Published online Feb 14, 2020. doi: 10.3748/wjg.v26.i6.614
Peer-review started: August 13, 2019
First decision: October 14, 2019
Revised: December 25, 2019
Accepted: January 11, 2020
Article in press: January 11, 2020
Published online: February 14, 2020
Processing time: 185 Days and 2.6 Hours
This study determined the composition and diversity of intestinal microflora in patients with colorectal adenoma (CRA), which may provide precedence for investigating the role of intestinal microflora in the pathogenesis of colorectal tumors, the composition of intestinal microflora closely related to CRA, and further validating the possibility of intestinal flora as a biomarker of CRA.
To study the relationship between intestinal microflora and CRA.
This is a prospective control case study from October 2014 to June 2015 involving healthy volunteers and patients with advanced CRA. High-throughput sequencing and bioinformatics analysis were used to investigate the composition and diversity of intestinal microflora in 36 healthy subjects and 49 patients with advanced CRA. Endpoints measured were operational taxonomic units of intestinal flora, as well as their abundance and diversity (α and β types).
In this study, the age, gender, body mass index, as well as location between controls and patients had no significant differences. The mucosa-associated gut microbiota diversity and bacterial distribution in healthy controls and colorectal adenomas were similar. The operational taxonomic unit, abundance, and α and β diversity were all reduced in patients with CRA compared to controls. At the phylum level, the composition of intestinal microflora was comparable between patients and controls, but the abundance of Proteobacteria was increased, and Firmicutes and Bacteroides were significantly decreased (P < 0.05). The increase in Halomonadaceae and Shewanella algae, and reduction in Coprococcus and Bacteroides ovatus, could serve as biomarkers of CRA. High-throughput sequencing confirms the special characteristics and diversity of intestinal microflora in healthy controls and patients with CRA.
The diversity of intestinal microflora was decreased in patients with CRA. An increase in Halomonadaceae and Shewanella algae are markers of CRA.
Core tip: Colorectal adenomas (CRAs), the most important pre-cancerous lesions in colorectal cancer, have a close relationship with intestinal microflora. However, the mechanism is not clear. Early detection of CRA can effectively reduce the morbidity and mortality of colorectal cancer. This study analyzes the composition and diversity of intestinal microflora in patients with CRA through 16s rDNA gene sequencing. It was proven that the diversity of intestinal microflora in patients with CRA decreased, and that an increase in the number of Halomonadaceae and Shewanella algae may be biomarkers of CRA, which provides a basis for future research.
- Citation: Wang WJ, Zhou YL, He J, Feng ZQ, Zhang L, Lai XB, Zhou JX, Wang H. Characterizing the composition of intestinal microflora by 16S rRNA gene sequencing. World J Gastroenterol 2020; 26(6): 614-626
- URL: https://www.wjgnet.com/1007-9327/full/v26/i6/614.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i6.614
Colorectal adenomas (CRAs) are the main pre-cancerous lesions in colorectal cancer (CRC). CRC is the second most common malignancy worldwide, and sporadic CRC exhibits a typical progression from adenoma to cancer. Advanced adenomas have a high risk for CRC[1], and the following characteristics are indicative of advanced CRA: Diameter of the polyp or lesion is ≥ 10 mm; villous structure (accounts for > 25% of the villous or mixed adenoma); and if high-grade intraepithelial neoplasia is present[2]. Previous studies have shown that carcinogenesis of the intestinal mucosa is not only a multi-step process and a result of accumulating multi-gene mutations, but also closely related to changes in intestinal microflora[3]. In the present study, 16S rDNA gene sequencing was used to analyze the composition of intestinal microflora in patients with advanced CRA, to investigate the composition of intestinal microflora, and to identify biomarkers of CRA. This knowledge may be helpful for elucidating CRC pathogenesis from a micro-ecological perspective.
Sample collection and processing: This study was approved by the Ethics Committee of the First People’s Hospital of Guangzhou, and informed consent was obtained from all patients. Biopsy samples from advanced CRA and adjacent normal mucosa were obtained from 49 patients who underwent colonoscopies at the First People’s Hospital of Guangzhou between October 2014 and June 2015. Six endoscopically-obtained biopsy samples were taken from each patient, three from advanced CRA and three from adjacent normal mucosa (5.0 cm beyond the CRA tissue). In addition, normal intestinal samples were collected from 36 healthy subjects as controls. The mean diameter of advanced CRA was ≥ 1 cm, and the advanced CRA were pathologically confirmed to be advanced. Tissues were stored in liquid nitrogen immediately after biopsy sampling, then transferred into a refrigerator at -80 degrees Celsius within 4 h.
Inclusion criteria: (1) Patients in generally good condition with stable vital signs; (2) Age: 18-80 years old, no gender restrictions; (3) Pathological diagnosis of intestinal adenoma; (4) No special diet (vegetarian food, etc.); and (5) No diabetes, history of liver cirrhosis, chronic renal failure, drug chemotherapy or surgical treatment.
Healthy volunteers: Criteria 3 in the inclusion criteria was changed to: No obvious lesions were found in the colonoscopy.
Exclusion criteria of research objects: (1) Antibiotics, probiotics or prebiotics (including yogurt) were used within four weeks before sampling; (2) Familial or hereditary colorectal adenoma or tumor, other system or organ tumors; and (3) Recent history of bleeding or taking anticoagulants.
Main reagents: Tiangen centrifugal columnar genomic DNA extraction kit, primers for polymerase chain reaction (PCR) (Shenzhen BGI Gene Company, Shenzhen, China), and Premixtaq (version 2.0; Takara, Dalian, Liaoning, China) were used in the present study.
Mucosal tissues (0.5 cm in diameter) were mixed with 200 μL of GA buffer in a 1.5 cm centrifuge tube, followed by addition of 3 mm beads. After homogenization by vortexing at 5000-6000 rpm (g values are preferred) for 3-4 min, the cell suspension was harvested. Genomic DNA was extracted with a Tiangen centrifugal columnar genomic DNA extraction kit according to the manufacturer’s instructions. DNA concentrations were measured with a microplate reader. After 1% agarose gel electrophoresis for 40 min at 150 v, gene integrity was confirmed. DNA was then stored at -20 degrees Celsius.
