalance in ions (e.g. calcium, potassium, sodium) due to dysregulated ionic transport and homeostasis as a potential underlying mechanism. Ionic imbalance can shift the osmotic pressure resulting in mitochondrial swelling and consequent dysfunction[39]. Moreover, ionic imbalance can have critical consequences on intracellular signaling and cellular morphology which can be another mechanism through which SARS-CoV-2 induces the observed transcriptomic shifts[49,50].
Clinical assessment of SARS-CoV-2 infected liver tissues revealed mitochondrial swelling which can be an underlying cause for the mitochondrial dysfunction observed in the infected cells. One of the potential mechanisms resulting in the mitochondrial swelling is the dysregulation of ions binding, transport and balance within the cell and its compartments. Our analysis showed a significant enrichment of downregulated transcripts implicated in iron, copper, sodium, potassium, zinc, and calcium binding, transport, homeostasis and signaling. Imbalance of some of these ions (e.g., calcium and potassium ions) have been shown to induce mitochondrial swelling through shifting osmotic pressure[39]. Ionic imbalances were similarly found to be induced by SARS-CoV and Influenza virus resulting in cellular morphology changes as well as stimulation of the inflammasome and subsequent inflammatory mechanisms such as IL-1β production and signaling[49,50]. Potassium levels imbalance and absorption dysregulation has been reported in Covid-19 patients in the form of hypokalemia and the patients were found to respond well to potassium supplements during recovery[51]. Taken together, we speculate that SARS-CoV-2 hijacks cellular metabolic and biosynthetic pathways by disrupting ion imbalance and mitochondrial function.
One of the main targets that appear to be suppressed in response to SARS-CoV-2 infection is the CYPs family which plays a central role in xenobiotic and drug metabolism in liver tissues. The suppression of the members of this family of metabolic enzymes may potentially result in hepatoxicity and acute liver injury upon the administration of treatments. Currently used therapeutic agents for the treatment of COVID-19 patients including chloroquine and hydroxychloroquine are metabolized by members of the CYPs family enzymes (e.g., CYP2C8 and CYP3A4/5)[40,41], which we observed to be downregulated in SARS-CoV-2 infected liver samples. Therefore, the downregulation of these enzymes might dysregulate the metabolism of these therapeutic agents resulting in hepatotoxicity, autoimmune hepatitis, and liver injury as observed histologically[44]. In concordance with these findings, metabolomic profiling of covid-19 patients’ sera suggested a decline in liver detoxification function [32]. Moreover, the downregulation of CYPs has been implicated in the dysregulation of metabolic pathways in cirrhotic patients[52]. Therefore, the downregulation of the CYPs may contribute to the liver injury through the dysregulation of drug metabolism as well as the activation of tissue remodeling mechanisms that ensure the pathogenesis of other liver diseases (e.g., cirrhosis).
Whilst the main limitation of this study is the lack of validation on patients’ liver tissue samples, we cross-validated and confirmed the dysregulation of some DEGs (DNAJB1, HSP90AB1, and CYP39A1) using blood plasma samples of severe COVID-19 patients with liver dysfunction. The systems biology analysis carried out in this study highlighted some molecular mechanisms which may potentially elucidate the clinically observed haptic dysfunction in severe COVID-19 patients. Moreover, the findings of this study shed the light on the potentially activation of fibrosis/ cirrhosis/steatosis patterns in response to the activation of wound healing and tissue remodeling mechanisms in severe COVID-19 patients which may require long-term follow up. These findings may prove to be critical in cases presenting with long COVID, as the chronic exposure to the infection and disease symptoms may induce irreversible tissue remodeling of the liver. Furthermore, based on the observed significant reduction in the metabolic pathways and hepatic detoxification in our study, serious consideration is required for the administered hepatotoxic treatments to severe COVID-19 patients (e.g., chloroquine, hydroxychloroquine, and remdesivir). However, it remains to be ascertained whether the observed transcriptional shifts observed in the liver tissue of severe COVID-19 patients are the result of viral infection, systemic inflammation, or drug induced liver injury and toxicity.
In conclusion, we aimed at getting some insight into the molecular mechanisms underlying liver dysfunction in severe COVID-19 patients using RNA-seq data of severe COVID-19 liver autopsy samples. Through systems biology analysis, we observed a significant upregulation in transcripts implicated in GPCRs signaling, tissue remodeling, and intracellular/transmembrane transport (e.g., DNAJB1, IGF2, EGFR, and HDGF). On the other hand, we observed a significant downregulation in transcripts implicated in metabolic pathways and mitochondrial function (e.g., ACAD11, CIDEB, GNMT, and GPAM). Moreover, we observed a significant suppres
The coronavirus disease 2019 (COVID-19), a pandemic contributing to more than 105 million cases and more than 2.3 million deaths worldwide is associated with extrapulmonary manifestations, including liver dysfunction. Liver dysfunction and elevated liver enzymes were observed in about 53% of COVID-19 patients. However, the molecular mechanistic aspect of this is not clear but can be of importance in understanding how the virus infects the hepatocytes and the infection leads to liver dysfunction. Understanding this can help in the diagnosis and treatment of liver dysfunction caused by COVID-19.
Liver is an important organ in human health as it is involved in the detoxification of many compounds accumulated through pollution or using the wrong diet. Liver dysfunction can lead to many systemic diseases in the body. One of the organs affected by SARS-CoV-2 infection is the liver and therefore understanding the mechanism of infection and the cause of liver dysfunction can help COVID-19 patients whose liver is affected but also the results can shed light on the molecular mechanism of liver dysfunction that may results from other liver diseases including cirrhosis, fibrosis, non-alcoholic fatty liver disease (NAFLD), and hepatitis A/B/C.
The main objectives of this research are: (1) Obtain genetic data from COVID-19 patients who had hepatic dysfunction; (2) Compare the genetic signature of COVID-19 patients with hepatic failure with that of other liver diseases including cirrhosis, fibrosis, NAFLD, and hepatitis A/B/C; and (3) Carry out in vitro validation of the biomarkers identified using blood samples from severe COVID-19 patients with liver dysfunction.
The transcriptome of liver autopsy samples from severe COVID-19 patients against those of non-COVID donors was analyzed. Differentially expressed genes were identified from normalized RNA-seq data and analyzed for the enrichment of functional clusters and pathways. The differentially expressed genes were then compared against the genetic signatures of liver diseases including cirrhosis, fibrosis, NAFLD, and hepatitis A/B/C. Gene expression of some differentially expressed genes was assessed in the blood samples of severe COVID-19 patients with liver dysfunction using qRT-PCR.
Analysis of the differential transcriptome of the liver tissue of severe COVID-19 patients revealed a significant upregulation of transcripts implicated in tissue remodeling including GPCRs, DNAJB1, IGF2, EGFR, and HDGF. Concordantly, the differential transcriptome of severe COVID-19 liver tissues substantially overlapped with the disease signature of liver diseases characterized with pathological tissue remodeling (liver cirrhosis, Fibrosis, NAFLD, and hepatitis A/B/C). Moreover, we observed a significant suppression of transcripts implicated in metabolic pathways as well as mitochondrial function, including cytochrome P450 family members, ACAD11, CIDEB, GNMT, and GPAM. Consequently, drug and xenobiotics metabolism pathways are significantly suppressed suggesting a decrease in liver detoxification capacity. In correspondence with the RNA-seq data analysis, we observed a significant upregulation of DNAJB1 and HSP90AB1 as well as significant downregulation of CYP39A1 in the blood plasma of severe COVID-19 patients with liver dysfunction.
Some insights into the molecular mechanisms underlying liver dysfunction in severe COVID-19 patients was obtained using RNA-seq data of severe COVID-19 liver autopsy samples. Through systems biology analysis, we observed a significant upregulation in transcripts implicated in GPCRs signaling, tissue remodeling, and intracellular/transmembrane transport (e.g., DNAJB1, IGF2, EGFR, and HDGF). On the other hand, we observed a significant downregulation in transcripts implicated in metabolic pathways and mitochondrial function (e.g., ACAD11, CIDEB, GNMT, and GPAM). Moreover, we observed a significant suppression of cytochrome P450 family members, which play central roles in the metabolism of drugs and xenobiotics metabolism suggesting a compromised hepatic detoxification capacity. Moreover, the dysregulation of some genes (e.g., DNAJB1, HSP90AB1, and CYP39A1) in the tissue autopsies correlated with their dysregulation in the blood plasma of severe COVID-19 patients with hepatic dysfunction suggesting their potential as putative biomarkers of liver dysfunction.
This study identified key cellular pathways involved in liver dysfunction in general but also liver dysfunction linked to COVID-19 patients. The analysis identified key molecular biomarkers which can be used in future to general a biomarker panel that can assess the degree of hepatic dysfunction in patients from a blood test since the biomarkers identified in this study was shown to follow the same trend when compared with blood samples from COVID-19 patients with hepatic dysfunction. Mechanistically, the study showed that severe COVID-19 patients appear to experience significant transcriptional shift that may ensue tissue remodeling, mitochondrial dysfunction and apparent hepatic toxification resulting in the clinically observed liver dysfunction. The finding from this study can be used in future to assess the degree of hepatic dysfunction but also to differentiate patients with COVID-19 related hepatic dysfunction from hepatic dysfunction caused by other liver diseases. This can be of importance in the management of COVID-19 patients including those with long COVID who have experienced hepatic dysfunction.
Manuscript source: Unsolicited manuscript
Specialty type: Medical informatics
Country/Territory of origin: United Arab Emirates
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Alberca RW S-Editor: Liu M L-Editor: A P-Editor: Ma YJ
| 1. | Utunen H, Ndiaye N, Piroux C, George R, Attias M, Gamhewage G. Global Reach of an Online COVID-19 Course in Multiple Languages on OpenWHO in the First Quarter of 2020: Analysis of Platform Use Data. J Med Internet Res. 2020;22:e19076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 2. | Lin L, Jiang X, Zhang Z, Huang S, Fang Z, Gu Z, Gao L, Shi H, Mai L, Liu Y, Lin X, Lai R, Yan Z, Li X, Shan H. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997-1001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 630] [Cited by in RCA: 657] [Article Influence: 131.4] [Reference Citation Analysis (0)] |
| 3. | Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2493] [Cited by in RCA: 2310] [Article Influence: 462.0] [Reference Citation Analysis (0)] |
| 4. | Weber S, Mayerle J, Irlbeck M, Gerbes AL. Severe liver failure during SARS-CoV-2 infection. Gut. 2020;69:1365-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 5. | Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, Ma K, Xu D, Yu H, Wang H, Wang T, Guo W, Chen J, Ding C, Zhang X, Huang J, Han M, Li S, Luo X, Zhao J, Ning Q. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368:m1091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2289] [Cited by in RCA: 2550] [Article Influence: 510.0] [Reference Citation Analysis (2)] |
| 6. | Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol Hepatol. 2020;5:428-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1348] [Cited by in RCA: 1295] [Article Influence: 259.0] [Reference Citation Analysis (4)] |
| 7. | Yip TC, Lui GC, Wong VW, Chow VC, Ho TH, Li TC, Tse YK, Hui DS, Chan HL, Wong GL. Liver injury is independently associated with adverse clinical outcomes in patients with COVID-19. Gut. 2021;70:733-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 8. | Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol. 2020;73:1231-1240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 358] [Article Influence: 71.6] [Reference Citation Analysis (1)] |
| 9. | Desai N, Neyaz A, Szabolcs A, Shih AR, Chen JH, Thapar V, Nieman LT, Solovyov A, Mehta A, Lieb DJ, Kulkarni AS, Jaicks C, Xu KH, Raabe MJ, Pinto CJ, Juric D, Chebib I, Colvin RB, Kim AY, Monroe R, Warren SE, Danaher P, Reeves JW, Gong J, Rueckert EH, Greenbaum BD, Hacohen N, Lagana SM, Rivera MN, Sholl LM, Stone JR, Ting DT, Deshpande V. Temporal and spatial heterogeneity of host response to SARS-CoV-2 pulmonary infection. Nat Commun. 2020;11:6319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 175] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 10. | Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755-W762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 227] [Cited by in RCA: 253] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 11. | Li J, Witten DM, Johnstone IM, Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics. 2012;13:523-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 224] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 12. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37449] [Article Influence: 1872.5] [Reference Citation Analysis (0)] |
| 13. | Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 998] [Cited by in RCA: 1558] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 14. | Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3766] [Cited by in RCA: 8765] [Article Influence: 1460.8] [Reference Citation Analysis (0)] |
| 15. | Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845-D855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 1078] [Article Influence: 215.6] [Reference Citation Analysis (0)] |
| 16. | Sodhi A, Montaner S, Gutkind JS. Viral hijacking of G-protein-coupled-receptor signalling networks. Nat Rev Mol Cell Biol. 2004;5:998-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 132] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 17. | Wang Z, Lu W, Zhang Y, Zou F, Jin Z, Zhao T. The Hippo Pathway and Viral Infections. Front Microbiol. 2019;10:3033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 18. | Ljungberg JK, Kling JC, Tran TT, Blumenthal A. Functions of the WNT Signaling Network in Shaping Host Responses to Infection. Front Immunol. 2019;10:2521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 19. | Smelkinson MG. The Hedgehog Signaling Pathway Emerges as a Pathogenic Target. J Dev Biol. 2017;5:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 20. | More S, Yang X, Zhu Z, Bamunuarachchi G, Guo Y, Huang C, Bailey K, Metcalf JP, Liu L. Regulation of influenza virus replication by Wnt/β-catenin signaling. PLoS One. 2018;13:e0191010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Cheng H, Lear-Rooney CM, Johansen L, Varhegyi E, Chen ZW, Olinger GG, Rong L. Inhibition of Ebola and Marburg Virus Entry by G Protein-Coupled Receptor Antagonists. J Virol. 2015;89:9932-9938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 22. | Graham RP, Jin L, Knutson DL, Kloft-Nelson SM, Greipp PT, Waldburger N, Roessler S, Longerich T, Roberts LR, Oliveira AM, Halling KC, Schirmacher P, Torbenson MS. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Mod Pathol. 2015;28:822-829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 120] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | Kessler SM, Laggai S, Van Wonterg E, Gemperlein K, Müller R, Haybaeck J, Vandenbroucke RE, Ogris M, Libert C, Kiemer AK. Transient Hepatic Overexpression of Insulin-Like Growth Factor 2 Induces Free Cholesterol and Lipid Droplet Formation. Front Physiol. 2016;7:147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Kao YH, Chen CL, Jawan B, Chung YH, Sun CK, Kuo SM, Hu TH, Lin YC, Chan HH, Cheng KH, Wu DC, Goto S, Cheng YF, Chao D, Tai MH. Upregulation of hepatoma-derived growth factor is involved in murine hepatic fibrogenesis. J Hepatol. 2010;52:96-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 25. | Venkataraman T, Frieman MB. The role of epidermal growth factor receptor (EGFR) signaling in SARS coronavirus-induced pulmonary fibrosis. Antiviral Res. 2017;143:142-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 26. | Enomoto Y, Orihara K, Takamasu T, Matsuda A, Gon Y, Saito H, Ra C, Okayama Y. Tissue remodeling induced by hypersecreted epidermal growth factor and amphiregulin in the airway after an acute asthma attack. J Allergy Clin Immunol 2009; 124: 913-20. e1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 27. | Corpechot C, Barbu V, Wendum D, Kinnman N, Rey C, Poupon R, Housset C, Rosmorduc O. Hypoxia-induced VEGF and collagen I expressions are associated with angiogenesis and fibrogenesis in experimental cirrhosis. Hepatology. 2002;35:1010-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 369] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 28. | Leclercq IA, Farrell GC, Schriemer R, Robertson GR. Leptin is essential for the hepatic fibrogenic response to chronic liver injury. J Hepatol. 2002;37:206-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 297] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 29. | Dong XC. PNPLA3-A Potential Therapeutic Target for Personalized Treatment of Chronic Liver Disease. Front Med (Lausanne). 2019;6:304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 30. | Cazanave S, Podtelezhnikov A, Jensen K, Seneshaw M, Kumar DP, Min HK, Santhekadur PK, Banini B, Mauro AG, M Oseini A, Vincent R, Tanis KQ, Webber AL, Wang L, Bedossa P, Mirshahi F, Sanyal AJ. The Transcriptomic Signature Of Disease Development And Progression Of Nonalcoholic Fatty Liver Disease. Sci Rep. 2017;7:17193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Perugorria MJ, Esparza-Baquer A, Oakley F, Labiano I, Korosec A, Jais A, Mann J, Tiniakos D, Santos-Laso A, Arbelaiz A, Gawish R, Sampedro A, Fontanellas A, Hijona E, Jimenez-Agüero R, Esterbauer H, Stoiber D, Bujanda L, Banales JM, Knapp S, Sharif O, Mann DA. Non-parenchymal TREM-2 protects the liver from immune-mediated hepatocellular damage. Gut. 2019;68:533-546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 116] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 32. | Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, Quan S, Zhang F, Sun R, Qian L, Ge W, Liu W, Liang S, Chen H, Zhang Y, Li J, Xu J, He Z, Chen B, Wang J, Yan H, Zheng Y, Wang D, Zhu J, Kong Z, Kang Z, Liang X, Ding X, Ruan G, Xiang N, Cai X, Gao H, Li L, Li S, Xiao Q, Lu T, Zhu Y, Liu H, Guo T. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020; 182: 59-72. e15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 728] [Cited by in RCA: 1091] [Article Influence: 218.2] [Reference Citation Analysis (0)] |
| 33. | Ben-Shachar D, Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS One. 2007;2:e817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | He M, Pei Z, Mohsen AW, Watkins P, Murdoch G, Van Veldhoven PP, Ensenauer R, Vockley J. Identification and characterization of new long chain acyl-CoA dehydrogenases. Mol Genet Metab. 2011;102:418-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 35. | Slayton M, Gupta A, Balakrishnan B, Puri V. CIDE Proteins in Human Health and Disease. Cells. 2019;8:238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 36. | Varela-Rey M, Martínez-López N, Fernández-Ramos D, Embade N, Calvisi DF, Woodhoo A, Rodríguez J, Fraga MF, Julve J, Rodríguez-Millán E, Frades I, Torres L, Luka Z, Wagner C, Esteller M, Lu SC, Martínez-Chantar ML, Mato JM. Fatty liver and fibrosis in glycine N-methyltransferase knockout mice is prevented by nicotinamide. Hepatology. 2010;52:105-114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Chen YQ, Kuo MS, Li S, Bui HH, Peake DA, Sanders PE, Thibodeaux SJ, Chu S, Qian YW, Zhao Y, Bredt DS, Moller DE, Konrad RJ, Beigneux AP, Young SG, Cao G. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. J Biol Chem. 2008;283:10048-10057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 38. | Pierce SB, Walsh T, Chisholm KM, Lee MK, Thornton AM, Fiumara A, Opitz JM, Levy-Lahad E, Klevit RE, King MC. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am J Hum Genet. 2010;87:282-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 191] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 39. | Javadov S, Chapa-Dubocq X, Makarov V. Different approaches to modeling analysis of mitochondrial swelling. Mitochondrion. 2018;38:58-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 105] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 40. | Kim KA, Park JY, Lee JS, Lim S. Cytochrome P450 2C8 and CYP3A4/5 are involved in chloroquine metabolism in human liver microsomes. Arch Pharm Res. 2003;26:631-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 95] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 41. | Wahie S, Daly AK, Cordell HJ, Goodfield MJ, Jones SK, Lovell CR, Carmichael AJ, Carr MM, Drummond A, Natarajan S, Smith CH, Reynolds NJ, Meggitt SJ. Clinical and pharmacogenetic influences on response to hydroxychloroquine in discoid lupus erythematosus: a retrospective cohort study. J Invest Dermatol. 2011;131:1981-1986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Adamek A, Kasprzak A. Insulin-Like Growth Factor (IGF) System in Liver Diseases. Int J Mol Sci. 2018;19:1308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 196] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 43. | Deinhardt-Emmer S, Wittschieber D, Sanft J, Kleemann S, Elschner S, Haupt KF, Vau V, Häring C, Rödel J, Henke A, Ehrhardt C, Bauer M, Philipp M, Gaßler N, Nietzsche S, Löffler B, Mall G. Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage. Elife. 2021;10:e60361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 44. | Abenavoli L, Gentile I, Maraolo AE, Negro F. SARS-CoV-2 and liver damage: a possible pathogenetic link. Hepatobiliary Surg Nutr. 2020;9:322-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Singh KK, Chaubey G, Chen JY, Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am J Physiol Cell Physiol. 2020;319:C258-C267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 255] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 46. | Wu KE, Fazal FM, Parker KR, Zou J, Chang HY. RNA-GPS Predicts SARS-CoV-2 RNA Residency to Host Mitochondria and Nucleolus. Cell Syst 2020; 11: 102-108. e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 112] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 47. | Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O'Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d'Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3145] [Cited by in RCA: 3186] [Article Influence: 637.2] [Reference Citation Analysis (0)] |
| 48. | Qiao X, Zhou ZC, Niu R, Su YT, Sun Y, Liu HL, Teng JL, Ye JN, Shi H, Yang CD, Cheng XB. Hydroxychloroquine Improves Obesity-Associated Insulin Resistance and Hepatic Steatosis by Regulating Lipid Metabolism. Front Pharmacol. 2019;10:855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 49. | Nieto-Torres JL, DeDiego ML, Verdiá-Báguena C, Jimenez-Guardeño JM, Regla-Nava JA, Fernandez-Delgado R, Castaño-Rodriguez C, Alcaraz A, Torres J, Aguilella VM, Enjuanes L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10:e1004077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 327] [Cited by in RCA: 375] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 50. | Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404-410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 521] [Cited by in RCA: 510] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 51. | Chen D, Li X, Song Q, Hu C, Su F, Dai J, Ye Y, Huang J, Zhang X. Assessment of Hypokalemia and Clinical Characteristics in Patients With Coronavirus Disease 2019 in Wenzhou, China. JAMA Netw Open. 2020;3:e2011122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 52. | Dietrich CG, Götze O, Geier A. Molecular changes in hepatic metabolism and transport in cirrhosis and their functional importance. World J Gastroenterol. 2016;22:72-88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |