Published online Dec 28, 2020. doi: 10.3748/wjg.v26.i48.7679
Peer-review started: October 16, 2020
First decision: November 3, 2020
Revised: November 15, 2020
Accepted: December 6, 2020
Article in press: December 6, 2020
Published online: December 28, 2020
Processing time: 70 Days and 1.9 Hours
Microbiota profiles differ between patients with pancreatic cancer and healthy people, and understanding these differences may help in early detection of pancreatic cancer. Saliva sampling is an easy and cost-effective way to determine microbiota profiles compared to fecal and tissue sample collection.
To investigate the saliva microbiome distribution in patients with pancreatic adenocarcinoma (PDAC) and the role of oral microbiota profiles in detection and risk prediction of pancreatic cancer.
We conducted a prospective study of patients with pancreatic cancer (n = 41) and healthy individuals (n = 69). Bacterial taxa were identified by 16S ribosomal ribonucleic acid gene sequencing, and a linear discriminant analysis effect size algorithm was used to identify differences in taxa. Operational taxonomic unit values of all selected taxa were converted into a normalized Z-score, and logistic regressions were used to calculate risk prediction of pancreatic cancer.
Compared with the healthy control group, carriage of Streptococcus and Leptotrichina (z-score) was associated with a higher risk of PDAC [odds ratio (OR) = 5.344, 95% confidence interval (CI): 1.282-22.282, P = 0.021 and OR = 6.886, 95%CI: 1.423-33.337, P = 0.016, respectively]. Veillonella and Neisseria (z-score) were considered a protective microbe that decreased the risk of PDAC (OR = 0.187, 95%CI: 0.055-0.631, P = 0.007 and OR = 0.309, 95%CI: 0.100-0.952, P = 0.041, respectively). Among the patients with PDAC, patients reporting bloating have a higher abundance of Porphyromonas (P = 0.039), Fusobacterium (P = 0.024), and Alloprevotella (P = 0.041); while patients reporting jaundice had a higher amount of Prevotella (P = 0.008); patients reporting dark brown urine had a higher amount of Veillonella (P = 0.035). Patients reporting diarrhea had a lower amount of Neisseria and Campylobacter (P = 0.024 and P = 0.034), and patients reporting vomiting had decreased Alloprevotella (P = 0.036).
Saliva microbiome was able to distinguish patients with pancreatic cancer and healthy individuals. Leptotrichia may be specific for patients living in Sichuan Province, southwest China. Symptomatic patients had different bacteria profiles than asymptomatic patients. Combined symptom and microbiome evaluation may help in the early detection of pancreatic cancer.
Core Tip: Pancreatic adenocarcinoma (PDAC) patients benefit from early detection. This study analyzed the composition and diversity of saliva microbiota in PDAC patients through 16S ribosomal ribonucleic acid sequencing. Normalized z-score of bacteria abundance associated clinical data were analyzed for PDAC risk prediction. Microbiome abundance differences were found between PDAC patients with symptoms and patients without symptoms. Combined symptom and microbiome evaluation may help in early detection and risk prediction of pancreatic cancer.
- Citation: Wei AL, Li M, Li GQ, Wang X, Hu WM, Li ZL, Yuan J, Liu HY, Zhou LL, Li K, Li A, Fu MR. Oral microbiome and pancreatic cancer. World J Gastroenterol 2020; 26(48): 7679-7692
- URL: https://www.wjgnet.com/1007-9327/full/v26/i48/7679.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i48.7679
Pancreatic cancer or pancreatic adenocarcinoma (PDAC) is a lethal disease with a 5-year survival rate of about 6%[1,2]. Early detection and diagnosis are essential for effective surgery treatment that improves cancer survival[3,4], yet these remain a great challenge. A variety of diagnostic methods are available. For example, deoxyrib-onucleic acid (DNA) sequencing for detecting and diagnosing pancreatic cancer are limited in clinical use due to the need for fresh, high-quality specimens, tumor content, and tumor heterogeneity[5,6]. Molecular markers, such as mutant DNA or DNA methylomes, are also limited in clinical use to enhance diagnostic sensitivity or early detection of pancreatic cancer recurrence[7,8]. Biomarker Ca19-9 has been commonly used for diagnosis and prognosis of pancreatic cancer with diagnostic sensitivity of 0.78 and specificity of 0.77, but this biomarker test has limited sensitivity among patients with jaundice, pancreatitis, enteritis, and elevated blood glucose, since such patients usually have elevated Ca19-9 concentrations[9-11]. In addition, 7%-10% Lewis (a-/b-) populations could not express Ca19-9[12].
The oral or fecal microbiota profile of gastrointestinal and colorectal cancer, oropharyngeal cancer, liver cancer, and lung cancer may be a novel and potential diagnostic biomarker[13-19]. Accumulated studies have revealed that oral and gastrointestinal microbiomes differ in abundance in patients with pancreatic cancer compared with healthy individuals[20-23]. Cancer risk increases with carriage of Porphyromonas gingivalis[21], Actinobacillus actinomycetemcomitans[21], and Alloprevotella[21], while Fusobacterium[21,24], Leptotrichia[21,25,26], Neisseria elongate[21,23], and Streptococcus mitis[21,23] might be a protective factor for having pancreatic cancer. However, Olson et al[22] did not find significant differences in the diversity of the oral microbiome among PDAC patients (n = 40), intraductal papillary mucinous neoplasms (IPMNs) (n = 39), and healthy participants (n = 58) in the United States[22]. The conflicting findings in the prior studies may be due to the differences in methodological approach and sample collection. For example, some studies performed real-time quantitative polymerase chain reaction (PCR) for validation of bacterial candidates[23], and some sequenced the microbiota profile in samples of tongue coating[20] or oral wash samples[21]. Tongue coating change is a major often-used approach of tongue diagnosis in traditional Chinese medicine, but tongue coating can only capture partial oral microbiota[27,28]. The oral wash method is more complicated and relatively expensive.
Oral cavity contains nearly 619 taxa in 13 phyla (Firmicutes, Proteobacteria, Actinobacteria, Fusobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Euryarchaeota, Spirochaetes, SR1, Synergistes, Tenericutes, and TM7), and 68% of these bacteria are uncultivated phylotypes[29,30]. Advanced genomic sequencing for human oral microbiome distribution makes it possible to measure the proportions of bacterial species without relying on traditional culture methods[31-33]. Saliva has been found to contain broad spectrum of bacteria with easy sampling method and is relatively cost-effective. Although there are some studies on oral flora and pancreatic cancer in non-Chinese population, the impact of geographical and medical factors, such as race and ethnicity, different dietary habits, antibiotic use, and cancer, may make the oral microbial profile differ among people from different geographic locations. In addition, there are few studies on oral saliva flora and pancreatic cancer in China. Thus, the purpose of our study was to: (1) Determine the saliva microbiome distribution of pancreatic cancer (including resectable PDAC and unresectable PDAC) among Chinese population using 16S rRNA sequencing; and (2) Select proper and specific microbiota for PDAC detecting.