Common primers with a barcode for the PCR of the 16S rDNA V4 variable region were as follows: 515F, GTGCCAGCMGCCGCGGTAA; 806R, GGACTA-CHVGGGTWTCTAAT (Shenzhen BGI Gene Company). The 20-μL mixture included 0.5 μL of forward primer, 0.5 μL of reverse primer, 1 μL of template, 10 μL of Premix Taq, and 8 μL of distilled water. PCR was performed under the following conditions: Pre-denaturation at 94 degrees Celsius for 2 min; 20 cycles of denaturation at 94 degrees Celsius for 30 s; annealing at 58 degrees Celsius for 30 s; extension at 72 degrees Celsius for 1 min; and a final extension at 72 degrees Celsius for 10 min.
PCR products of the 16S rDNA V4 variable region were subjected to paired-end sequencing in an Illumina platform Miseq PE 250 (Shenzhen BIG Gene Company). Tags of the variable region were obtained with Fast Length Adjustment of Short reads (FLASH; v1.2.11). The connected tags were clustered as an operational taxonomic unit (OTU) with USEARCH (v7.0.1090). Then, the species classification of the OTU was performed by OTU noting. The OTU and its abundance analysis, sample diversity analysis, principal component analysis of samples, differential analysis, and analysis of biomarkers were performed using bioinformatics analysis.
Clinical data were analyzed with IBM SPSS 21.0 version. Quantitative data with a normal distribution are expressed as the mean ± SD. Data with a normal distribution were compared with an independent t-test. Categorical data were compared with a χ2 test. P < 0.05 was considered statistically significant.
Clinical characteristics were compared between controls and patients with CRA. There were no significant differences in age, gender, body mass index, or site of CRA between the two groups (P > 0.05) (Table 1).
| Characteristic | Normal mucosa | Advanced CRA | P value |
| Age in yr | 58.9 ± 7.2 | 62.9 ± 9.9 | 0.058 |
| Gender | |||
| Male | 18 | 28 | 0.448 |
| Female | 18 | 20 | |
| BMI | 25.2 ± 3.6 | 26.3 ± 3.9 | 0.229 |
| Site | |||
| Right colon | 9 | 10 | 0.651 |
| Left colon | 27 | 39 | |
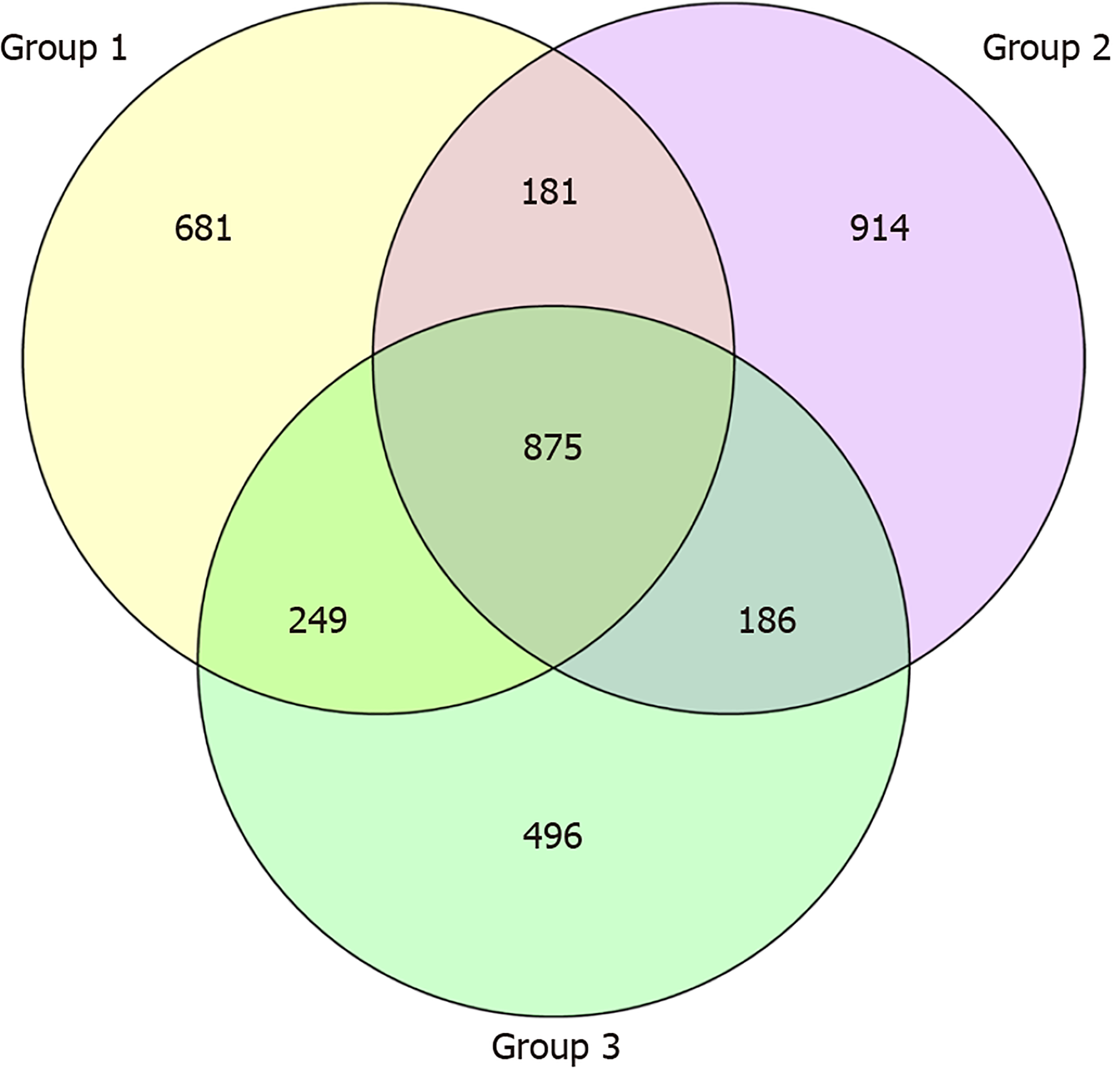
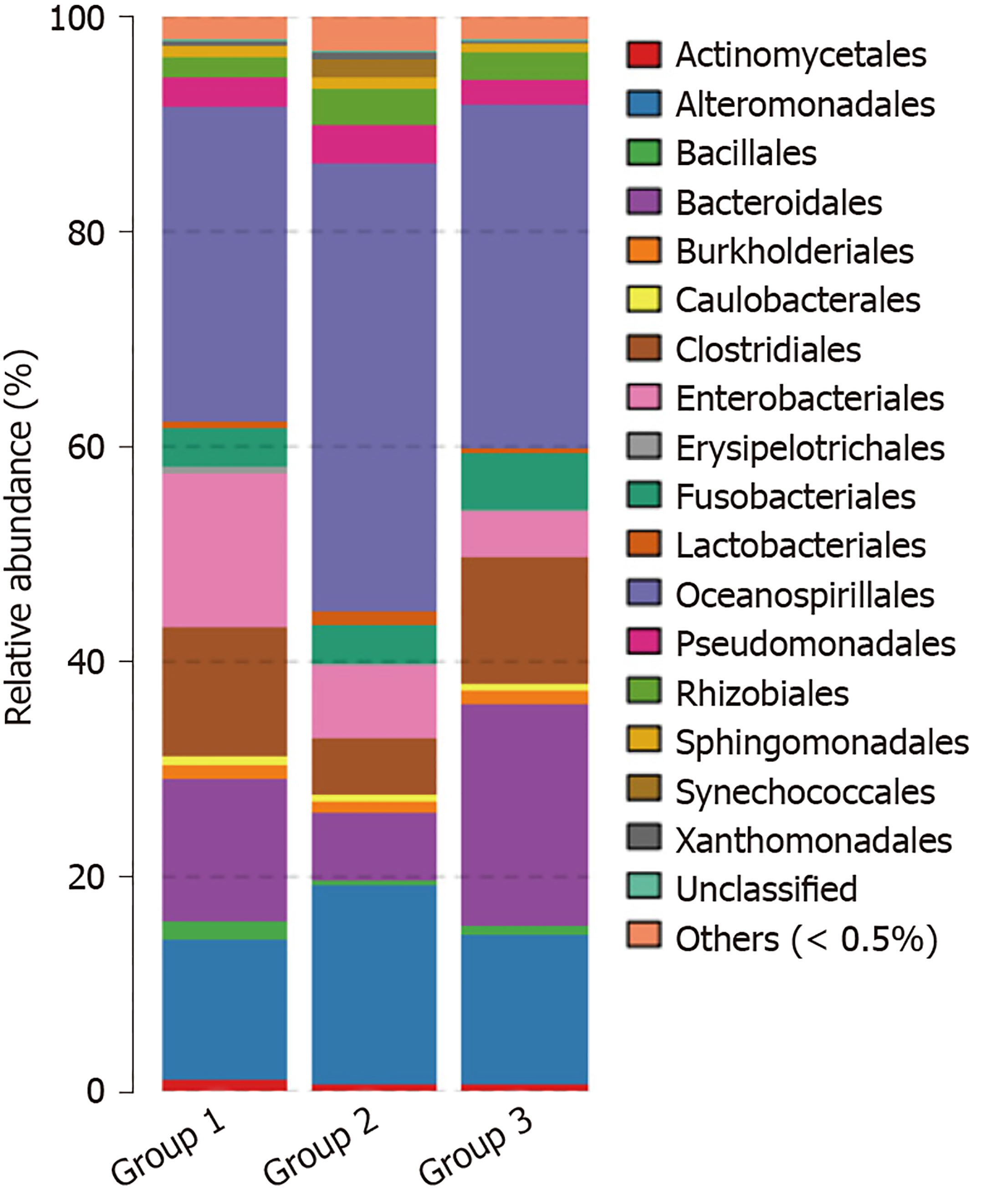
OTU and its abundance: (1) Group 1: Normal mucosa; (2) Group 2: CRA mucosa; and (3) Group 3: Adjacent normal mucosa of CRA specimens (Figure 1).
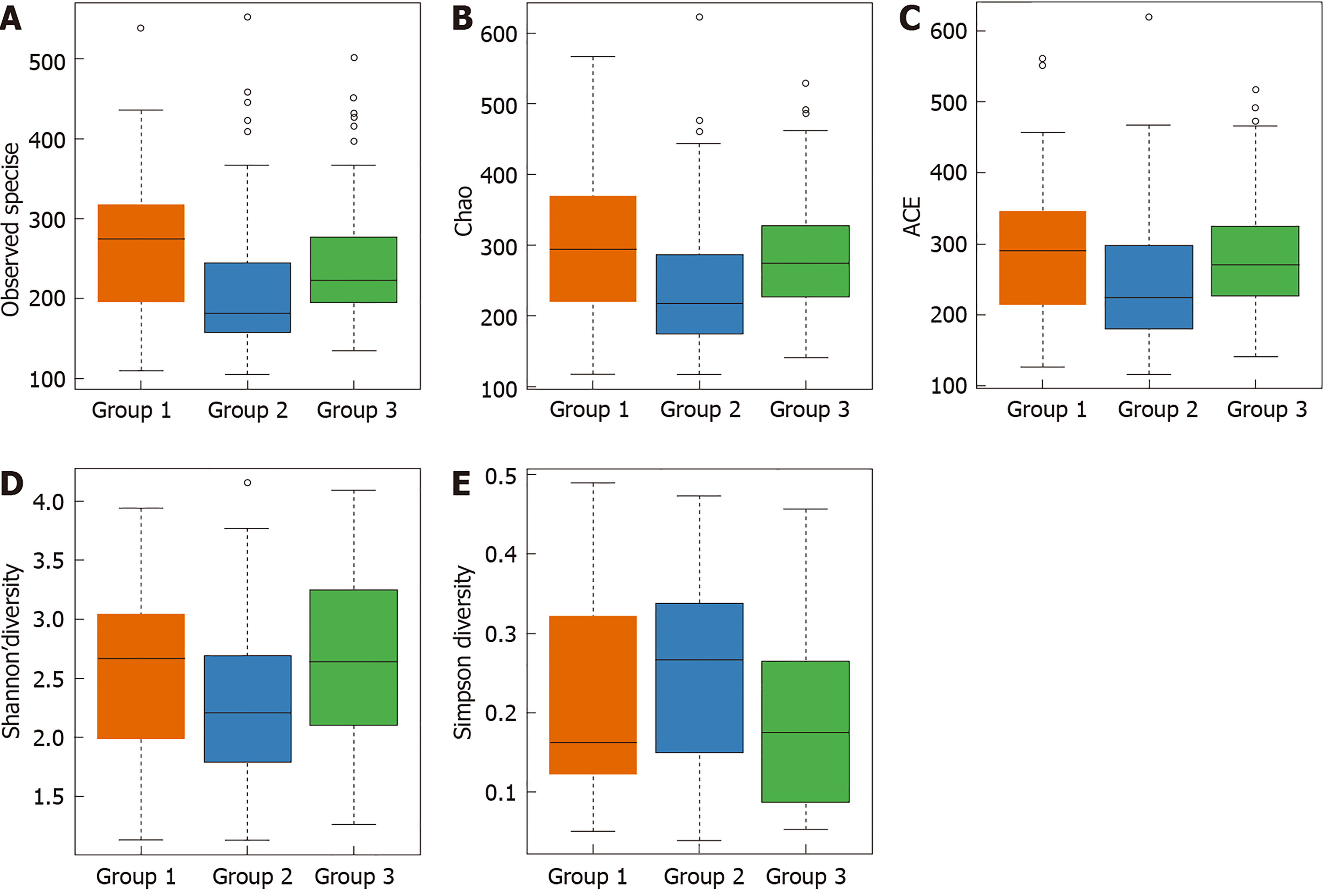
Analysis of alpha diversity: Species diversity was compared among normal mucosa (group 1), CRA mucosa (group 2), and adjacent normal mucosa of CRA specimens (group 3). There were significant differences in the observed OTU, Chao, the index used to estimate the number of OTU in the community (ACE), Shannon index, and Simpson diversity index among these tissues (P < 0.05). The OTU in the CRA group was significantly reduced compared with the control group, but the OTU was comparable between adjacent normal mucosa and normal mucosa specimens. This finding suggests that the species diversity of microflora was reduced in patients with CRA, and that species diversity was similar between CRA patients and controls (Table 2 and Figure 2).
| α | Normal mucosa | Advanced CRA | Adjacent normal mucosa | P value |
| Sobs | 271 ± 90 | 215 ± 86 | 253 ± 91 | < 0.001a |
| Chao | 301 ± 102 | 249 ± 104 | 291 ± 95 | 0.008 |
| ACE | 296 ± 102 | 253 ± 105 | 291 ± 93 | 0.020 |
| Shannon | 2.56 ± 0.68 | 2.30 ± 0.71 | 2.68 ± 0.72 | 0.028 |
| Simpson | 0.21 ± 0.12 | 0.26 ± 0.11 | 0.20 ± 0.12 | 0.031 |
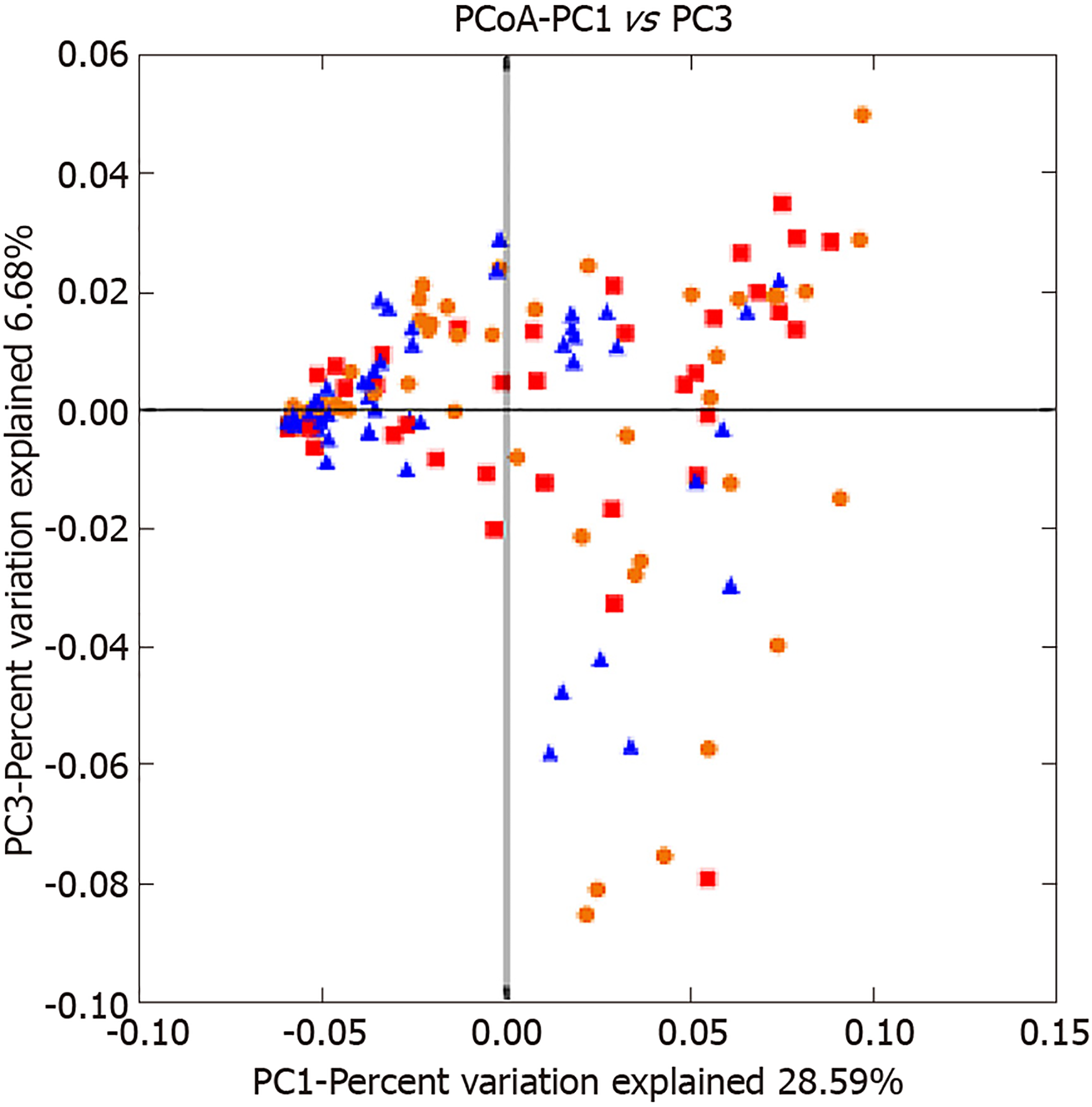
Analysis of beta diversity: Beta diversity reflects differences in species diversity among samples. As shown in the principal coordinates analysis figure, the distance between samples was short and aggregated in the CRA group, but the distance was relatively long and displayed a diffuse distribution (Figure 3). In the heat map figure, all samples were classified as 1 of 2 types, and 90% of the polyps were classified as the low abundance group, but samples were evenly distributed in the control group (Figure 4). Thus, the species have small differences, high similarities, and favorable clustering.
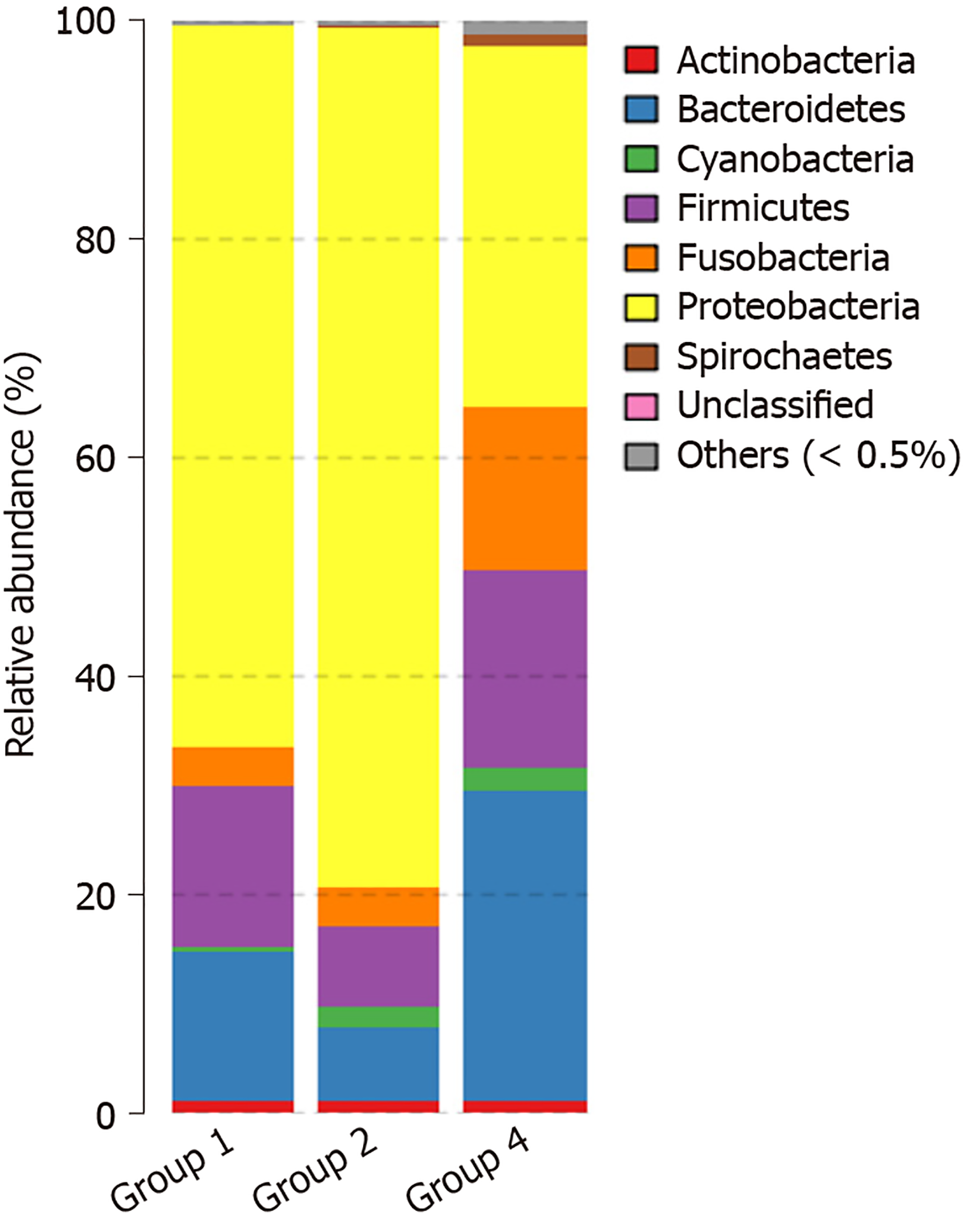
Species noting, principal component analysis, and differential analysis: The OTU was compared with the database, and species were noted at the phylum, class, order, family, genus, and species levels for delineation of the profiling histogram. In this study, analysis was mainly performed at the phylum, order, genus, and species levels. Results showed that the species were mainly distributed into the following seven types of phyla: Proteobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Actinobacteria, Spirochaetes, and Cyanobacteria. The principal component distribution is shown in Figure 5. When compared with normal mucosa, the abundance of Proteobacteria increased from 66.0% to 77.4%, the abundance of Firmicutes was reduced from 14.8% to 7.8%, and the abundance of Bacteroidetes was reduced from 13.7% to 7.0% in CRA mucosa (P < 0.05).
At the order level, microflora in the advanced CRA group mainly included Oceanospirillales (40%) and Alteromonadales (13%) of gamma Proteobacteria of Proteobacteria, and abundance increased significantly in the CRA group compared to the control group (P < 0.05). In the advanced CRA group, the abundances of Clostridiales of Firmicutes (5.7%) and Bacteroidales of Bacteroidetes (6.6%) were markedly reduced compared with the control group (P < 0.05). The principal component distribution is shown in Figure 6.
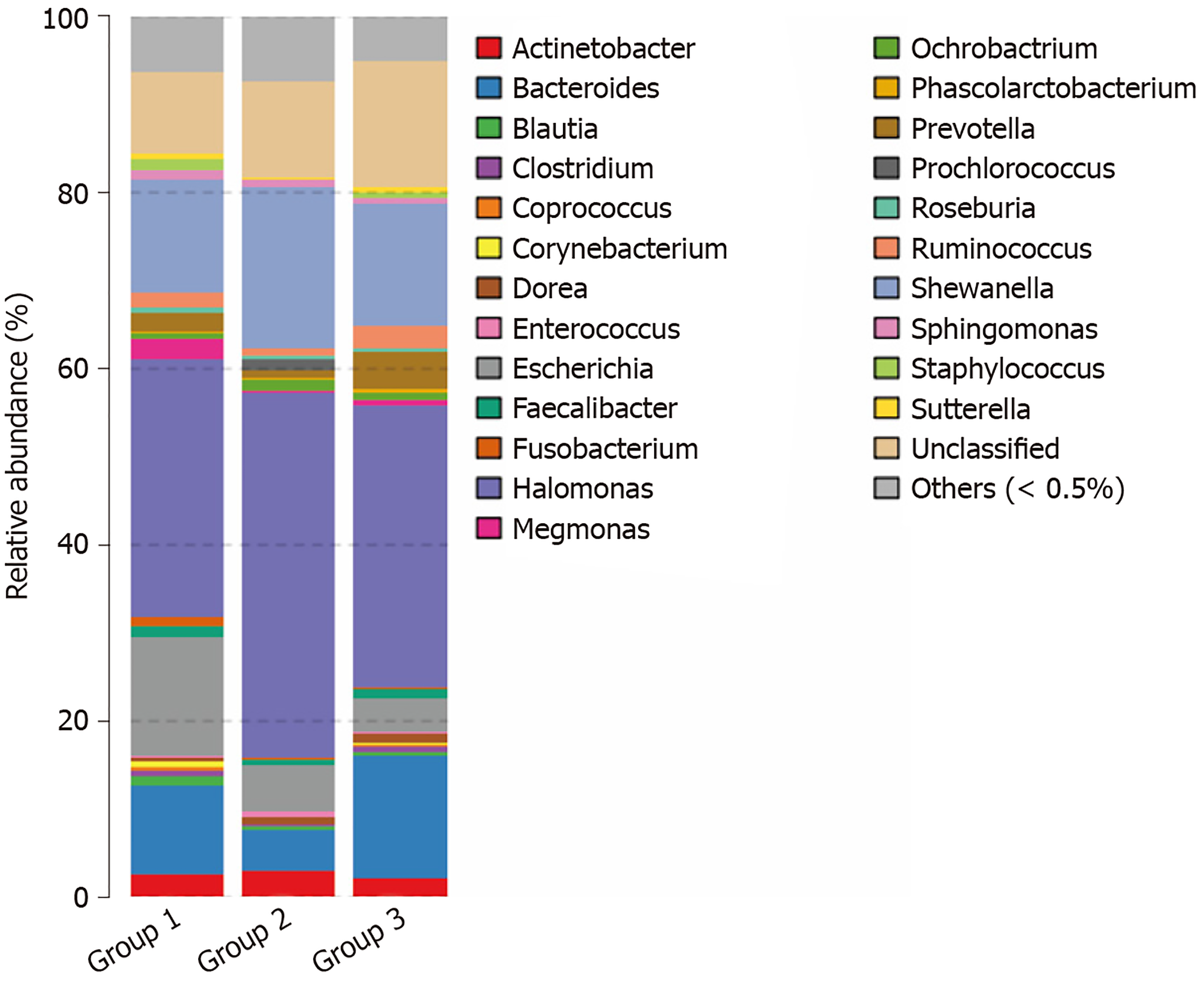
At the family level, microflora in advanced CRA mainly included Halomonas and Oceanospirillales of Proteobacteria (40.5%) and Shewanella of Alteromonadales (18.4%), and abundances increased significantly in the CRA group compared to the control group. The abundance of Blautia (0.38%), Coprococcus (0.08%), Bacteroides (5.1%), and Fusobacterium (0.08%) of Lachnospiraceae, Clostridiales, and Firmicutes in the CRA group were markedly decreased compared with the control group (P < 0.05). The principal component distribution is shown in Figure 7.
At the species level, the abundance of Shewanella_algae (18.1%) increased significantly (P < 0.05) in the CRA group, but the abundance of Bacteroides_fragilis, Escherichia_coli, and Prevotella_copri was slightly reduced (P > 0.05) compared to the control group. In addition, the abundance of Faecalibacterium prausnitzii of Clostridiales (0.7%) was dramatically reduced in the CRA group compared with the control group (P < 0.05). The principal component distribution is shown in Figure 8.
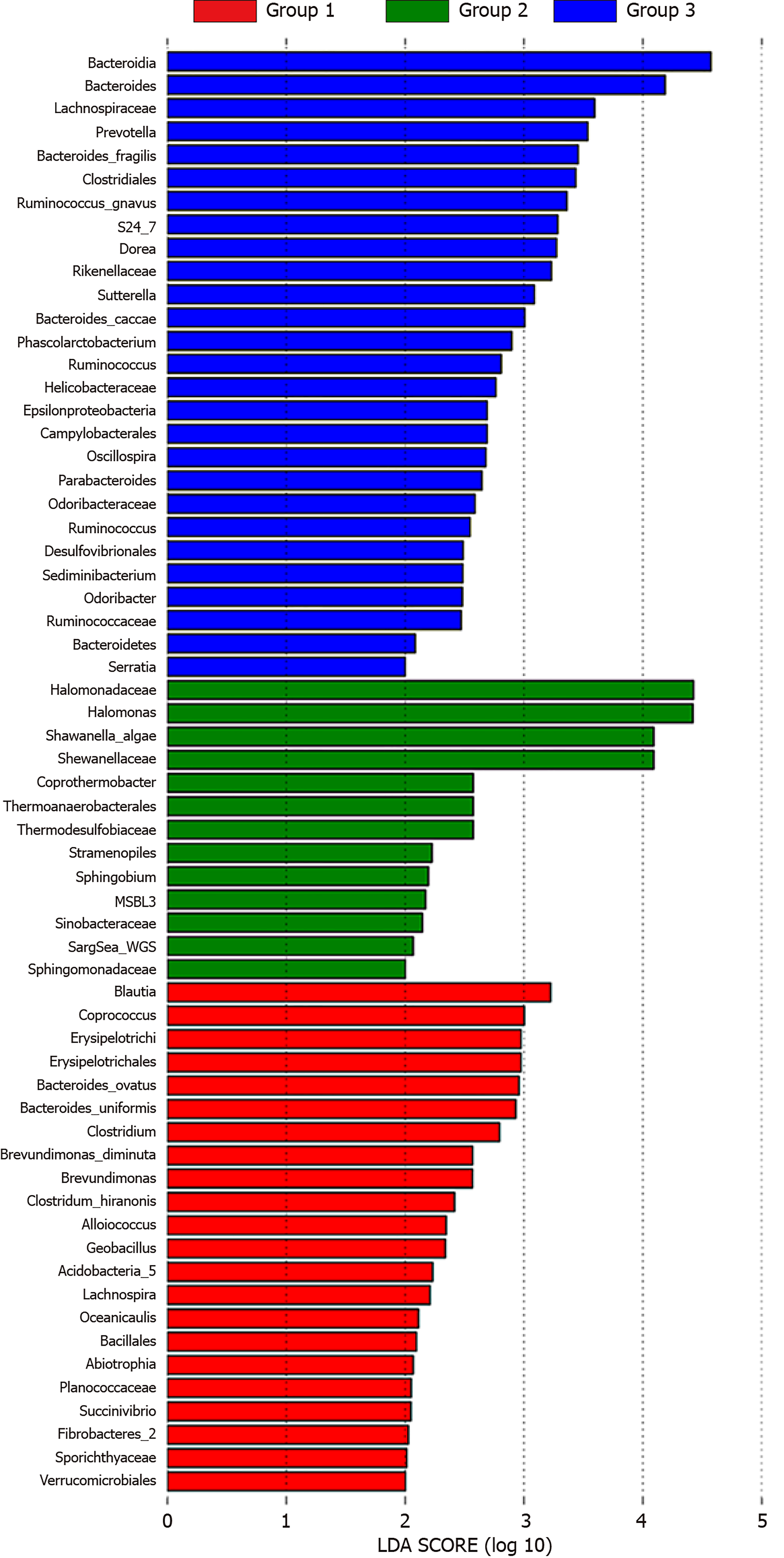
Metagenome biomarkers: Linear discriminant analysis effect size, also known as the metagenome biomarker, was used to identify the microbioflora with significant differences between the two groups. Biomarkers in the advanced CRA group were mainly found in Oceanospirillales and Alteromonadales. The abundance of Halomonadaceae, Halomonas, Shewanellaceae, and Shewanella algae also increased in the CRA group. In addition, the abundance of Blautia, Coprococcus of Lachnospiraceae and Bacteroidetes of Bacteroides ovatus was reduced significantly in the CRA group compared to the control group (Figure 9).
In recent years, intestinal microflora has been a major focus of research. Intestinal micro-ecology is composed of intestinal microbiota and their metabolites. Intestinal microbiota includes intestine flora and mucosa-related microflora[4]. Under the influence of the environment and dietary fiber, mucosa-related microflora may interact with intestinal mucosa, leading to epithelial transformation of the mucosa, and causing mucosal pathology. In this study, the mucosa-related microbiota was investigated in normal and CRA mucosa specimens. Microbiota diversity was reduced in CRA mucosa, and the abundance of Halomonadaceae, Shewanella algae, and Lachnospiraceae was reduced in CRA patients.
Shewanella are gram-negative saprophytic bacteria and secrete a tetrodotoxin-like toxin. It is widely accepted that Shewanella are the major bacterial contributor to the decay of high-protein foods and aquatic products. The Centers for Disease Control in the United States has indicated that only Shewanella algae and Shewanella putrefaciens in Shewanella are pathogenic for humans. Individuals with immune dysfunction, seawater exposure, or intake of seafood will develop zoonotic infectious diseases caused by Shewanella. Available case reports have shown that early-onset neonatal sepsis, acute hemorrhagic gastroenteritis, spontaneous bacterial peritonitis in cirrhosis, and squamous cell carcinoma are related to Shewanella infection[5-7]. In vitro experiments also indicate that Shewanella is able to inhibit the growth of plant pathogenic fungi, such as aflatoxin and Botrytis cinerea[8]. Telke et al[9] also separated the MARS 14 strain of Shewanella, and found that these bacteria could secrete ethanolamine phosphotransferase, which mediates polymyxin resistance. Liang et al[10] used 16S rDNA sequencing, and reported that patients with bile duct stones had Oddi sphincter relaxation and an abundance of Bilophila and Shewanella algae in the bile. Srinivas et al[11] conducted a 5-year study, and found that patients with concomitant diabetes mellitus, hypertension, or hepatobiliary disorders were more susceptible to Shewanella algae-related skin and soft tissue infections. In the current study, we showed that the abundance of Shewanella algae increased in the lesioned mucosa of patients with CRA, but the mechanism underlying the increased abundance of Shewanella algae in these patients is still poorly understood. We speculate that these bacteria may invade intestinal epithelial cells to interact with host tissues, or secrete some toxins to cause intestinal infections, but further molecular biological studies are required to confirm our hypothesis.
Studies have shown that intestinal Lachnospiraceae is significantly reduced in patients with inflammatory diseases[12]. Lachnospiraceae may antagonize the implantation of Clostridium difficile in the intestine after antibiotic therapy[13]. In addition, Lachnospiraceae is closely related to reduced carcinogenesis of the intestine[14]. Thus, the reduced abundance of Lachnospiraceae in patients with CRA may be associated with intestinal mucosal inflammation and the immune response. Different dietary components may change intestinal microbial composition and inflammatory markers[15]. There is evidence that CRP and IL-6 levels are different, not only between colorectal adenoma and cancer, but also between cancer stages[16]. Probiotics may have preventive effects on CRC, but the specific mechanism is still unclear. The binding or degradation of heterocyclic aromatic amines by probiotics may be an effective mechanism for removing carcinogens from the human body[17].
In the current study, CRA biomarkers were screened, but the relationship with the host requires further confirmation. The selective microbiota in this study also warrant further study in future clinical trials, and their role in the occurrence and development of CRA needs validation. With the development of intestinal microecological detection technology, changes in intestinal flora structure are expected to become an index to predict and evaluate the risk of CRC in hosts. At the same time, the use of microecological agents will provide new ideas and methods to prevent and treat CRC.
Colorectal adenomas (CRAs) are major precancerous lesions in colorectal cancer (CRC), which can greatly reduce the incidence and mortality of CRC and improve the quality of patient life. At present, many scholars have begun to study the relationship between intestinal flora and CRA. This paper provides a future direction for the prevention and treatment of by measuring the composition and diversity of intestinal flora in CRA patients, as well as the biomarkers related to it.
In order to further study the mechanism of CRC and its precancerous lesion CRA, the composition and diversity of intestinal flora in patients with CRA were determined by 16S rRNA gene sequencing, and the related biomarkers were also determined. However, the results obtained in this study need to be further verified in the population, and the relationship between the screened biomarkers and the host needs to be further confirmed.
This study found that the diversity of intestinal flora decreased in patients with CRA, and an increase in the number of Halomonadaceae and Shewanella algae may be a marker of CRA, a finding that provides new insights into the mechanism of CRA and CRC, and their development in the future.
The intestinal flora composition of 36 healthy people and 49 patients with advanced CRA in Guangzhou First People’s Hospital was studied by 16s rRNA gene sequencing. The operational taxonomic unit and its abundance analysis, sample diversity analysis, principal component analysis of samples, differential analysis, and analysis of biomarkers were performed via bioinformatics analysis.
This study found that the diversity of intestinal flora decreased in patients with CRA, and that an increase in the number of Halomonadaceae and Shewanella algae may be a marker of CRA. However, the results of this study need to be further verified in the population, and the relationship between the selected biomarkers and the host needs to be further confirmed.
CRA is the main precancerous lesion in CRC. In this study, the composition and diversity of intestinal flora in CRA patients were measured by 16s rRNA gene sequencing and bioinformatics. It was found that the diversity of intestinal flora in CRA patients decreased, and that an increase in Halomonadaceae and Shewanella algae may be a biomarker of CRA. However, the results obtained in this study need to be further verified in the population, and the relationship between the selected biomarkers and the host also needs to be further confirmed. This finding provides new ideas for the future study of the mechanism of CRA and CRC.
This study confirmed the difference in intestinal flora between normal people and CRA patients, and identified possible biomarkers. On this basis, future studies will further confirm the correlation, expand the sample size, and further study the interaction between biomarkers and hosts.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Biondi A, Uppara M S-Editor: Zhang L L-Editor: Filipodia E-Editor: Liu JH
| 1. | Brenner H, Hoffmeister M, Stegmaier C, Brenner G, Altenhofen L, Haug U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: estimates based on 840,149 screening colonoscopies. Gut. 2007;56:1585-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 304] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 2. | Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, Li X, Li X, Li J, Xiao L, Huber-Schönauer U, Niederseer D, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Arumugam M, Tilg H, Datz C, Wang J. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 968] [Article Influence: 96.8] [Reference Citation Analysis (0)] |
| 3. | Yu YN, Fang JY. Gut Microbiota and Colorectal Cancer. Gastrointest Tumors. 2015;2:26-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 788] [Cited by in RCA: 735] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 5. | Charles MV, Srirangaraj S, Kali A. Neonatal sepsis caused by Shewanella algae: A case report. Australas Med J. 2015;8:64-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 6. | Dey S, Bhattacharya D, Roy S, Nadgir SD, Patil A, Kholkute SD. Shewanella algae in acute gastroenteritis. Indian J Med Microbiol. 2015;33:172-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 7. | Sumathi BG, Kumarswamy SR, Amritam U, Arjunan R. Shewanella algae: First case report of the fast-emerging marine pathogen from squamous cell carcinoma patient in India. South Asian J Cancer. 2014;3:188-189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Gong AD, Li HP, Shen L, Zhang JB, Wu AB, He WJ, Yuan QS, He JD, Liao YC. The Shewanella algae strain YM8 produces volatiles with strong inhibition activity against Aspergillus pathogens and aflatoxins. Front Microbiol. 2015;6:1091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Telke AA, Rolain JM. Functional genomics to discover antibiotic resistance genes: The paradigm of resistance to colistin mediated by ethanolamine phosphotransferase in Shewanella algae MARS 14. Int J Antimicrob Agents. 2015;46:648-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Liang T, Su W, Zhang Q, Li G, Gao S, Lou J, Zhang Y, Ma T, Bai X. Roles of Sphincter of Oddi Laxity in Bile Duct Microenvironment in Patients with Cholangiolithiasis: From the Perspective of the Microbiome and Metabolome. J Am Coll Surg. 2016;222:269-280.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Srinivas J, Pillai M, Vinod V, Dinesh RK. Skin and Soft Tissue Infections due to Shewanella algae - An Emerging Pathogen. J Clin Diagn Res. 2015;9:DC16-DC20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J; MetaHIT Consortium, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M'rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5822] [Cited by in RCA: 5036] [Article Influence: 359.7] [Reference Citation Analysis (2)] |
| 13. | Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun. 2012;80:3786-3794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 231] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 14. | Mai V, Colbert LH, Perkins SN, Schatzkin A, Hursting SD. Intestinal microbiota: a potential diet-responsive prevention target in ApcMin mice. Mol Carcinog. 2007;46:42-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Telle-Hansen VH, Holven KB, Ulven SM. Impact of a Healthy Dietary Pattern on Gut Microbiota and Systemic Inflammation in Humans. Nutrients. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 16. | Godos J, Biondi A, Galvano F, Basile F, Sciacca S, Giovannucci EL, Grosso G. Markers of systemic inflammation and colorectal adenoma risk: Meta-analysis of observational studies. World J Gastroenterol. 2017;23:1909-1919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 17. | Uccello M, Malaguarnera G, Basile F, D'agata V, Malaguarnera M, Bertino G, Vacante M, Drago F, Biondi A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012;12 Suppl 1:S35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 147] [Article Influence: 11.3] [Reference Citation Analysis (0)] |