Published online Feb 14, 2020. doi: 10.3748/wjg.v26.i6.670
Peer-review started: November 8, 2019
First decision: December 23, 2019
Revised: January 6, 2020
Accepted: January 11, 2020
Article in press: January 11, 2020
Published online: February 14, 2020
Esophageal carcinoma is a malignant gastrointestinal tumor with a very poor prognosis. MicroRNA (miR)-1304 is a newly discovered non-coding RNA, which shows differential expression in other cancers, and its clinical value in esophageal carcinoma remains unclear.
To explore the expression of miR-1304 in patients with esophageal carcinoma and its clinical value.
The expression of miR-1304 in patients with esophageal carcinoma was analyzed based on the data on miR in esophageal carcinoma downloaded from The Cancer Genome Atlas database. Quantitative real-time polymerase chain reaction was adopted to determine the expression of miR-1304 in the tissues and serum of patients. The clinical diagnostic value of miR-1304 and independent factors for recurrence and prognosis of esophageal carcinoma were then analyzed. The potential target genes of miR-1304 were predicted, and then analyzed based on gene ontology, Kyoto Encyclopedia of Genes, and Genomes, and protein-protein interaction.
The expression of miR-1304 in the tissues and serum of patients with esophageal carcinoma increased, and was also increased according to the database. Patients with high expression of miR-1304 suffered increased rates of tumor ≥ 3 cm, low differentiation and stage II + III. miR-1304 had a diagnostic value in identifying esophageal carcinoma, tumor size, differentiation and TNM stage. Tumor size, differentiation, TNM stage, and miR-1304 were independent risk factors for recurrence of esophageal carcinoma, and they had certain predictive and diagnostic value for the recurrence of esophageal carcinoma. Seventy-eight patients showed a 3-year survival rate of 38.46%, and patients with high expression of miR-1304 had a relatively lower survival rate. Multivariate analysis revealed that tumor size, differentiation, recurrence and miR-1304 were independent factors for the prognosis of patients. MiRTarBase, miRDB, and Targetscan predicted 20 target genes in total. Gene ontology enrichment analysis found 18 functions with aP < 0.05, and Kyoto Encyclopedia of Genes, and Genomes analysis found 11 signal pathways with aP < 0.05. String analysis of protein co-expression found 269 relationship pairs, of which co-expression with epidermal growth factor was the most common.
miR-1304 can be used as a potential indicator for the diagnosis and recurrence of esophageal carcinoma and for survival of patients with this disease.
Core tip: In recent years, the morbidity and mortality of esophageal carcinoma have increased significantly. However, there are few clinical tumor markers related to esophageal carcinoma. miRNA has been a hot research topic in recent years. This study analyzed the expression of miR-1304 in patients with esophageal carcinoma, and found that miR-1304 was highly expressed in these patients, and was an independent factor for recurrence and prognosis of esophageal carcinoma. miR-1304 is expected to become a potential index for predicting prognosis and recurrence of esophageal carcinoma.
- Citation: Luo YG, Duan LW, Ji X, Jia WY, Liu Y, Sun ML, Liu GM. Expression of miR-1304 in patients with esophageal carcinoma and risk factors for recurrence. World J Gastroenterol 2020; 26(6): 670-685
- URL: https://www.wjgnet.com/1007-9327/full/v26/i6/670.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i6.670
Esophageal carcinoma is one of the common digestive tract cancers, with morbidity ranking eighth and mortality ranking sixth of all cancers[1]. According to the 2015 cancer statistics, there were 477900 new cases of esophageal carcinoma and 375000 deaths due to esophageal carcinoma in China[2]. Such high mortality and morbidity causes a huge problem in terms of medical care. Patients with esophageal carcinoma in the early stage have insidious symptoms, and patients in the intermediate stage suffer from the typical symptom of dysphagia[3]. At present, although there are patients with early esophageal carcinoma in hospitals, most patients with esophageal carcinoma are already in the intermediate or late stage when admitted to hospital[4]. The optimal treatment plan for esophageal carcinoma in clinical practice is surgical resection, but its long-term efficacy is unsatisfactory, which is mainly due to the following factors: Esophageal carcinoma has the characteristic of adjacent lymph nodes which are prone to skipping metastasis, and leads to recurrence of esophageal carcinoma; thus, resulting in failure of surgical treatment[5,6]. If the recurrence of esophageal carcinoma can be predicted by detecting relevant indices in patients, this could further prevent metastasis during treatment[7]. However, there are few clinical indices for the detection of esophageal carcinoma; therefore, it is particularly important and urgent to find a potential biomarker.
Non-coding RNA is a transcript substance without any encoded protein[8]. To date, several types of non-coding RNA have been identified by RNA sequencing and bioinformatics methods, of which long-chain non-coding RNA and short-chain non-coding RNA (miR) have attracted much attention in various fields[9-11]. miR is a highly conserved short-chain non-coding RNA about 21-25 nt long. Studies have revealed that miR is disordered in patients with diseases such as cardiovascular diseases and cancer[12,13]. Many studies have confirmed that miR can suppress the translation of target genes after transcription by binding to untranslated regions[14]. The study by Li et al[15] found that miR-377 could suppress the occurrence and development of esophageal carcinoma by mediating CD133 and vascular endothelial growth factor. Some studies showed that miR-506 could be used as a biomarker for the prognosis of esophageal squamous cell carcinoma[16]. miR-1304 is a newly discovered miR. A previous study indicated that miR-1304 was in expression imbalance in nasopharyngeal cancer patients treated with paclitaxel[17], but there is no related study on the expression of miR-1304 in patients with esophageal carcinoma. This study analyzed the expression of miR in patients with esophageal carcinoma based on The Cancer Genome Atlas (TCGA) database and found that miR-1304 was highly expressed in these patients, which indicated that miR-1304 is expected to be a potential indicator of esophageal carcinoma.
Therefore, this study determined the expression of miR-1304 in patients with esophageal carcinoma and its clinical value, in order to provide a reference for clinicians.
Manifest, Cart, and Metadata were downloaded by logging into https://portal.gdc.cancer.gov/, selecting Repository-Cases-Esophagus-The Cancer Genome Atlas (TCGA)-ESCA-File-Transcriptome Profiling-miRNA Expression Quantification-HTSeq-Counts, and adding all Files to the cart. A total of 198 specimens were collected, including 185 cancer tissue specimens and 13 tumor-adjacent tissue specimens. The files were converted into a matrix to extract data on miR-1304 for analysis.
A total of 78 patients with esophageal carcinoma treated in the Second Hospital of Jilin University from March 2015 to March 2018 were enrolled as the patient group, and another 50 healthy subjects during the same period were enrolled as the normal group. The patient group consisted of 44 males and 34 females, with an average age of 58.4 ± 5.9 years, and the normal group consisted of 30 males and 20 females, with an average age of 59.4 ± 4.8 years. Inclusion criteria were as follows: Patients diagnosed with esophageal squamous cell carcinoma based on pathology; patients meeting the 8th edition of TNM stage criteria for esophageal carcinoma released by the American Joint Committee on Cancer in 2017[18]; patients who had not undergone cancer therapy; patients who and whose families signed an informed consent form after understanding the study, and patients with detailed clinical data and willing to cooperate during follow-up. Exclusion criteria were as follows: Patients comorbid with other tumors; patients with infection before admission, severe cardiac or cerebral function injury, or immune deficiency; patients unable to receive the full treatment, or unwilling to cooperate during follow-up, and those with expected survival time less than 3 mo.
All 78 patients were treated by resection of esophageal carcinoma and lymph node dissection, and received auxiliary therapy after surgery. Twenty-six patients were treated with radiotherapy, and fifty-two patients with chemotherapy. Radiotherapy was performed mainly by three-dimensional conformal radiation therapy and intensity-modulated radiation therapy[19], and chemotherapy was mainly performed using fluorouracil and cisplatin as follows: 500 mg/m2 fluorouracil (Hainan Choitec Pharmaceuticals Co., Ltd., Hainan, China) was administered by intravenous drip for 1-5 d, and cisplatin (Guizhou Hanfang Pharmaceutical Co., Ltd., Guizhou, China) was administered in the same way for 1-5 d. Patients received the two drugs for at least 2 cycles, 28 d a cycle.
Cancer tissues and tumor-adjacent tissues were sampled from the patients during surgery, and sent to a laboratory for subsequent analysis in liquid nitrogen. Peripheral fasting venous blood (5 mL) was obtained from each patient in the morning of the day before surgery and each person undergoing physical examination in the morning of the day of physical examination, allowed to stand for 30 min, and then centrifuged at 3000 rpm for 10 min to collect the supernatant. The total RNA in the collected serum and tissues was extracted with TRIzol reagent (Carlsbad Invitrogen Company, CA, United States, 15596018), and the purity, concentration and integrity of the total RNA were determined using ultraviolet spectrophotometry and agarose gel electro-phoresis. Reverse transcription was performed using the TransScript® miRNA RT Enzyme Mix and 2 × TS miRNA Reaction Mix in TransScript Green miRNA Two-Step Quantitative real-time polymerase chain reaction (qRT-PCR) SuperMix kit (TransGen Biotech, Beijing, China, AQ202-01) in strict accordance with the original kit instructions. The amplification system of miR-1304 consisted of 1 μL of cDNA, 0.4 μL of upstream and downstream primers, respectively, 10 μL of 2 × TransScript® Tip Green qPCR SuperMix, 0.4 μL of Passive Reference Dye (50 ×) and ddH2O (added to make up to 20 μL in total). The amplification conditions were as follows: Pre-denaturation at 94°C for 30 s, denaturation at 94°C for 5 s, and annealing and extension at 60°C for 30 s, 40 cycles in total. Three repeated wells were set for each specimen, and the experiment was repeated three times. U6 was used as an internal reference, and 2-ΔΔct was used to analyze the data[20]. Experiments were carried out using the 7500 PCR instrument from ABI, United States. The upstream primer and downstream primer of the primer sequence miR-1304 were 5'-3' and 5'-3', respectively, and those of U6 were 5'-3' and 5'-3', respectively.
If the lymph node short diameter was found to be longer than or equal to 10 mm in terms of pathology or cytology based on puncture, and in terms of imaging, this was considered regional lymph node recurrence.
The patients were followed up mainly by telephone as well as reexamination or review of an electronic archive of them at the 3rd, 6th, 9th and 12th mo in the first year, and every 4 mo in the 2nd and 3rd years.
Target genes were predicted on three online target gene prediction websites for miR, namely, MiRTarBase, miRDB, and TargetScan, respectively, and a Venn diagram was drawn. The signal pathways of potential mRNAs were analyzed based on the gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) online websites, DAVID and KOBAS, and a network map for protein co-expression was drawn on the online website, String.
In this study, the SPSS 20.0 software package was used to analyze the collected data statistically, and the GraphPad 7 software package was used to draw the required figures. The K-S test was used to analyze the distribution of measurement data. The measurement data with normal distribution were expressed by mean ± SD, and those with non-normal distribution were expressed by median and interquartile range P50 (P25-P75), analyzed using the non-parametric test, and then expressed by Z. Comparisons between groups were analyzed using the independent-samples t test, and comparisons within groups were analyzed using the paired t test, and expressed by t. Enumeration data were analyzed using the χ2 test, and expressed by χ2. Comparisons among multiple groups were analyzed using one-way ANOVA, and expressed by F. Post-hoc pairwise comparisons were analyzed using the LSD-t test. Receiver operating characteristic (ROC) curves were drawn for the diagnostic value of miR-1304 in esophageal carcinoma. Spearman’s correlation analysis was adopted to analyze the correlation between miR-1304 in the tissues and serum of patients. A Kaplan-Meier curve of the 5-year survival of patients was drawn, and was analyzed using the Log-rank test. Multivariate Cox regression was adopted to analyze independent risk factors for the prognosis of patients and Logistic regression was used to analyze independent risk factors for recurrence of esophageal carcinoma in patients. aP < 0.05 indicated a significant difference.
Clinical baseline data in the normal group and patient group were compared, and no significant differences were found between them in terms of age, gender, smoking history and history of alcoholism (Table 1).
| Factors | Normal group (n = 50) | Patient group (n = 78) | t / χ2 | P value |
| Gender | 0.161 | 0.688 | ||
| Male | 30 (60.00) | 44 (56.41) | ||
| Female | 20 (40.00) | 34 (43.59) | ||
| Age (yr) | 60.4 ± 4.8 | 58.7 ± 5.4 | 0.310 | 0.757 |
| Smoking history | 0.295 | 0.587 | ||
| Yes | 30 (60.00) | 43 (55.13) | ||
| No | 20 (40.00) | 35 (44.87) | ||
| History of alcoholism | 0.081 | 0.776 | ||
| Yes | 8 (16.00) | 14 (17.95) | ||
| No | 42 (84.00) | 64 (82.05) | ||
| Location | ||||
| The upper thoracic area | 19 (24.36) | |||
| The middle thoracic area | 34 (43.59) | |||
| The lower thoracic area | 25 (32.05) | |||
| Tumor size | ||||
| ≥ 3 cm | 45 (57.69) | |||
| < 3 cm | 33 (42.31) | |||
| Differentiation | ||||
| Low differentiation | 25 (32.05) | |||
| Moderate + high differentiation | 53 (67.95) | |||
| TNM stage | ||||
| Stage I | 27 (34.62) | |||
| Stage II + III | 51 (65.38) |
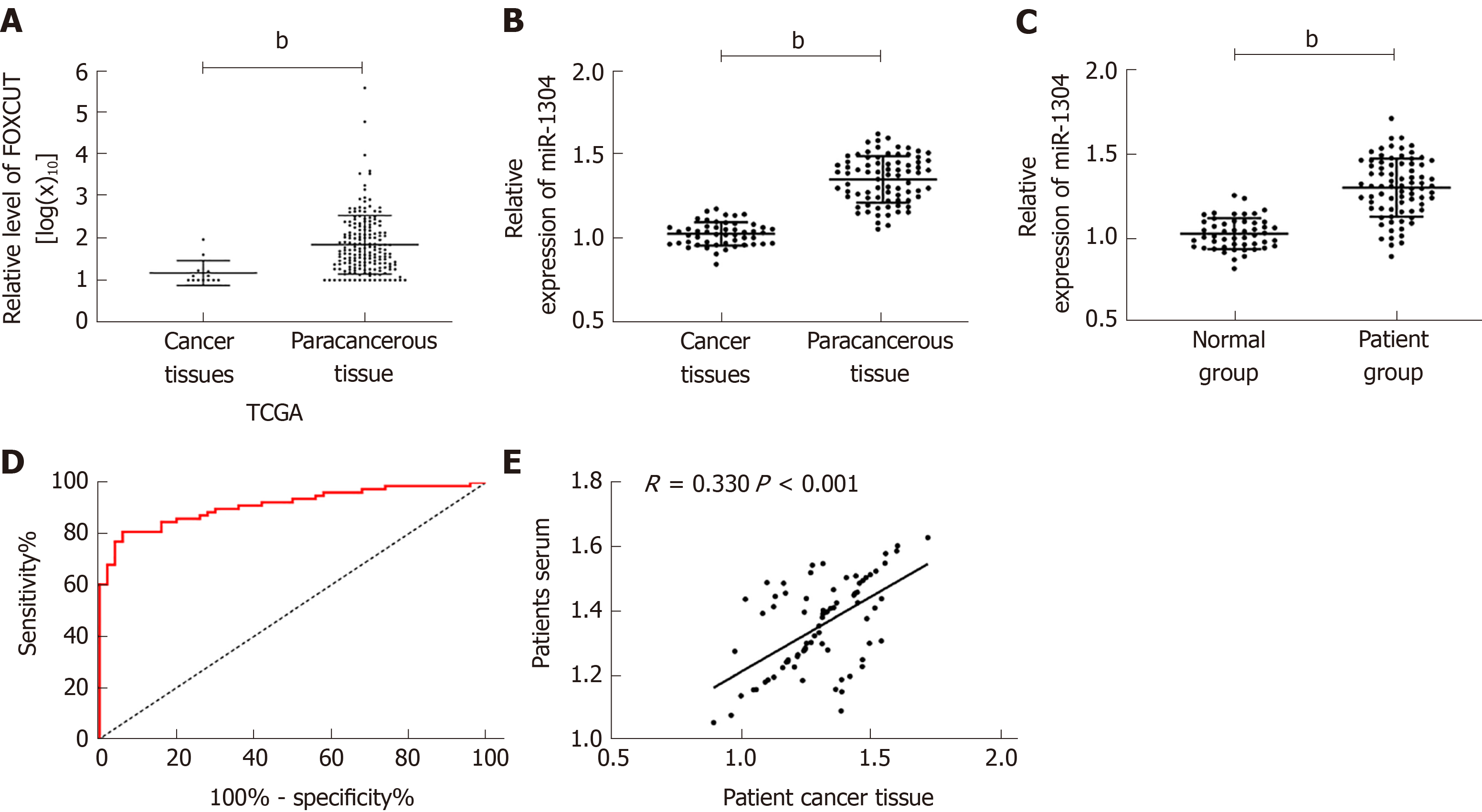
Firstly, analysis of the expression of miR-1304 from TCGA database revealed that cancer tissues showed significantly higher expression of miR-1304 than normal tissues (bP < 0.001). Analysis of the expression of miR-1304 in the serum and tissues of patients showed that the expression of miR-1304 was significantly higher than that in tumor-adjacent tissues and normal serum. Correlation analysis revealed that the expression of miR-1304 in the tissues of patients was positively correlated with that in serum (R = 0.330, bP < 0.001). ROC curve analysis revealed that the area under the curve (AUC) of miR-1304 for the diagnosis of esophageal carcinoma was 0.912; thus, miR-1304 had high diagnostic value (Figure 1).
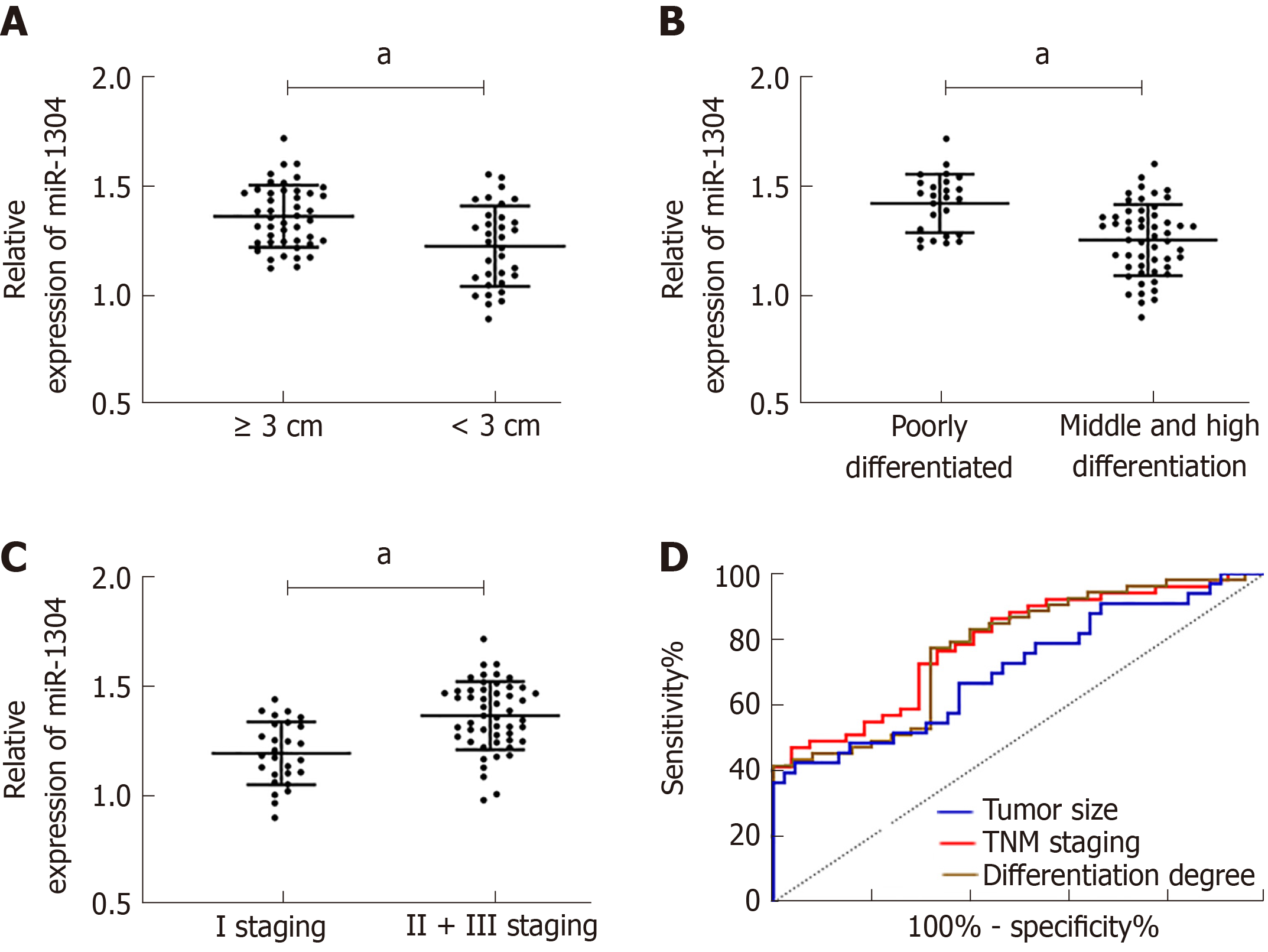
The patients were divided into a high miR-1304 expression group and a low miR-1304 expression group according to the median expression, and the pathological data were compared. No significant differences in gender, age, and lesion location were found between the two groups, and the high miR-1304 expression group had significantly higher rates of tumor size ≥ 3 cm, low differentiation and stage II + III than the low miR-1304 expression group (aP < 0.05). Therefore, we further analyzed the correlation between miR-1304 expression and tumor size, differentiation, and TNM stage, and found that patients with tumor size ≥ 3 cm and low differentiation at stage II + III, and showed that the expression of miR-1304 was different to other patients in their group. ROC curves revealed that the AUCs of miR-1304 for identifying tumor size, differentiation and TNM stage were 0.710, 0.773, and 0.788, respectively; therefore, miR-1304 had a high diagnostic value (Figure 2, Tables 2 and 3).
| Factors | High expression (n = 39) | Low expression (n = 39) | χ2 | P value |
| Gender | 0.834 | 0.361 | ||
| Male (n = 44) | 20 (51.28) | 24 (61.54) | ||
| Female (n = 34) | 19 (48.72) | 15 (38.46) | ||
| Age (yr) | 0.821 | 0.365 | ||
| ≥ 60 (n = 38) | 17 (43.59) | 21 (53.85) | ||
| < 60 (n = 40) | 22 (56.41) | 18 (46.15) | ||
| Lesion location | 1.523 | 0.467 | ||
| The upper thoracic area (n = 19) | 10 (25.64) | 9 (23.08) | ||
| The middle thoracic area (n = 34) | 19 (48.72) | 15 (38.46) | ||
| The lower thoracic area (n = 25) | 10 (25.64) | 15 (38.46) | ||
| Tumor size | 6.356 | 0.001 | ||
| ≥ 3 cm (n = 45) | 28 (71.79) | 17 (43.59) | ||
| < 3 cm (n = 33) | 11 (28.21) | 22 (56.41) | ||
| Differentiation | 4.768 | 0.029 | ||
| Low differentiation (n = 25) | 17 (43.59) | 8 (20.51) | ||
| Moderate + high differentiation (n = 53) | 22 (56.41) | 31 (79.49) | ||
| TNM stage | 6.854 | 0.009 | ||
| Stage I (n = 27) | 8 (20.51) | 19 (48.72) | ||
| Stage II + III (n = 51) | 31 (79.49) | 20 (51.28) |
| Parameters | AUC | 95%CI | P value | Sensitivity (%) | Specificity (%) | Cut-off |
| Tumor size | 0.710 | 0.591-0.829 | 0.002 | 42.42 | 95.56 | < 1.158 |
| Differentiation | 0.773 | 0.665-0.881 | 0.001 | 77.36 | 68.00 | < 1.363 |
| TNM staging | 0.788 | 0.687-0.889 | < 0.001 | 47.06 | 96.30 | > 1.393 |
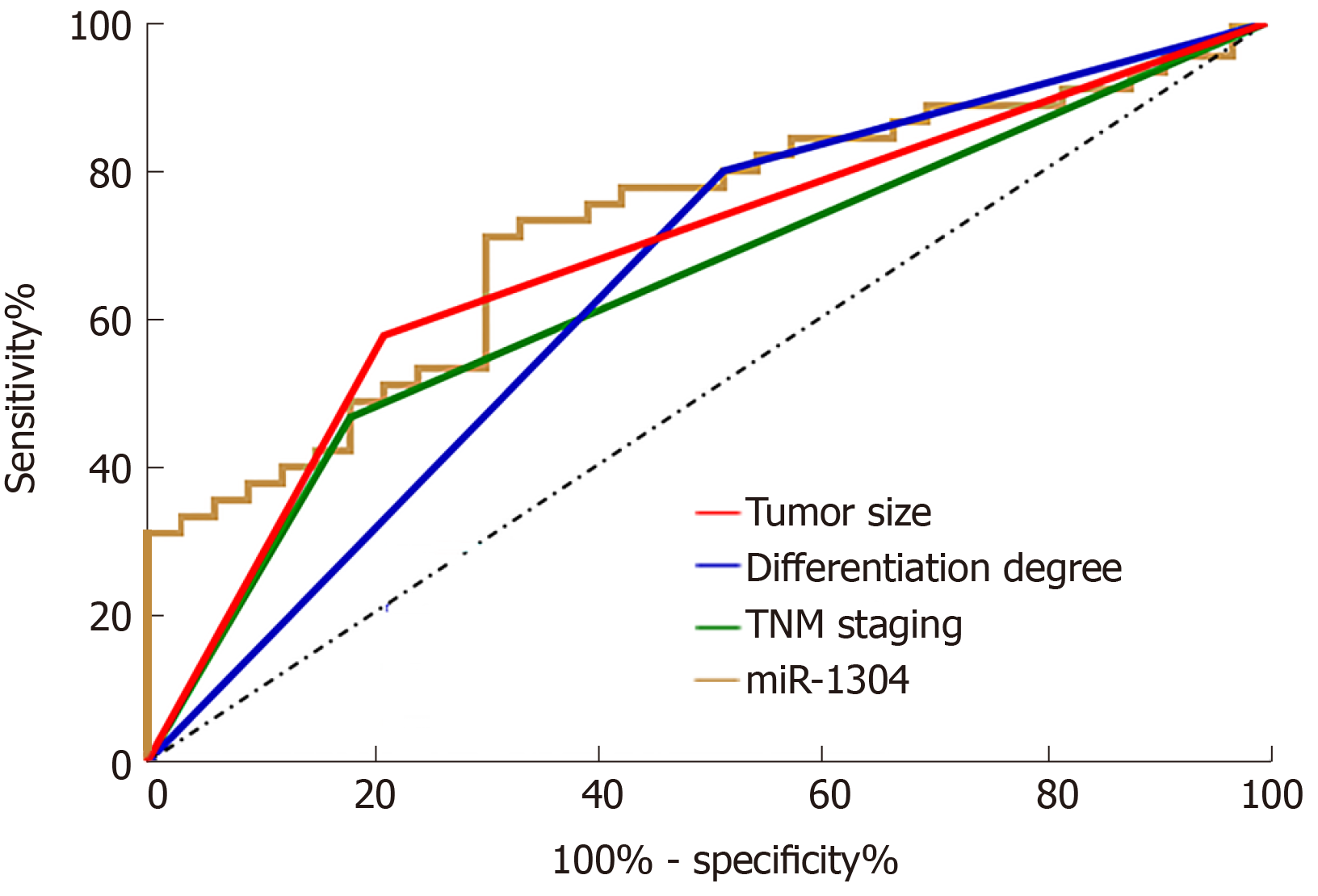
The statistics on recurrence of esophageal carcinoma in patients after treatment revealed that the 78 patients showed a recurrence rate of 42.31% with recurrence in 33 patients. Univariate analysis was performed on the collected clinical data, and it was found that there was no significant differences between the two groups in terms of gender, age, smoking history, history of alcoholism, lesion location and adjuvant therapy (all P > 0.05), while there were differences between patients in terms of tumor size, differentiation, TNM stage, and expression of miR-1304 (all aP < 0.05) (Table 4). Assignment of these factors was carried (Table 5) using Logistic regression analysis and backward LR was selected for statistics. It was found that tumor size, differentiation, TNM stage, and expression of miR-1304 were independent risk factors for recurrence of esophageal carcinoma (Table 6). In addition, ROC curves were drawn for these independent risk factors, and tumor size, differentiation, TNM stage, and miR-1304 were found to have certain clinical value in predicating recurrence (Figure 3).
| Factors | Recurrence group (n = 33) | Non-recurrence group (n = 45) | t /χ2 | P value |
| Gender | 2.792 | 0.095 | ||
| Male (n = 44) | 15 (45.45) | 29 (64.44) | ||
| Female (n = 34) | 18 (54.55) | 16 (35.56) | ||
| Age (yr) | 0.179 | 0.672 | ||
| ≥ 60 (n = 38) | 17 (51.52) | 21 (46.67) | ||
| < 60 (n = 40) | 16 (48.48) | 24 (53.33) | ||
| Smoking history | 0.694 | 0.405 | ||
| Yes (n=43) | 20 (60.61) | 23 (51.11) | ||
| No (n=35) | 13 (39.39) | 22 (48.89) | ||
| History of alcoholism | 1.319 | 0.251 | ||
| Yes (n=14) | 4 (12.12) | 10 (22.22) | ||
| No (n=64) | 29 (87.88) | 35 (77.78) | ||
| Lesion location | 0.602 | 0.740 | ||
| The upper thoracic area (n = 19) | 7 (21.21) | 12 (26.67) | ||
| The middle thoracic area (n = 34) | 16 (48.49) | 18 (40.00) | ||
| The lower thoracic area (n = 25) | 10 (30.30) | 15 (33.33) | ||
| Tumor size | 7.648 | 0.006 | ||
| ≥ 3 cm (n = 45) | 25 (75.76) | 20 (44.44) | ||
| < 3 cm (n = 33) | 8 (24.24) | 25 (55.56) | ||
| Differentiation | 7.093 | 0.008 | ||
| Low differentiation (n = 25) | 16 (48.48) | 9 (20.00) | ||
| Moderate + high differentiation (n = 53) | 17 (51.52) | 36 (80.00) | ||
| TNM staging | 6.825 | 0.009 | ||
| Stage I (n = 27) | 6 (18.18) | 21 (46.67) | ||
| Stage II + III (n = 51) | 27 (81.82) | 24 (53.33) | ||
| Adjuvant therapy | 0.946 | 0.331 | ||
| Chemotherapy (n = 52) | 24 (72.73) | 28 (62.22) | ||
| Radiotherapy (n = 26) | 9 (27.27) | 17 (37.78) | ||
| Expression of miR-1304 | 8.877 | 0.003 | ||
| High expression (n = 39) | 23 (69.70) | 16 (35.56) | ||
| Low expression (n = 39) | 10 (30.30) | 29 (64.44) |
| Factors | Assignment |
| Tumor size | ≥ 3 cm = 1, < 3 cm = 0 |
| Differentiation | Low differentiation = 1, Moderate + high differentiation = 0 |
| TNM stage | Stage I = 1, stage II + III = 0 |
| Expression of miR-1304 | High differentiation = 1, low differentiation = 0 |
| Recurrence | Recurrence = 1, non-recurrence = 0 |
| Factors | β | SD | χ2 | P value | Exp (HR) | HR 95%CI | |
| Up | Down | ||||||
| Tumor size | 1.861 | 0.646 | 8.297 | 0.004 | 6.429 | 1.812 | 22.803 |
| Differentiation | 1.811 | 0.665 | 7.414 | 0.006 | 6.114 | 1.661 | 22.506 |
| TNM stage | -1.801 | 0.675 | 7.118 | 0.008 | 0.165 | 0.044 | 0.620 |
| Expression of miR-1304 | 1.838 | 0.627 | 8.586 | 0.003 | 6.286 | 1.838 | 21.501 |
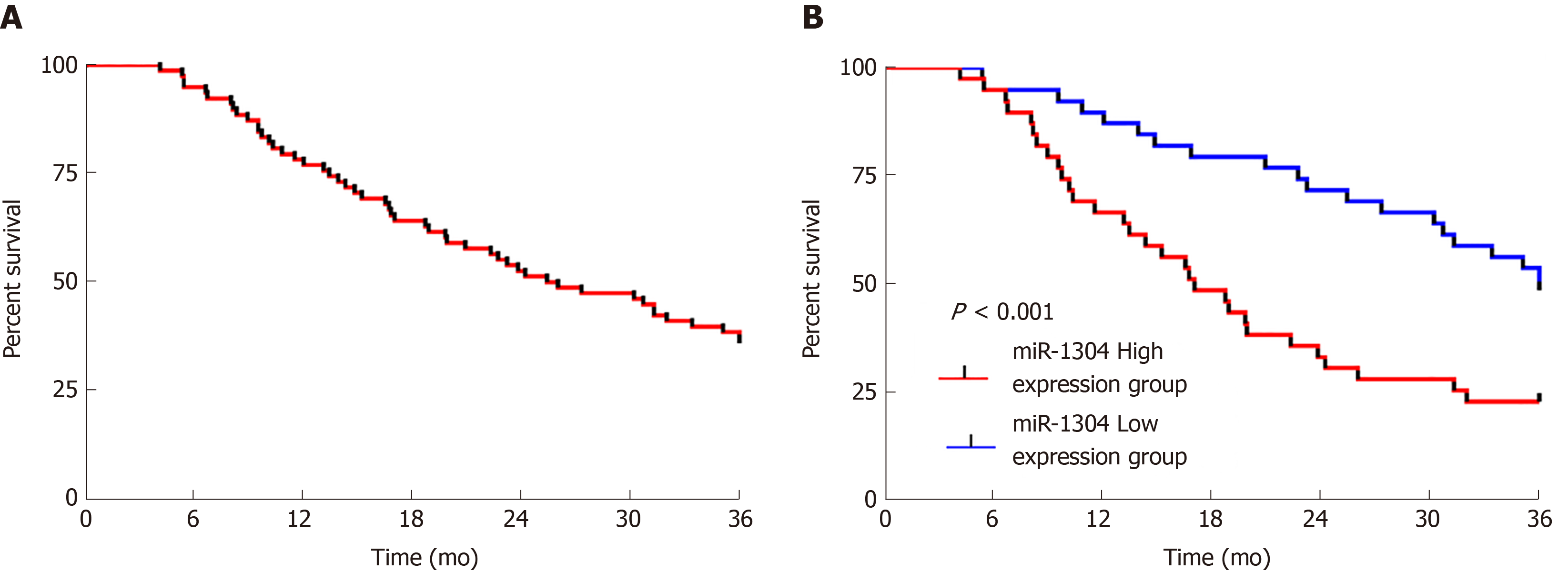
The patients were followed-up for 3 years. All 78 patients were successfully followed-up with no patients lost to follow-up, 30 of whom survived for 3 years, showing a 3-year survival rate of 38.46%. Survival curves were drawn according to the expression of miR-1304, which revealed that patients with low expression of miR-1304 showed a significantly higher survival rate than those with high expression (bP < 0.001). Univariate Cox regression analysis of the collected pathological data revealed that tumor size, differentiation, recurrence, and miR-1304 were prognostic factors for patients, and multivariate analysis revealed that these factors were independent predictors of prognosis (Figure 4 and Table 7).
| Factors | 3-yr univariate Cox | 3-yr multivariate Cox | ||
| P value | H (95%CI) | P value | HR (95%CI) | |
| Gender (male vs female) | 0.803 | 1.075 (0.608-1.903) | ||
| Age (≥ 60 yr vs < 65 yr) | 0.534 | 0.835 (0.473-1.474) | ||
| Lesion location (upper vs middle vs lower) | 0.427 | 1.166 (0.798-1.705) | ||
| Tumor size (≥ 3 cm vs < 3 cm) | 0.000 | 3.473 (1.827-6.601) | 0.010 | 2.402 (1.237-4.665) |
| Differentiation (low differentiation vs moderate + high differentiation) | 0.000 | 3.09 (1.724-5.539) | 0.015 | 2.153 (1.159-4.002) |
| TNM stage (stage I vs stage II + III) | 0.060 | 0.543 (0.287-1.027) | ||
| Adjuvant therapy plan (chemotherapy vs radiotherapy) | 0.939 | 1.025 (0.549-1.911) | ||
| Recurrence (recurrence vs non-recurrence) | 0.002 | 2.561 (1.408-4.657) | 0.034 | 1.949 (1.05-3.619) |
| miR-1304 (high expression vs low expression) | 0.001 | 2.614 (1.448-4.717) | 0.036 | 1.93 (1.044-3.565) |
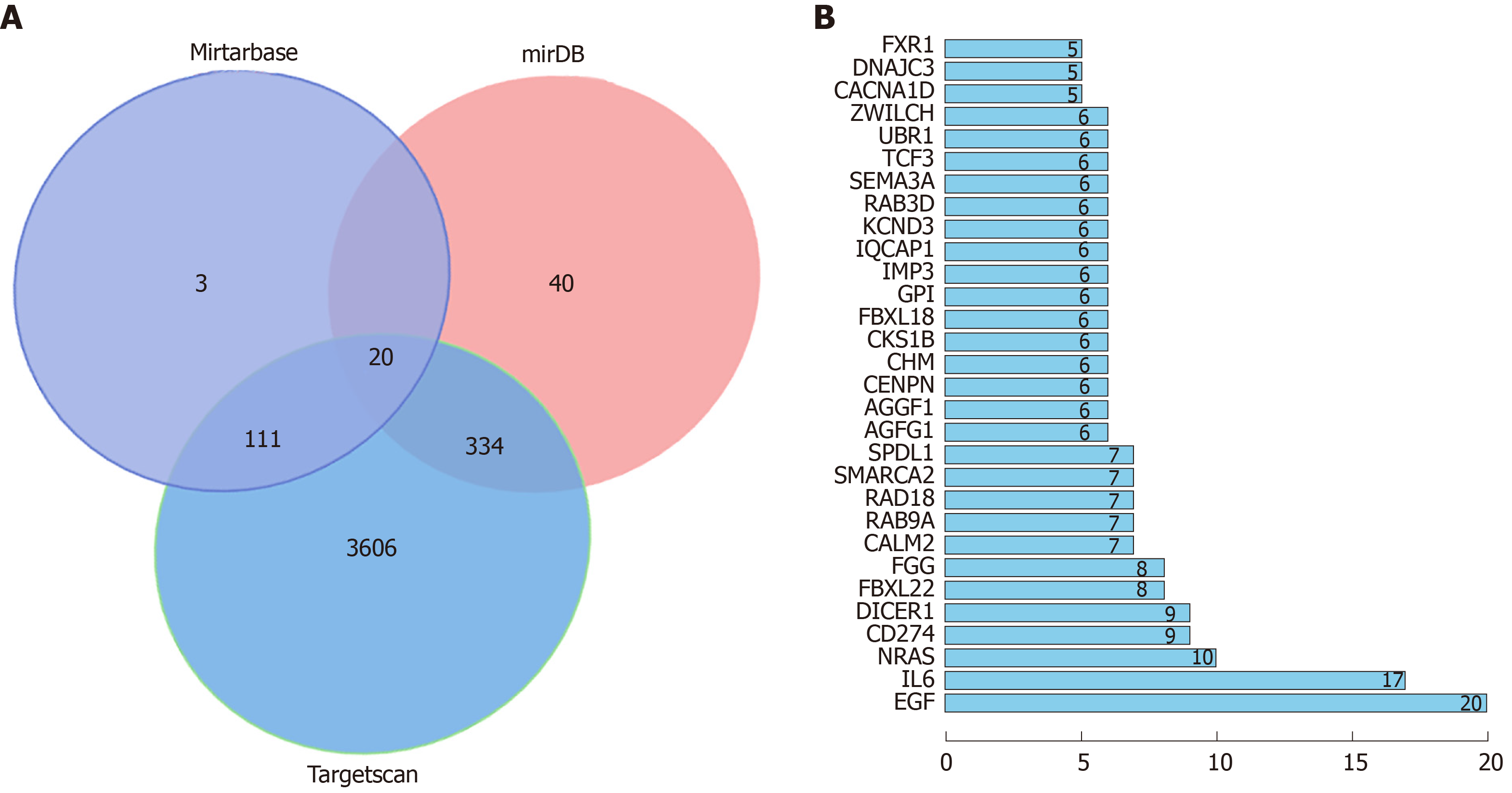
Three online target gene prediction websites for miR, namely, MiRTarBase, miRDB, and Targetscan, were used to predict target genes of miR-1304, and 20 target genes were predicted in total. GO enrichment was performed on target genes predicted through pairwise websites based on DAVID, and 18 functions with aP < 0.05 were found. KEGG analysis of the target genes was performed, and 11 signal pathways with aP < 0.05 were found. String analysis of protein co-expression found 269 relationship pairs, of which co-expression with epidermal growth factor (EGF) was the most common (Figure 5 and Tables 8-10).
| Gene | Target gene |
| miR-1304 | LYPD3, MYC, LMNB1, KLHL15, ZNF99, SPRYD4, CCNT2, PLEKHF2, DSEL, ADIPOR2, CBX5, DDX3X, SMAD5, DARS, PCGF3, FAM83H, RRAS, PDE3A, CALM2, and TMBIM6 |
| ID | Term | Gene | Count | P value |
| GO: 0043392 | Negative regulation of DNA binding | WFIKKN2, SP100, HAND2, HMOX1, HMGA2 | 5 | 0.001 |
| GO: 0051091 | Positive regulation of sequence-specific DNA binding transcription factor activity | IL6, SP100, HIPK2, PYCARD, HMGA2, TRIM22, TCF3, ATF2 | 8 | 0.001 |
| GO: 0000122 | Negative regulation of transcription from RNA polymerase II promoter | ATF7IP, SATB1, BACH2, SP100, GABPA, DICER1, YBX3, KLF16, ZEB2, HMGA2, ZNF345, ATF2, SUV39H2, VDR, OLIG3, MLX, PRMT6, HIPK2, ZNF431, TCF3, SMARCA2, SUDS3 | 22 | 0.002 |
| GO: 0006351 | Transcription, DNA-templated | XRCC5, ZNF555, IRX5, CCNT1, YBX3, BBX, TTLL5, ZNF250, ZEB2, ZNF652, ZNF184, VDR, OLIG3, PRMT6, ZNF426, KDM3B, BAZ2A, TCF3, MAP2K6, ATF7IP, SATB1, SP100, ZNF620, ZNF92, KLF16, SCAI, TRIM22, ATMIN, ZNF585B, FAM208A, SUV39H2, ZBTB25, EYA4, ZNF439, MLX, PARP14, HIPK2, ZNF431, ZNF432, SMARCA2, FOXD4L6, SUDS3, ZNF573, ZBTB8A | 44 | 0.002 |
| GO: 0006355 | Regulation of transcription, DNA-templated | ZNF555, IRX5, YBX3, BBX, ZNF250, ZNF652, ATF2, VDR, ZNF184, ZNF426, KDM3B, BAZ2A, INSR, TCF3, MAP2K6, SATB1, ZNF620, ZNF92, SCAI, AFF1, HMGA2, TRIM22, ZNF585B, FAM208A, ZBTB25, EYA4, TULP4, ZNF439, PARP14, CDKN2AIP, MLX, ZNF431, ZNF432, SMARCA2, ZBTB8A, ZNF573 | 36 | 0.003 |
| GO: 0005138 | Interleukin-6 receptor binding | IL6, PYCARD, ADAM17 | 3 | 0.004 |
| GO: 0015031 | Protein transport | SLC7A6OS, RAB9A, RAB3D, KIF17, DUOXA1, RAB39A, VPS52, PKDCC, HOOK3, GOPC, SNX30, SNX20, EXOC6B | 13 | 0.011 |
| GO: 0050892 | Intestinal absorption | F11R, VDR, KCNQ1 | 3 | 0.016 |
| GO: 0042147 | Retrograde transport, endosome to Golgi | SPAG9, RAB9A, STX16, VPS52, VPS26A | 5 | 0.017 |
| GO: 0032259 | Methylation | METTL8, TRMT10B, SETD9, CARNMT1, NNMT | 5 | 0.020 |
| GO: 0032290 | Peripheral nervous system myelin formation | NCMAP, DICER1 | 2 | 0.028 |
| GO: 0005509 | Calcium ion binding | PCDH11Y, PCDH11X, RPH3AL, DAG1, STIM1, MMP17, FSTL4, TTN, IQGAP1, CALU, ITPR2, PLSCR1, CALML4, TPT1, SYTL2, EGF, CALM2, CACNA1B | 18 | 0.029 |
| GO: 0008544 | Epidermis development | SATB1, STS, ELF3, INSR, SCEL | 5 | 0.033 |
| GO: 0003677 | DNA binding | XRCC5, ZNF555, AGFG1, KIAA1958, CCNT1, YBX3, BBX, ZNF250, ZEB2, ZNF345, ZNF652, ZNF184, VDR, OLIG3, HIST1H4C, ZNF426, BAZ2A, TCF3, SATB1, SP100, ZNF620, CHTF8, GABPA, HMGA2, ZBTB25, PLSCR1, ZNF439, RFC2, MLX, HIPK2, RAD18, ZNF432, ZNF573, ZBTB8A | 34 | 0.035 |
| GO: 0046872 | Metal ion binding | ZNF555, MGAT5B, AGFG1, DICER1, ZNF250, ZEB2, ZNF345, ZNF652, ATF2, ZFC3H1, ZNF184, CLEC17A, FGG, HMOX1, ZC3H12B, ZNF426, KDM3B, STS, KCND3, NRXN3, ZNF620, ZNF92, KLF16, RPH3AL, ATMIN, ZNF585B, ZBTB25, EYA4, ZNF439, GNAQ, ADAM17, ZNF431, YME1L1, ZNF432, ADAM12, ZNHIT6, CACNA1D, UGP2, ZBTB8A, ZNF573 | 40 | 0.042 |
| GO: 0070644 | Vitamin D response element binding | VDR, TCF3 | 2 | 0.042 |
| GO: 0005794 | Golgi apparatus | STS, RAB39A, FGFRL1, VPS52, PKDCC, PRKG1, SART1, CALU, HOOK3, PLSCR1, NRAS, COPB1, STX16, GOPC, CNTNAP2, CAND1, B4GALT7, TRAPPC3, SLC30A7 | 19 | 0.044 |
| GO: 0007173 | Epidermal growth factor receptor signaling pathway | NRAS, ADAM17, EGF, IQGAP1 | 4 | 0.045 |
| ID | Term | Genes | Count | P value |
| hsa04912 | GnRH signaling pathway | NRAS, GNAQ, CACNA1D, MAP2K6, CALM2, ITPR2 | 6 | 0.007 |
| hsa04725 | Cholinergic synapse | NRAS, GNAQ, CACNA1D, KCNQ1, ITPR2, CACNA1B | 6 | 0.015 |
| hsa04022 | cGMP-PKG signaling pathway | GNAQ, PRKG1, CACNA1D, INSR, CALM2, ITPR2, ATF2 | 7 | 0.017 |
| hsa04925 | Aldosterone synthesis and secretion | GNAQ, CACNA1D, CALM2, ITPR2, ATF2 | 5 | 0.021 |
| hsa04728 | Dopaminergic synapse | GNAQ, CACNA1D, CALM2, ITPR2, ATF2, CACNA1B | 6 | 0.026 |
| hsa05164 | Influenza A | IL6, AGFG1, PYCARD, DNAJC3, MAP2K6, HLA-DRA, ATF2 | 7 | 0.026 |
| hsa04540 | Gap junction | NRAS, GNAQ, PRKG1, EGF, ITPR2 | 5 | 0.028 |
| hsa04066 | HIF-1 signaling pathway | IL6, PFKFB3, HMOX1, EGF, INSR | 5 | 0.037 |
| hsa04915 | Estrogen signaling pathway | NRAS, GNAQ, CALM2, ITPR2, ATF2 | 5 | 0.040 |
| hsa04922 | Glucagon signaling pathway | GNAQ, CALM2, G6PC2, ITPR2, ATF2 | 5 | 0.040 |
| hsa04730 | Long-term depression | NRAS, GNAQ, PRKG1, ITPR2 | 4 | 0.043 |
Esophageal carcinoma, a malignant gastrointestinal tumor, is the 8th most common malignant tumor in the world[21]. In 2018, there were more than 500000 new and deceased esophageal carcinoma patients worldwide, and the morbidity and mortality of esophageal carcinoma showed an upward trend[22]. The main reasons for this phenomenon are as follows: Firstly, the diet structure of patients has changed. Secondly, esophageal carcinoma has no obvious clinical characteristics in the early stage, so almost all patients are already in the middle or late stage when admitted to hospital, and have missed the best treatment opportunity. Finally, there is a lack of tumor markers with high specificity for esophageal carcinoma. These factors eventually lead to a rise in mortality and morbidity[23,24]. Therefore, the identification of a clinical diagnostic marker with high specificity is essential to solve this problem.
In this study, we found, for the first time, that miR-1304 was highly expressed in patients with esophageal carcinoma. miR has been a hot research topic in various fields in recent years. As a short-chain non-coding RNA, it can inhibit transcription and the expression of downstream target genes by regulating them through the 3-untranslated regions[25]. miR-1304 is an important member of the miR family. Previous studies have found that miR-1304 shows differential expression in patients with lung cancer, and can inhibit the growth of non-small cell lung cancer by regulating heme oxygenase-1[26]. However, there are few studies on esophageal carcinoma. miR-1304 was first found to be highly expressed in patients with esophageal carcinoma based on TCGA, which indicated that miR-1304 is expected to become a potential diagnostic indicator for esophageal carcinoma. Therefore, we conducted a clinical experiment, to determine if the expression of miR-1304 in the serum and tissues of patients was the same as that in the database; the expression of miR-1304 in tissues was positively correlated with that in serum, and the AUC of expression in the ROC curve was greater than 0.9. This further confirmed the role of miR-1304 in esophageal carcinoma, and indicated that miR-1304 could be a potential diagnostic indicator for esophageal carcinoma. Moreover, we analyzed the correlation between high and low expression of miR-1304 and pathological data, and found that patients with high expression of miR-1304 had significantly higher rates of tumor size ≥ 3 cm, low differentiation, and stage II + III disease, and the expression of miR-1304 had certain diagnostic value.
At present, the best treatment for esophageal carcinoma is radical resection, which can effectively improve the prognosis of patients to a certain extent together with postoperative radiotherapy and chemotherapy[27]. However, treatment of patients with esophageal carcinoma is prone to failure due to micrometastasis of some lesions and limited lymph node dissection; thus, resulting in local regional recurrence or distal metastasis[28]. At present, recurrence of esophageal carcinoma in patients is mainly judged by the identification of metastasis through imaging after treatment. If indices were available to predict metastasis in patients before treatment, this would allow changes in the treatment of patients and avoid the recurrence of esophageal carcinoma[29]. In this study, 33 of 78 patients developed recurrence of esophageal carcinoma, a recurrence rate of 42.31%, which was consistent with national and international studies[30,31]. We collected clinical data, grouped the patients, performed Logistic regression analysis, and found that tumor size, differentiation, TNM stage, and miR-1304 were independent risk factors for prognosis in these patients. In addition, we drew ROC curves and found that the AUC of miR-1304 for predicting recurrence of esophageal carcinoma was larger than 0.7, and larger than that for tumor size, differentiation, and TNM stage. Many studies have confirmed that tumor size, differentiation and TNM stage are independent risk factors for recurrence in patients[32,33], but this is the first report to discover that miR-1304 may be an independent risk factor for the recurrence of esophageal carcinoma, which suggests that the expression of miR-1304 has certain predicative value in recurrence in these patients.
In addition, this study followed patients to assess their 3-year survival rate, and found that the 3-year overall survival rate was 38.46%, which was consistent with national and international studies[34,35]. We analyzed the 3-year survival of patients according to the expression of miR-1304, and patients with low expression of miR-1304 showed a significantly higher 3-year survival rate than those with high expression. Prognostic analysis revealed that miR-1304 may be an independent prognostic factor for 3-year survival in patients with esophageal carcinoma. Based on the above studies, we can confirm the clinical diagnostic and prognostic value of miR-1304 for esophageal carcinoma, but the specific mechanism of miR-1304 remains unclear. Therefore, we conducted a bioinformatics analysis, which revealed that the three predictive networks predicted a total of 20 target genes of miR-1304. GO and KEGG enrichment analysis based on DAVID and KOBAS found 18 functions with aP < 0.05 and 11 signal pathways with aP < 0.05, respectively. It is worth mentioning that previous reports indicated that HIF-1 and GnRH signaling pathways were involved in the occurrence and development of esophageal carcinoma[36,37], which may be our main research direction in the future. Finally, we plotted a protein-protein interaction co-expression spectrum, and found that relationship pairs with the EGF gene were the most common. EGF, a member of epidermal growth factor super family, is a powerful mitogenic factor with important functions in growth, proliferation and differentiation of various cells, and early studies have shown that EGF is closely related to the prognosis of esophageal carcinoma. Whether miR-1304 can regulate EGF and suppress the occurrence and development of esophageal carcinoma is the main direction of our future research.
This study has preliminarily proved the clinical value of miR-1304 in esophageal carcinoma, but it has certain limitations. Firstly, we did not carry out basic experiments, and did not clarify the relevant mechanisms of miR-1304 in esophageal carcinoma. Secondly, the specimens in this study were basic, and the study only compared the difference in expression of serum miR-1304 between patients with esophageal carcinoma and normal subjects. Whether differences in miR-1304 expression exist between patients with esophageal carcinoma and patients with benign esophageal lesion requires further investigation. Therefore, we hope to carry out basic experiments in future research, use diverse specimens and to compare miR-1304 and common tumor markers in esophageal carcinoma to further confirm the role of miR-1304 in esophageal carcinoma to supplement the present findings.
In conclusion, miR-1304 can be used as a potential indicator for the diagnosis and recurrence of esophageal carcinoma, and for survival of these patients.
Esophageal carcinoma is a common digestive tract cancer, which frequently recurs after treatment. MiR-1304 is a newly discovered non-coding RNA, which shows differential expression in other cancers, but its clinical value in esophageal carcinoma remains unclear.
To identify potential diagnostic and prognostic indicators of esophageal cancer recurrence.
To determine the diagnostic and prognostic value of miR-1304 in esophageal carcinoma recurrence.
Data on miRs with potential differences in esophageal carcinoma were screened from the Cancer Genome Atlas. A quantitative real-time polymerase chain reaction was employed to determine the expression of miR-1304 in esophageal carcinoma patients, and the clinicopathological features of these patients were analyzed. Based on the analysis of screened data and the expression of miR-1304 in esophageal carcinoma patients, the function of miR-1304 was evaluated. Moreover, the patients were followed-up to analyze prognosis. Target genes of miR-1304 were predicted, and the functions of these genes were analyzed.
The expression of miR-1304 in the tissues and serum of patients increased, similar to that seen in the database. Patients with high expression of miR-1304 had increased rates of tumor ≥ 3 cm, low differentiation and stage II + III disease. MiR-1304 had diagnostic value in identifying esophageal carcinoma, tumor size, differentiation and TNM staging. Tumor size, differentiation, TNM staging, and miR-1304 were independent risk factors for recurrence of esophageal carcinoma, and had certain predictive and diagnostic value for recurrence of this disease. Patients with high expression of miR-1304-3p had a lower survival rate. Multivariate analysis revealed that tumor size, differentiation, recurrence and miR-1304 were independent factors for prognosis. Furthermore, the target genes had 18 functions with aP < 0.05 according to gene ontology enrichment analysis and 11 signal pathways with aP < 0.05 according to the Kyoto Encyclopedia of Genes and Genomes. In addition, there were 269 relationship pairs according to String analysis of protein co-expression, of which the co-expression pairs with epidermal growth factor were the most common.
MiR-1304 can be used as a potential indicator for the diagnosis and recurrence of esophageal carcinoma, and survival of patients.
In future research, the molecular mechanism of miR-1304 in esophageal carcinoma will be studied.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lee CL, Mohamed SY, S-Editor: Zhang L L-Editor: Webster JR E-Editor: Ma YJ
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Chen WQ, Zheng RS, Baade PD, Zhang SW, Zeng HM, Bray FB, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Probst A, Aust D, Märkl B, Anthuber M, Messmann H. Early esophageal cancer in Europe: endoscopic treatment by endoscopic submucosal dissection. Endoscopy. 2015;47:113-121. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Alsop BR, Sharma P. Esophageal Cancer. Gastroenterol Clin North Am. 2016;45:399-412. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Rubenstein JH, Shaheen NJ. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology. 2015;149:302-17.e1. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Parry K, Visser E, van Rossum PS, Mohammad NH, Ruurda JP, van Hillegersberg R. Prognosis and Treatment After Diagnosis of Recurrent Esophageal Carcinoma Following Esophagectomy with Curative Intent. Ann Surg Oncol. 2015;22 Suppl 3:S1292-S1300. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Sun Z, Liu X, Song JH, Cheng Y, Liu Y, Jia Y, Meltzer SJ, Wang Z. TNFAIP8 overexpression: a potential predictor of lymphatic metastatic recurrence in pN0 esophageal squamous cell carcinoma after Ivor Lewis esophagectomy. Tumour Biol. 2016;37:10923-10934. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Wang J, Chen J, Sen S. MicroRNA as Biomarkers and Diagnostics. J Cell Physiol. 2016;231:25-30. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-463. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321-333. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer--a brief overview. Adv Biol Regul. 2015;57:1-9. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Romaine SP, Tomaszewski M, Condorelli G, Samani NJ. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015;101:921-928. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee NPY, Law S, Xu LY, Li EM, Chan KW, Qin YR, Guan XY, He QY, Cheung ALM. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene. 2017;36:3986-4000. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Li SP, Su HX, Zhao D, Guan QL. Plasma miRNA-506 as a Prognostic Biomarker for Esophageal Squamous Cell Carcinoma. Med Sci Monit. 2016;22:2195-2201. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Xu CZ, Shi RJ, Chen D, Sun YY, Wu QW, Wang T, Wang PH. Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int J Clin Exp Pathol. 2013;6:2745-2756. [PubMed] [Cited in This Article: ] |
| 18. | Zhang D, Zheng Y, Wang Z, Huang Q, Cao X, Wang F, Liu S. Comparison of the 7th and proposed 8th editions of the AJCC/UICC TNM staging system for esophageal squamous cell carcinoma underwent radical surgery. Eur J Surg Oncol. 2017;43:1949-1955. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Freilich J, Hoffe SE, Almhanna K, Dinwoodie W, Yue B, Fulp W, Meredith KL, Shridhar R. Comparative outcomes for three-dimensional conformal versus intensity-modulated radiation therapy for esophageal cancer. Dis Esophagus. 2015;28:352-357. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Ljvak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Zeng HM, Zheng RS, Zhang SW, Zuo TT, Xia CF, Zou XN, Chen WQ. Esophageal cancer statistics in China, 2011: Estimates based on 177 cancer registries. Thorac Cancer. 2016;7:232-237. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Yang CS, Chen X, Tu S. Etiology and Prevention of Esophageal Cancer. Gastrointest Tumors. 2016;3:3-16. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | López-Lázaro M. A local mechanism by which alcohol consumption causes cancer. Oral Oncol. 2016;62:149-152. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Zhang L, Yang CS, Varelas X, Monti S. Altered RNA editing in 3' UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci Rep. 2016;6:23226. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Li CG, Pu MF, Li CZ, Gao M, Liu MX, Yu CZ, Yan H, Peng C, Zhao Y, Li Y, Ma ZL, Qi XM, Wang YZ, Miao LL, Ren J. MicroRNA-1304 suppresses human non-small cell lung cancer cell growth in vitro by targeting heme oxygenase-1. Acta Pharmacol Sin. 2017;38:110-119. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Pasquali S, Yim G, Vohra RS, Mocellin S, Nyanhongo D, Marriott P, Geh JI, Griffiths EA. Survival After Neoadjuvant and Adjuvant Treatments Compared to Surgery Alone for Resectable Esophageal Carcinoma: A Network Meta-analysis. Ann Surg. 2017;265:481-491. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Hiyoshi Y, Morita M, Kawano H, Otsu H, Ando K, Ito S, Miyamoto Y, Sakamoto Y, Saeki H, Oki E, Ikeda T, Baba H, Maehara Y. Clinical significance of surgical resection for the recurrence of esophageal cancer after radical esophagectomy. Ann Surg Oncol. 2015;22:240-246. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Goense L, van Rossum PS, Reitsma JB, Lam MG, Meijer GJ, van Vulpen M, Ruurda JP, van Hillegersberg R. Diagnostic Performance of ¹⁸F-FDG PET and PET/CT for the Detection of Recurrent Esophageal Cancer After Treatment with Curative Intent: A Systematic Review and Meta-Analysis. J Nucl Med. 2015;56:995-1002. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Liu Q, Cai XW, Wu B, Zhu ZF, Chen HQ, Fu XL. Patterns of failure after radical surgery among patients with thoracic esophageal squamous cell carcinoma: implications for the clinical target volume design of postoperative radiotherapy. PLoS One. 2014;9:e97225. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Xu Y, Chen Q, Yu X, Zhou X, Zheng X, Mao W. Factors influencing the risk of recurrence in patients with esophageal carcinoma treated with surgery: A single institution analysis consisting of 1002 cases. Oncol Lett. 2013;5:185-190. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Ninomiya I, Okamoto K, Tsukada T, Kinoshita J, Oyama K, Fushida S, Osugi H, Ohta T. Recurrence patterns and risk factors following thoracoscopic esophagectomy with radical lymph node dissection for thoracic esophageal squamous cell carcinoma. Mol Clin Oncol. 2016;4:278-284. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Kofoed SC, Calatayud D, Jensen LS, Helgstrand F, Achiam MP, De Heer P, Svendsen LB; Danish Esophageal, Cardia and Stomach Cancer Group. Intrathoracic anastomotic leakage after gastroesophageal cancer resection is associated with increased risk of recurrence. J Thorac Cardiovasc Surg. 2015;150:42-48. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Larue RTHM, Klaassen R, Jochems A, Leijenaar RTH, Hulshof MCCM, van Berge Henegouwen MI, Schreurs WMJ, Sosef MN, van Elmpt W, van Laarhoven HWM, Lambin P. Pre-treatment CT radiomics to predict 3-year overall survival following chemoradiotherapy of esophageal cancer. Acta Oncol. 2018;57:1475-1481. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Yerokun BA, Sun Z, Yang CJ, Gulack BC, Speicher PJ, Adam MA, D'Amico TA, Onaitis MW, Harpole DH, Berry MF, Hartwig MG. Minimally Invasive Versus Open Esophagectomy for Esophageal Cancer: A Population-Based Analysis. Ann Thorac Surg. 2016;102:416-423. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Zhu Y, Zang Y, Zhao F, Li Z, Zhang J, Fang L, Li M, Xing L, Xu Z, Yu J. Inhibition of HIF-1α by PX-478 suppresses tumor growth of esophageal squamous cell cancer in vitro and in vivo. Am J Cancer Res. 2017;7:1198-1212. [PubMed] [Cited in This Article: ] |
| 37. | Lee G, Cheung AP, Ge B, Zhu M, Giolma B, Li B, Wong E, Li Y, Wang Y, Chen Z, Gu J. CA215 and GnRH receptor as targets for cancer therapy. Cancer Immunol Immunother. 2012;61:1805-1817. [PubMed] [DOI] [Cited in This Article: ] |