The Institutional Review Board of the West China Hospital, Sichuan University approved this prospective study. All participants signed written informed consent.
This was a prospective study. We consecutively recruited 80 patients who were over age 18 years and suspected to have pancreatic tumor prior to biopsy or surgery. Histopathological results confirmed 45 patients with primary PDAC and 35 patients with non-cancer pancreatic tumors, including 9 IPMN, 11 pancreatic serous cystadenoma, 5 solid pseudopapillary neoplasm, and 10 neuroendocrine tumors. We also recruited 69 healthy participants from the community as a comparison group. Healthy adults had normal liver and renal function, normal cardio-pulmonary function, no history of cancer, and no viral infection. Participants were excluded if they had: (1) A history of prior malignancy and chemotherapy or radiotherapy; (2) Metastatic PDAC or PDAC with other cancer; (3) A history of viral infection (i.e. hepatitis B virus, hepatitis C virus, human immunodeficiency virus); (4) Use of antibiotics (including oral, intravenous, or intramuscular) and probiotics within 4 wk prior to enrollment; and (5) Use of corticosteroids (nasal or inhaled) or other immunosuppressants. In addition, we excluded participants with insufficient saliva sample (n = 12) for sequencing analysis and patients with non-cancer pancreatic tumors (n = 35).
The demographic information collected included age, gender, body mass index (BMI), smoking history, alcohol consumption, dietary habit, and chronic diseases (hypertension and type II diabetes). Clinical information was also collected to include cancer site, surgery type, and cancer stages using the American Joint Commission on Cancer, seventh edition staging manual[34].
Since there is no measure or checklist for symptoms specific to pancreatic cancer, we developed a checklist based on literature review to assess symptoms specific to pancreatic cancer, such as bloating, jaundice, nausea, vomiting, dark brown urine, diarrhea, constipation, pale stools, pruritus, lack of appetite, pain, fatigue, and disturbed sleeping. Patients reported the presence and absence of symptoms by checking “Yes” or “No.”
Before the patients had surgery to confirm pancreatic cancer diagnosis, saliva samples were collected by trained professionals (Wang X and Li GQ). All the participants were instructed to not eat and drink for 0.5 h prior to saliva sample collection. Participants were also instructed not to brush their teeth at least 8 h prior to saliva sample collection, since brushing teeth may remove part of the oral flora. Participants were asked to rinse their mouths to remove debris from the oral cavity before saliva collection. To ensure all sample collection was at a similar time period in a day, we collected patient samples around 4:00 pm on the day of admission prior to biopsy or surgery for cancer diagnosis. Healthy subjects’ saliva samples were also collected around 4:00 pm in the afternoon. About 3 mL saliva was collected in a sterile tube after it accumulated on the mouth floor. The fresh samples were placed on ice and transported to the laboratory. Samples were divided into 1.5 mL aliquots and stored immediately at -80 °C.
We used the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, United States) to extract bacterial genomic DNA from saliva samples. DNA concentration and purity was quantified by Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Wilmington, DE, United States) and agarose gel electrophoresis. Genome DNA with strong smear or with concentration lower than 5 μg/mL (by Qubit) was excluded for library construction.
The third and fourth hypervariable regions (V3-V4) of the 16S rRNA gene of bacteria were amplified by PCR with a domain-specific primer: 341F (5'-CCTACGGGNGGCWGCAG-3') and 805R (5'- GACTACHVGGGTATCTA ATCC-3'). PCR reactions were performed with a 15 µL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, United States), 0.2 μmol/L of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98 °C for 1 min, followed by 30 cycles of denaturation at 98 °C for 10 s, annealing at 50 °C for 30 s, and elongation at 72 °C for 30 s. Finally, samples were incubated at 72 °C for 5 min. The library quality was assessed by Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). Sequencing was performed on an Illumina Novaseq6000 sequencing platform (Illumina, San Diego, CA, United States), and 250 bp paired-end reads were generated.
Phenotype data analysis: Statistical analyses were performed using SPSS (v23.0, SAGE IBM, Armonk, NY, United States). Continuous variables (age and BMI) were estimated as average ± standard error, and categorical variables were analyzed in terms of frequencies and percentages. Chi square analysis and Fisher’s exact tests were used for categorical variables; t-test and Mann-Whitney U test were used for continuous variables. All tests were two-sided, and P values < 0.05 were considered statistically significant with 95% confidence interval (CI).
Profile and quality assurance: Raw sequences were denoised via FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/)[35]. Quality filtering was performed on raw sequences using QIIME quality control process (v1.9.1_http://qiime.org/index.html) and then high quality clean tags were obtained[36]. Tags were compared with gold database (http://drive5.com/uchime/uchime_download.html), and chimeras were removed with the UCHIME algorithm (v11.0, http://www.drive5.com/usearch/manual/uchime_algo.html)[37]. Effective Tags were finally obtained. All effective sequence analysis was performed by Uparse software (v7.0.1001, http://drive5.com/uparse/)[38]. The optimized, high-quality sequences were clustered into operational taxonomic units (OTUs) at 97% sequence identity.
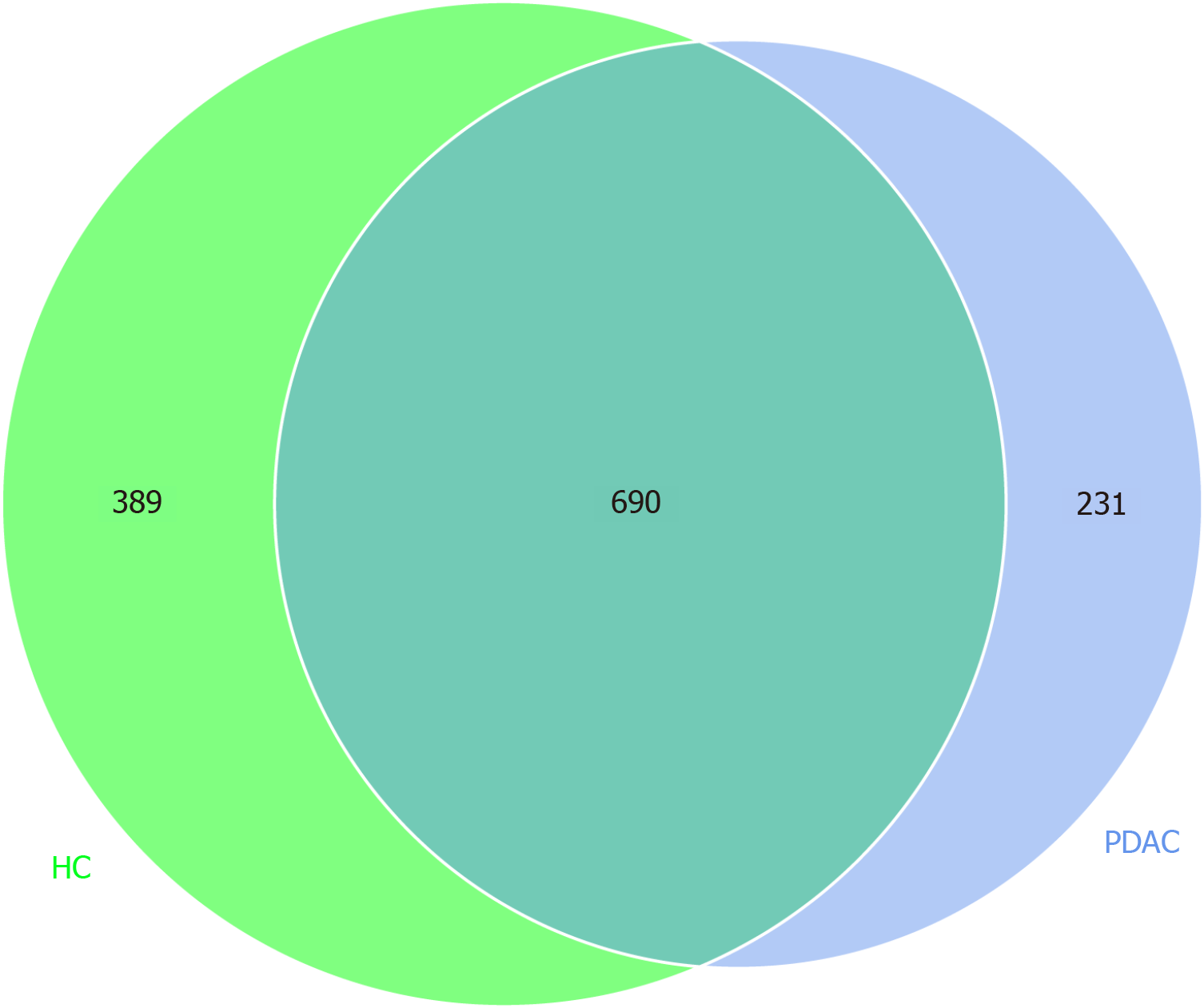
Microbiome diversity: According to the results of OTUs clustering analysis and the research requirements, the Venn diagram was constructed to illustrate the number of unique and shared species in saliva samples between PDAC and healthy groups. The Venn diagram was made using R program (Package_VennDiagram). We applied alpha diversity to analyze complexity of species diversity for a sample. Four indices were used: “Chao1” and “Abundance-based coverage estimator (ACE)” estimate the species abundance; “Shannon index ” and “Simpson” account for the richness and evenness. The value of Simpson index was calculated as Simpson’s index of diversity 1-D. Thus, higher Shannon and Simpson indices mean higher species diversity. All indices were calculated with QIIME (v1.9.1) and R software (V2.15.3, Auckland, New Zealand). We compared four indices between PDAC and healthy control group using Mann-Whitney U test. Mann-Whitney U test was used to compare the alpha diversity indices between groups of resectable PDAC (rPDAC) and unresectable PDAC (unrPDAC). The bacterial taxonomic compositions were evaluated with a linear discriminant analysis effect size algorithm (https://huttenhower.sph.harvard.edu/).
Abundance of bacteria and symptom: We used Wilcoxon rank-sum test to compare the abundance of bacteria (top 10 positively expressed flora) in PDAC patients with and without typical symptoms of PDAC, including bloating, jaundice, nausea, vomiting, dark brown urine, diarrhea, constipation, pale stools, pruritus, lack of appetite, pain, fatigue, and disturbed sleeping.
Logistic regressions were used to explore the association of significant taxa with clinical covariates (age, BMI, smoking status, alcohol consumption status, history of blood hypertension, and eating habits). To avoid the occurrence of false negative diagnosis, we focused on the top 20 species (OTUs abundance) and the flora associated with PDAC that has been reported[20-23]. Finally, Streptococcus, Prevotella, Porphyromonas, Neisseria, Veillonella, Leptotrichia, Lactobacillus, Actinomyces, Haemophilus, Rothia, and Fusobacterium were selected for analysis. To make the values comparable, we converted the OTU values of all selected taxa into a normalized z-score. The tetranucleotide-derived z-score, superior to (G + C) content differences, was calculated according to the previous methods[39,40]. Odds ratio with 95%CIs were calculated.
Between November 2017 and December 2018, a total of 157 participants were enrolled in this study; four PDAC patients and eight healthy participants were eventually excluded due to the insufficient saliva sample for sequencing analysis. A final sample of 110 included patients in PDAC (n = 41) and healthy individuals (n = 69). Table 1 shows the demographic characteristics of PDAC patients and healthy participants. Compared with the healthy group, the PDAC had lower BMIs (22.76 vs 24.44, P < 0.0001). As for eating habits, more PDAC patients (61%) preferred oily and fatty foods compared to the healthy control group (P = 0.002). More healthy control participants had hypertension (P = 0.006). Among the 41 patients with PDAC, 31 (76%) had head pancreatic cancer, and 20 (49%) patients had resectable pancreatic cancer.
| Variables | PDAC group, n = 41 | Healthy control group, n = 69 | P value |
| Age, average ± standard error | 61.17 ± 1.79 | 64.64 ± 1.04 | 0.098 |
| Gender, n (%) | |||
| Male | 24 (59) | 50 (72) | 0.132 |
| Female | 17 (41) | 19 (28) | |
| BMI, average ± standard error | 22.76 ± 0.94 | 24.44 ± 0.39 | < 0.0001 |
| Smoking history, n (%) | 17 (41) | 37 (54) | 0.217 |
| Alcohol consumption, n (%) | 16 (39) | 30 (43) | 0.647 |
| Dietary habit, n (%) | |||
| Oily and fatty food | 25 (61) | 21 (31) | 0.002 |
| Salty food | 6 (15) | 8 (11) | 0.664 |
| Light diet | 10 (24) | 40 (58) | 0.001 |
| Chronic disease, n (%) | |||
| Hypertension | 1 (2) | 15 (22) | 0.006 |
| Type II diabetes | 2 (5) | 6 (9) | 0.714 |
| Both | 3 (7) | 5 (7) | 1.000 |
| Loss of weight, n (%) | 23 (56) | 3 (4) | 0.0001 |
| Primary cancer site, n (%) | |||
| Head | 31 (76) | NA | NA |
| Body and tail | 10 | ||
| Surgery, n (%) | |||
| Pancreaticoduodenectomy | 14 (34) | NA | NA |
| Distal pancreatectomy | 6 (15) | ||
| Palliative intervention techniques | 21 (51) | ||
| AJCC staging | NA | NA | |
| I-IIB | 20 (49) | ||
| III-IV | 21 |
Alpha-diversity analysis of the study participant groups: From 110 samples, we filtered 6356399 qualified reads. We randomly chose 2235200 reads (110 samples multiplied by 20320 reads/sample, the minimum number of reads/sample). Finally, we obtained 1975 OTUs for further analysis. A Venn diagram (Figure 1) shows the details of the OTUs at 97% identity for PDAC patients and healthy participants. The two groups had 690 shared species, 231 unique species for PDAC patient, and 389 for healthy control group. As Table 2 shows, compared with the healthy group, the PDAC group had significantly increased microbial abundance estimated by the Chao1 index and ACE index while decreased microbial diversity estimated by Shannon and Simpson indices (P < 0.0001). Patients with rPDAC had lower bacteria abundance and diversity than patients with unrPDAC estimated by Chao1, ACE, Shannon indices, and Simpson indices. However, Shannon (P = 0.273), Simpson (P = 0.715), Chao1 (P = 0.159), and ACE (P = 0.137) were not able to distinguish rPDAC and unrPDAC.
| PDAC group, n = 41 | Healthy control group, n = 69 | P value | |
| Shannon | 5.14 ± 0.67 | 5.67 ± 0.51 | 0.0001 |
| Simpson | 0.90 ± 0.08 | 0.95 ± 0.02 | 0.0001 |
| Chao1 | 423.48 ± 55.69 | 295.00 ± 54.05 | 0.0001 |
| ACE | 424.00 ± 55.72 | 293.97 ± 50.09 | 0.0001 |
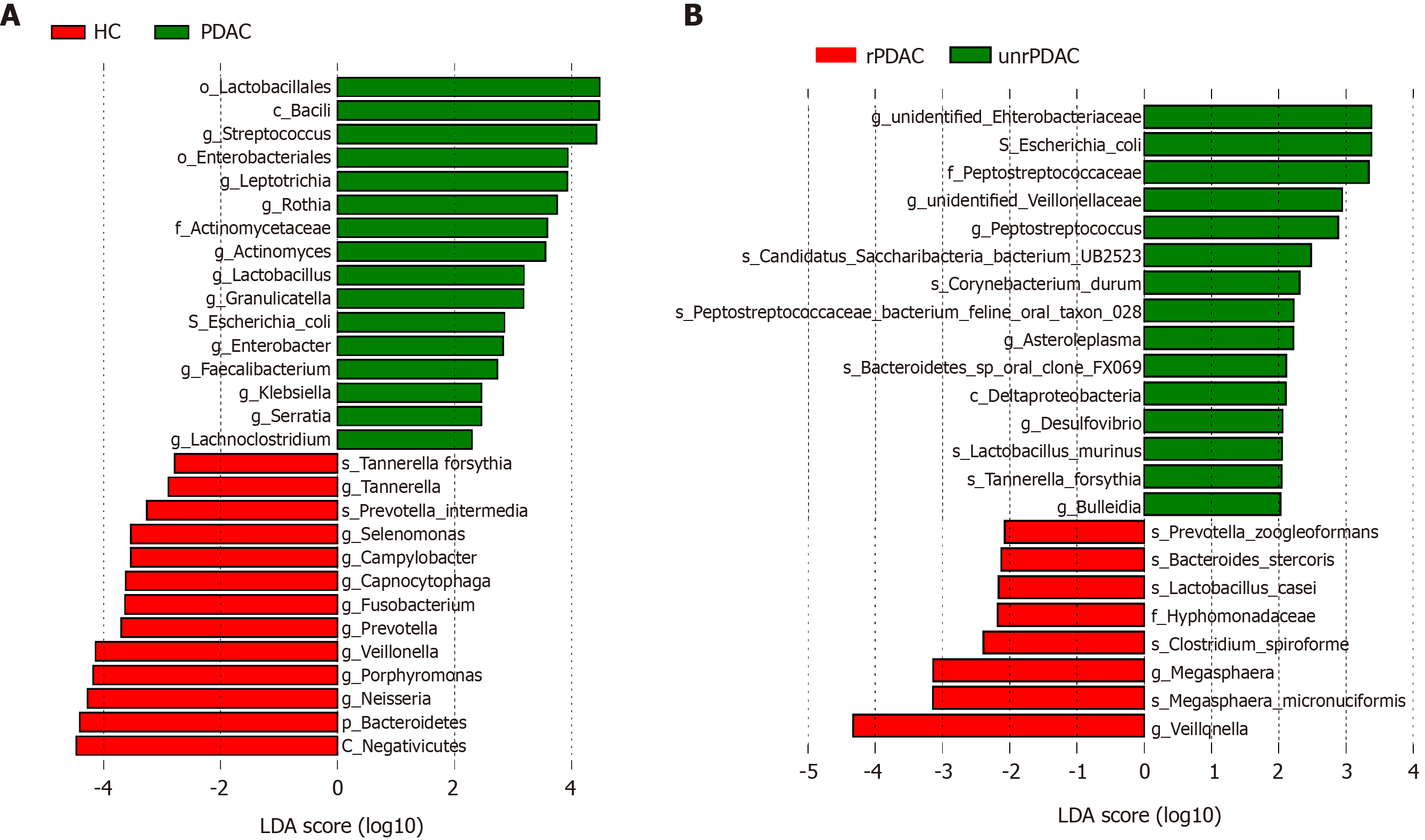
We used a linear discriminant analysis effect size algorithm to assess the bacterial taxonomic compositions and differences between PDAC group and healthy control subjects. Compared with the healthy group, PDAC patients were significantly enriched in order_Lactobacillales, class_Bacilli, genus_Streptococcus, phylum_Firmicutes, genus_Actinomyces, genus_Rothia, genus_Leptotrichia, genus_Lactobacillus, species_Escherichia_coli, and order_Enterobacteriales (Figure 2A). Conversely, PDAC patients had significantly reduced abundances of Selenomonas, Porphyromnas, Prevotella, Capnocytophaga, Alloprevotella, Tannerella, and Neisseria at genus level. We also compared the bacterial distributions between rPDAC and unrPDAC patients. Figure 2B shows that species_Escherichia coli, genus_Peptostreptococcus, genus_Asteroleplasma, and species_Tannerella forstythia were more prevalent in the unrPDAC group, whereas we found reduced occurrence of species_Bacteroides stercoris, genus_Megasphaera, and genus_Veillonella (Figure 2).
Table 3 presented flora abundance differences between the PDAC patients with symptoms and without symptoms. Patient reporting bloating had greater abundance of Porphyromonas (660.4 ± 461.0, P = 0.039), Fusobacteria (490.0 ± 186.6, P = 0.024), and Alloprevotella (155.4 ± 124.1, P = 0.041) compared to those without bloating (412.0 ± 394.3, 361.8 ± 184.4 and 99.3 ± 81.9, respectively). Prevotella presented greater abundance in patients without jaundice (669.4 ± 384.3, P = 0.008) compared to those with jaundice (403.2 ± 310.8). Veillonella presented greater abundance in patients without dark brown urine (1863.8 ± 1449.2, P = 0.035) compared to those with dark brown urine (1018.6 ± 766.7). Alloprevotella presented greater abundance in patients without vomiting (130.3 ± 100.9, P = 0.036) compared to those with vomiting (91.8 ± 134.4), while Neisseria presented greater abundance in patients with vomiting (3343.3 ± 1829.9, P = 0.024) compared to those without vomiting (1360.3 ± 1256.6). Campylobacter presented greater abundance in patients with diarrhea (130.5 ± 59.7, P = 0.034) compared to those without diarrhea (74.9 ± 87.2).
| Symptoms | Microbiome | Without symptoms | With symptoms | P value |
| Bloating | Porphyromonas | 412.0 ± 394.3 | 660.4 ± 461.0 | 0.039 |
| Fusobacteria | 361.8 ± 184.4 | 490.0 ± 186.6 | 0.024 | |
| Alloprevotella | 99.3 ± 81.9 | 155.4 ± 124.1 | 0.041 | |
| Jaundice | Prevotella | 669.4 ± 384.3 | 403.2 ± 310.8 | 0.008 |
| Dark brown urine | Veillonella | 1863.8 ± 1449.2 | 1018.6 ± 766.7 | 0.035 |
| Vomiting | Alloprevotella | 130.3 ± 100.9 | 91.8 ± 134.4 | 0.036 |
| Diarrhea | Neisseria | 1360.3 ± 1256.6 | 3343.3± 1829.9 | 0.024 |
| Campylobacter | 74.9 ± 87.2 | 130.5 ± 59.7 | 0.034 |
We explored the PDAC risk in relation to selected bacteria abundances (normalized z-score). As shown in Table 4, compared with healthy control group, carriage of Streptococcus (OR = 5.344, 95%CI: 1.282-22.282, P = 0.021) and Leptotrichina (OR = 6.886, 95%CI: 1.423-33.337, P = 0.016) were associated with a higher risk of PDAC. With each increase of z-score of Streptococcus and Leptotrichina in PDAC patients, the risk of pancreatic cancer increased by 5.344 odds and 6.886 odds, respectively. Carriage of Veillonella and Neisseria were protective factors of having PDAC (OR = 0.187, 95%CI: 0.055-0.631, P = 0.007 and OR = 0.309, 95%CI: 0.100-0.952, P = 0.041, respectively). With each decrease of z-score of Veillonella and Neisseria in PDAC patients, the risk of pancreatic cancer decreased by 0.187 odds and 0.309 odds, respectively.
| Odds ratio | 95%CI | P value | ||
| Healthy control group | Base outcome | |||
| PDAC group | ||||
| Age | 0.956 | 0.875 | 1.046 | 0.327 |
| BMI | 0.973 | 0.708 | 1.338 | 0.866 |
| Oily and fatty food | 0.759 | 0.122 | 4.730 | 0.768 |
| Streptococcus | 5.344 | 1.282 | 22.282 | 0.021 |
| Veillonella | 0.187 | 0.055 | 0.631 | 0.007 |
| Neisseria | 0.309 | 0.100 | 0.952 | 0.041 |
| Lactobacillus | 0.713 | 0.357 | 1.425 | 0.338 |
| Leptotrichia | 6.886 | 1.423 | 33.337 | 0.016 |
| Actinomyces | 4.515 | 0.444 | 45.887 | 0.203 |
| Haemophilus | 1.185 | 0.513 | 2.738 | 0.691 |
| Prevotella | 0.673 | 0.298 | 1.519 | 0.341 |
| Porphyromonas | 0.294 | 0.084 | 1.033 | 0.056 |
| Rothia | 1.257 | 0.467 | 3.384 | 0.650 |
| Fusobacterium | 1.006 | 0.335 | 3.017 | 0.576 |
This prospective study found dysbacteriosis of the oral microbiota existed in patients with PDAC. Fecal bacteria flora has been the main sample method for research on pancreatic cancer[41,42]. Our study used saliva sample method, which is convenient and the quality of sample is easy to control during sample collection. When comparing bacteria profiles from our saliva samples and fecal samples from other research on Chinese PDAC patients[20,42], salivary and intestinal bacteria flora consistently had low Shannon index and high Chao1 index, and Lactobacillus, Enterobacter, and Leptotrichia at the genus level was significantly increased. This provides supporting evidence that saliva sample method yields similar bacteria flora profiles compared to the fecal sample method, which is very often difficult to collect the samples. Findings of our study also provided additional evidence to confirm that Neisseria and Streptococcaceae are risk factors for pancreatic cancer[21,23]. Currently, no studies have focused on comparing the advantages and disadvantages of using different sample collection techniques, and studies are necessary to compare the effectiveness of using different sample collection techniques, such as saliva, tongue coating, and oral wash, on sample quality for microbiota profiles and preference of patients.
In terms of microbiota abundance and species diversity, our study found that the PDAC group had significantly increased microbial abundance as estimated by the Chao1 and ACE indices and decreased microbial diversity as estimated by Shannon and Simpson indices. Lu et al[20] also had similar findings from a study on Chinese pancreatic cancer patients using tongue coating samples[20]. However, studies of non-Chinese population did not find any differences of alpha diversity indices of oral microbiota composition between pancreatic cancer patients and healthy individuals[22,23]. Findings of our study and Lu et al[20] demonstrated that seven of fourteen bacterial families (Leptotrichiaceae, Actinomycetaceae, Lachnospiraceae, Micrococcaceae, Erysipelotrichaceae, Coriobacteriaceae, Moraxellaceae) were consistently significantly increased, and Porphyromonadaceae was significantly decreased in Chinese PDAC patients. However, our study found that the abundance of three of fourteen bacterial families (Fusobacteriaceae, Campylobacteraceae, Spirochaetaceae) were significantly decreased in PDAC patients, while Lu et al[20] found significantly more abundance[20]. Both our study and the study by Lu et al[20] found significant increase in the genus of Leptotrichia, Actinomyces, Rothia, Rothia, Solobacterium, Peptostreptococcus, and Oribacterium. Yet, decreased abundance in Selenomona, Tannerella, and Campylobacter was found in our study using saliva sample method but was increased in the study by Lu et al[20] using tongue coating sample method[20]. There are four known main periodontal disease contributors: Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia were more prevalent in PDAC patients in Fan et al[21]. However, except for Actinomyces, Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia were significantly reduced in our study. Leptotrichia also showed different distribution in our study comparing to Fan et al[21]. Torres et al[43] found higher Leptotrichia and lower Porphyromonas in the saliva of patients with pancreatic cancer, but no significant differences were found in the expression of Streptococcus mitis and Granulicatella adiacens. The conflicting findings between our study and other studies may due to different sample collection methods, e.g., saliva vs tongue coating method. Future research should compare different sample collection methods for microbiome research, e.g., saliva vs tongue coating method. The other factor for the conflicting findings may geographic food preferences of Chinese population. For example, subjects in Lu et al[20]’s study were enrolled from Zhejiang University, which is located in Hangzhou (southeast of China). Generally, people in Hangzhou have different diet preferences, such as preferences for milder taste and more sugar. Subjects in our study from Sichuan Province preferred adding a large amount of different herbs and spices and more fat and salt in food, which may lead to a high incidence of digestive system tumors[44,45]. Future research should focus on the effects of geographical location, race, diet, antibiotic usage (including consuming meat products containing antibiotics), injury, illness, and hormonal change on flora analysis[46].
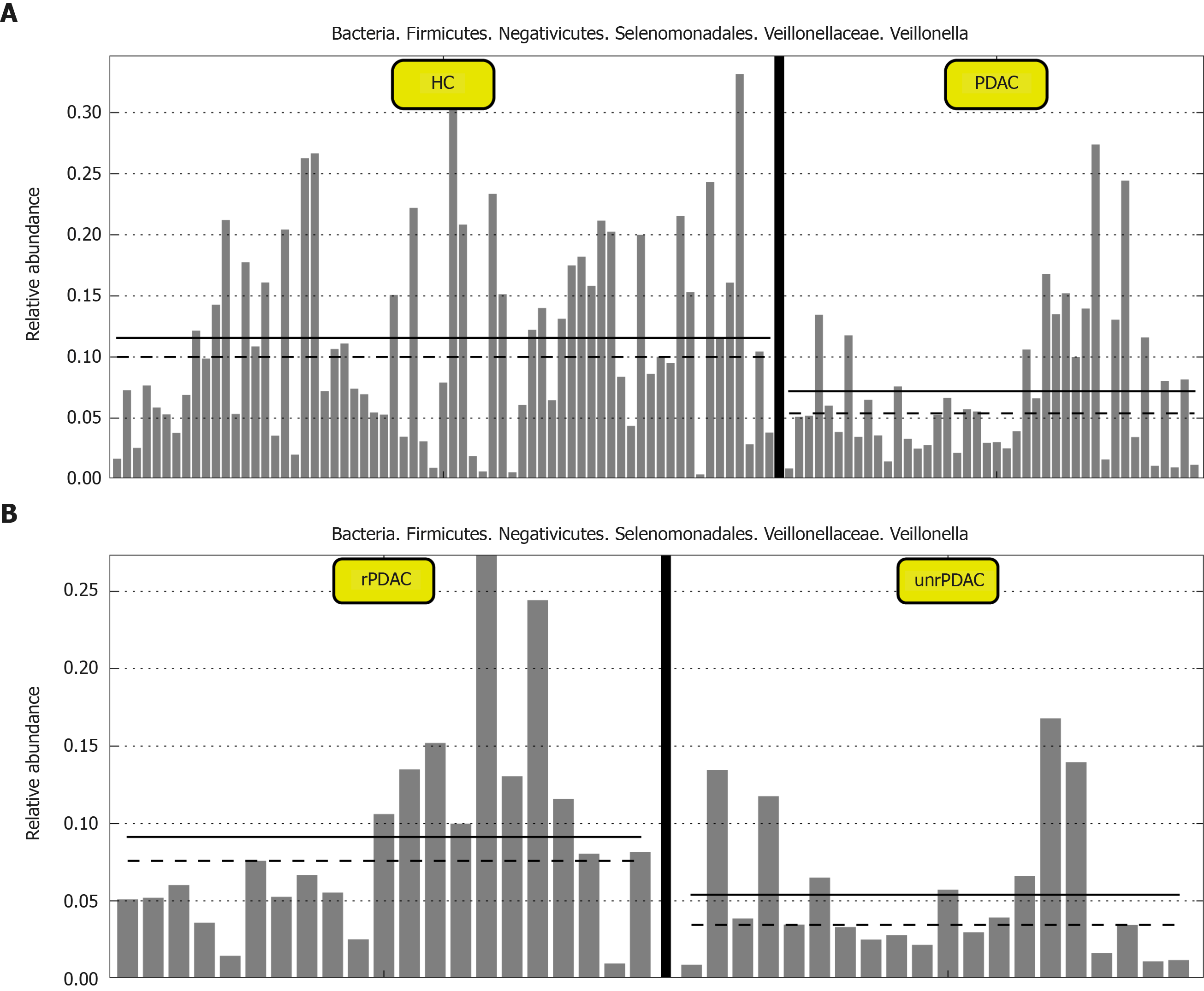
One important finding of our study was that bacteria flora was able to differentiate patients with rPDAC and unrPDAC. This is important because patients with rPDAC usually have better prognosis with timely surgical treatment. We found that species_Escherichia coli and species_Tannerella forstythia were increased significantly in unrPDAC, and these bacteria may be able to predict a tumor that is already advanced. In contrast, the expression of Veillonella demonstrated a gradual decline in saliva samples from healthy people, rPDAC, and advanced PDAC (Figure 3), which indicates that Veillonella may be protective bacteria for PDAC development.
Our study is the first to investigate the associations between bacteria profiles and symptoms related to pancreatic cancer. Symptomatic patients had different bacteria profiles than asymptomatic patients in our study. For examples, PDAC with bloating have a higher abundance of Porphyromonas, Fusobacterium, and Alloprevotella, and Alloprevotella is decreased in patients with vomiting. In addition, PDAC with jaundice had a higher amount of Prevotella compared with the PDAC without jaundice. There was a higher amount of Veillonella in patients with dark brown urine. PDAC with diarrhea had a lower amount of Neisseria and Campylobacter compared with PDAC without diarrhea. One benefit of having symptoms is that patients will seek medical help earlier, leading to the diagnosis of early PDAC and improved survival. The exact microbiome mechanism for symptoms is unknown, and more studies are needed to investigate the associations between microbiota and symptoms. Perhaps, combined symptom and microbiome evaluation may help in early detection of pancreatic cancer.
Our study had limitations. We did not include the data of other pancreatic diseases because the sample size was very small. Second, we used only 16S rRNA sequencing to analyze bacterial distributions; future research should include metagenomic sequencing to enhance accuracy of bacterial distributions. It should be noted that the rapid, inexpensive tests of 16S rRNA sequencing can have advantages for clinical implementation by using bacterial distribution test for early detection or prevention of PDAC. Some studies found the association between microbiome profile and dental disease[47]. Another limitation of our study was that we were not able to exclude participants with dental disease since our participants were not able to provide accurate history of dental disease, and there were no medical record regarding dental disease for us to verify participant dental disease status. In the future, it may be beneficial to have a professional dentist examine participant’s oral health status so as to ascertain the potential impact of oral health status on microbiome flora profile among patients with pancreatic cancer. One strength of the study is that we compared the bacteria abundances in patients with positive symptoms to find the relative association between the occurrence of symptoms and potential functions of flora.
Saliva microbiome are able to distinguish PDAC and healthy individuals. Higher Streptococcus and Leptotrichia abundances were associated with increased risk of PDAC. Veillonella and Neisseria were protective factors for detecting PDAC. Neisseria was recognized by all studies to reduce the risk of pancreatic cancer while Leptotrichia was identified in our study as a potential specific detector of PDAC in patients living in Sichuan Province, southwest China. Symptomatic patients had different bacteria profiles than asymptomatic patients. As symptoms of PDAC are usually nonspecific, combined symptom and microbiome evaluation may help in early detection of pancreatic cancer. Understanding the distribution of bacteria flora is essential step for developing probiotic treatment plans for reducing the risk of pancreatic cancer.
Understanding the distribution of bacteria flora is essential step for developing probiotic treatment plans for reducing the risk of pancreatic cancer.
The impact of geographical and medical factors, such as race and ethnicity, different dietary habits, antibiotic use, and cancer, may make the oral microbial profile differ among people from different geographic locations.
To investigate the saliva microbiome distribution in patients with pancreatic adenocarcinoma and the role of oral microbiota profiles in detection and risk prediction of pancreatic cancer.
A prospective design was utilized with 16S ribosomal ribonucleic acid gene sequencing to identify differences in bacterial taxa using a linear discriminant analysis effect size algorithm. Operational taxonomic unit values of all selected taxa were converted into a normalized Z-score, and logistic regressions were used to calculate risk prediction of pancreatic cancer.
Saliva microbiome was able to distinguish patients with pancreatic cancer and healthy individuals. Symptomatic patients had different bacteria profiles than asymptomatic patients.
Combined symptom and microbiome evaluation may help in early detection of pancreatic cancer.
Further work may focus on specific microbiota verification and diagnostic ability via large sample studies.
STROBE Statement: The authors have read the STROBE Statement—checklist of items, and the manuscript was prepared and revised according to the STROBE Statement—checklist of items.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Falasca M, Tsiaoussis J, Zhao Y S-Editor: Fan JR L-Editor: Filipodia P-Editor: Li JH
| 1. | McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. 2018;24:4846-4861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1338] [Cited by in RCA: 1262] [Article Influence: 180.3] [Reference Citation Analysis (39)] |
| 2. | Rawla P, Sunkara T, Gaduputi V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J Oncol. 2019;10:10-27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 893] [Cited by in RCA: 1520] [Article Influence: 253.3] [Reference Citation Analysis (1)] |
| 3. | Neal RD, Tharmanathan P, France B, Din NU, Cotton S, Fallon-Ferguson J, Hamilton W, Hendry A, Hendry M, Lewis R, Macleod U, Mitchell ED, Pickett M, Rai T, Shaw K, Stuart N, Tørring ML, Wilkinson C, Williams B, Williams N, Emery J. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Br J Cancer. 2015;112 Suppl 1:S92-107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 576] [Cited by in RCA: 725] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 4. | Matsubayashi H, Ishiwatari H, Sasaki K, Uesaka K, Ono H. Detecting Early Pancreatic Cancer: Current Problems and Future Prospects. Gut Liver. 2020;14:30-36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 5. | Rothwell DG, Ayub M, Cook N, Thistlethwaite F, Carter L, Dean E, Smith N, Villa S, Dransfield J, Clipson A, White D, Nessa K, Ferdous S, Howell M, Gupta A, Kilerci B, Mohan S, Frese K, Gulati S, Miller C, Jordan A, Eaton H, Hickson N, O'Brien C, Graham D, Kelly C, Aruketty S, Metcalf R, Chiramel J, Tinsley N, Vickers AJ, Kurup R, Frost H, Stevenson J, Southam S, Landers D, Wallace A, Marais R, Hughes AM, Brady G, Dive C, Krebs MG. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25:738-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 211] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 6. | Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, Kis O, Zhao Z, Spreafico A, Medina TDS, Wang Y, Roulois D, Ettayebi I, Chen Z, Chow S, Murphy T, Arruda A, O'Kane GM, Liu J, Mansour M, McPherson JD, O'Brien C, Leighl N, Bedard PL, Fleshner N, Liu G, Minden MD, Gallinger S, Goldenberg A, Pugh TJ, Hoffman MM, Bratman SV, Hung RJ, De Carvalho DD. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature. 2018;563:579-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 617] [Article Influence: 88.1] [Reference Citation Analysis (0)] |
| 7. | Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr, Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1752] [Cited by in RCA: 1885] [Article Influence: 269.3] [Reference Citation Analysis (0)] |
| 8. | Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, Anagnostou V, Parpart-Li S, Murphy D, Kay Li Q, Hruban CA, Scharpf R, White JR, O'Dwyer PJ, Allen PJ, Eshleman JR, Thompson CB, Klimstra DS, Linehan DC, Maitra A, Hruban RH, Diaz LA Jr, Von Hoff DD, Johansen JS, Drebin JA, Velculescu VE. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 368] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 9. | Gu J, Wang D, Huang Y, Lu Y, Peng C. Diagnostic value of combining CA 19-9 and K-ras gene mutation in pancreatic carcinoma: a meta-analysis. Int J Clin Exp Med. 2014;7:3225-3234. [PubMed] |
| 10. | Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 352] [Reference Citation Analysis (3)] |
| 11. | Zhang D, Hou W, Liu F, Yin J, Lu W, Li M, Zheng T, Lu F, Bao Y, Jia W. Metformin reduces serum CA199 levels in type 2 diabetes Chinese patients with time-effect and gender difference. Diabetes Technol Ther. 2015;17:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 880] [Article Influence: 146.7] [Reference Citation Analysis (0)] |
| 14. | Lu H, Ren Z, Li A, Zhang H, Jiang J, Xu S, Luo Q, Zhou K, Sun X, Zheng S, Li L. Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci Rep. 2016;6:33142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 15. | Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel Doeberitz M, Sobhani I, Bork P. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 651] [Cited by in RCA: 859] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 16. | Börnigen D, Ren B, Pickard R, Li J, Ozer E, Hartmann EM, Xiao W, Tickle T, Rider J, Gevers D, Franzosa EA, Davey ME, Gillison ML, Huttenhower C. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci Rep. 2017;7:17686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 17. | Gao S, Li S, Ma Z, Liang S, Shan T, Zhang M, Zhu X, Zhang P, Liu G, Zhou F, Yuan X, Jia R, Potempa J, Scott DA, Lamont RJ, Wang H, Feng X. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer. 2016;11:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 221] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 18. | Zhang W, Luo J, Dong X, Zhao S, Hao Y, Peng C, Shi H, Zhou Y, Shan L, Sun Q, Li Y, Zhao X. Salivary Microbial Dysbiosis is Associated with Systemic Inflammatory Markers and Predicted Oral Metabolites in Non-Small Cell Lung Cancer Patients. J Cancer. 2019;10:1651-1662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 19. | Yan X, Yang M, Liu J, Gao R, Hu J, Li J, Zhang L, Shi Y, Guo H, Cheng J, Razi M, Pang S, Yu X, Hu S. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111-3122. [PubMed] |
| 20. | Lu H, Ren Z, Li A, Li J, Xu S, Zhang H, Jiang J, Yang J, Luo Q, Zhou K, Zheng S, Li L. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J Oral Microbiol. 2019;11:1563409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 21. | Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, Purdue MP, Abnet CC, Stolzenberg-Solomon R, Miller G, Ravel J, Hayes RB, Ahn J. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67:120-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 552] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 22. | Olson SH, Satagopan J, Xu Y, Ling L, Leong S, Orlow I, Saldia A, Li P, Nunes P, Madonia V, Allen PJ, O'Reilly E, Pamer E, Kurtz RC. The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: a pilot study. Cancer Causes Control. 2017;28:959-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 23. | Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DT. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61:582-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 487] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 24. | Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjønneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, Boutron-Ruault MC, Clavel-Chapelon F, Racine A, Kaaks R, Boeing H, Foerster J, Trichopoulou A, Lagiou P, Trichopoulos D, Sacerdote C, Sieri S, Palli D, Tumino R, Panico S, Siersema PD, Peeters PH, Lund E, Barricarte A, Huerta JM, Molina-Montes E, Dorronsoro M, Quirós JR, Duell EJ, Ye W, Sund M, Lindkvist B, Johansen D, Khaw KT, Wareham N, Travis RC, Vineis P, Bueno-de-Mesquita HB, Riboli E. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764-1770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 310] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 25. | Maisonneuve P, Lowenfels AB. Risk factors for pancreatic cancer: a summary review of meta-analytical studies. Int J Epidemiol. 2015;44:186-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 291] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 26. | Wei MY, Shi S, Liang C, Meng QC, Hua J, Zhang YY, Liu J, Zhang B, Xu J, Yu XJ. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 193] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 27. | Zhao Y, Mao YF, Tang YS, Ni MZ, Liu QH, Wang Y, Feng Q, Peng JH, Hu YY. Altered oral microbiota in chronic hepatitis B patients with different tongue coatings. World J Gastroenterol. 2018;24:3448-3461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Hu J, Han S, Chen Y, Ji Z. Variations of Tongue Coating Microbiota in Patients with Gastric Cancer. Biomed Res Int. 2015;2015:173729. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 29. | Kilian M, Chapple IL, Hannig M, Marsh PD, Meuric V, Pedersen AM, Tonetti MS, Wade WG, Zaura E. The oral microbiome - an update for oral healthcare professionals. Br Dent J. 2016;221:657-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 714] [Article Influence: 102.0] [Reference Citation Analysis (0)] |
| 30. | Dawes C, Wong DTW. Role of Saliva and Salivary Diagnostics in the Advancement of Oral Health. J Dent Res. 2019;98:133-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 123] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 31. | Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 527] [Cited by in RCA: 1123] [Article Influence: 187.2] [Reference Citation Analysis (0)] |
| 32. | Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621-1624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5437] [Cited by in RCA: 5652] [Article Influence: 434.8] [Reference Citation Analysis (0)] |
| 33. | Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112-5120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4352] [Cited by in RCA: 4597] [Article Influence: 383.1] [Reference Citation Analysis (0)] |
| 34. | Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077-3079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 702] [Cited by in RCA: 814] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 35. | Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614-620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2616] [Cited by in RCA: 2763] [Article Influence: 230.3] [Reference Citation Analysis (0)] |
| 36. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23748] [Article Influence: 1583.2] [Reference Citation Analysis (0)] |
| 37. | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194-2200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9953] [Cited by in RCA: 9751] [Article Influence: 696.5] [Reference Citation Analysis (0)] |
| 38. | Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8951] [Cited by in RCA: 9979] [Article Influence: 831.6] [Reference Citation Analysis (0)] |
| 39. | Schbath S, Prum B, de Turckheim E. Exceptional motifs in different Markov chain models for a statistical analysis of DNA sequences. J Comput Biol. 1995;2:417-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 56] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 40. | Teeling H, Meyerdierks A, Bauer M, Amann R, Glöckner FO. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol. 2004;6:938-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 258] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 41. | Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 397] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 42. | Ren Z, Jiang J, Xie H, Li A, Lu H, Xu S, Zhou L, Zhang H, Cui G, Chen X, Liu Y, Wu L, Qin N, Sun R, Wang W, Li L, Wang W, Zheng S. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget. 2017;8:95176-95191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 43. | Torres PJ, Fletcher EM, Gibbons SM, Bouvet M, Doran KS, Kelley ST. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ. 2015;3:e1373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 44. | Tian D, Mo SJ, Han LK, Cheng L, Huang H, Hao S, Guan YL, Jiang KY, Deng JY, Feng HH, Wen HY, Fu MY. Investigation of Dietary Factors and Esophageal Cancer Knowledge: Comparison of Rural Residents in High- and Low-incidence Areas. Sci Rep. 2018;8:4914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Chen YH, Zou XN, Zheng TZ, Zhou Q, Qiu H, Chen YL, He M, Du J, Lei HK, Zhao P. High Spicy Food Intake and Risk of Cancer: A Meta-analysis of Case-control Studies. Chin Med J (Engl). 2017;130:2241-2250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 46. | Mills S, Stanton C, Lane JA, Smith GJ, Ross RP. Precision Nutrition and the Microbiome, Part I: Current State of the Science. Nutrients. 2019;11:923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 47. | Bracci PM. Oral Health and the Oral Microbiome in Pancreatic Cancer: An Overview of Epidemiological Studies. Cancer J. 2017;23:310-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |