Published online Jun 14, 2020. doi: 10.3748/wjg.v26.i22.2889
Peer-review started: December 28, 2019
First decision: March 21, 2020
Revised: March 26, 2020
Accepted: May 26, 2020
Article in press: May 26, 2020
Published online: June 14, 2020
Processing time: 169 Days and 5.1 Hours
Exosomes, a class of extracellular vesicles, are small membrane-bound vesicles derived from almost all cell types that can play important roles in intercellular communication. Exosomes contain proteins, lipids, and nucleic acids that are obtained from the parental cells and participate in various pathophysiological processes, including cell growth, migration, inflammation, immune regulation, and tumor pathogenesis. Moreover, exosomes might be applied in clinical settings, such as diagnosis, treatment, and outcome prediction of diseases, including various cancers. The incidence rates of Barrett's esophagus (BE) and esophageal adenocarcinoma (EAC) have increased in recent decades, and studies have proposed specific factors that may contribute to the development and progression of these diseases. However, how exosomes play a role in this pathological process needs to be clarified. Studies have identified candidate microRNAs (miRNAs) that might be related to BE/EAC. Further studies are needed to ascertain whether circulating exosomal miRNAs are altered before or after disease onset, which could also help understand the pathophysiology of and find potential targets for prevention, diagnosis, and therapy in BE/EAC. This review summarizes recent findings on the features of circulating exosomal miRNAs in BE/EAC, which could be valuable for the early diagnosis, therapeutic approaches, and outcome prediction of BE/EAC.
Core tip: Barrett's esophagus (BE) is the only recognized precursor of esophageal adenocarcinoma (EAC), which is a common malignancy with a poor prognosis. Exosomes contain proteins, lipids, and nucleic acids and participate in various pathophysiological processes. The role of exosomes in BE/EAC progression cannot be ignored. Current studies on circulating exosomal microRNAs (miRNAs) in BE/EAC have mainly focused on miRNA profiling, and little is known about the associations between circulating exosomes and the pathogenesis of BE/EAC. This review focuses on the features of circulating exosomal miRNAs in BE/EAC, which might be potential biomarkers and play a causative role in BE/EAC pathogenesis.
- Citation: Lv J, Zhao HP, Dai K, Cheng Y, Zhang J, Guo L. Circulating exosomal miRNAs as potential biomarkers for Barrett's esophagus and esophageal adenocarcinoma. World J Gastroenterol 2020; 26(22): 2889-2901
- URL: https://www.wjgnet.com/1007-9327/full/v26/i22/2889.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i22.2889
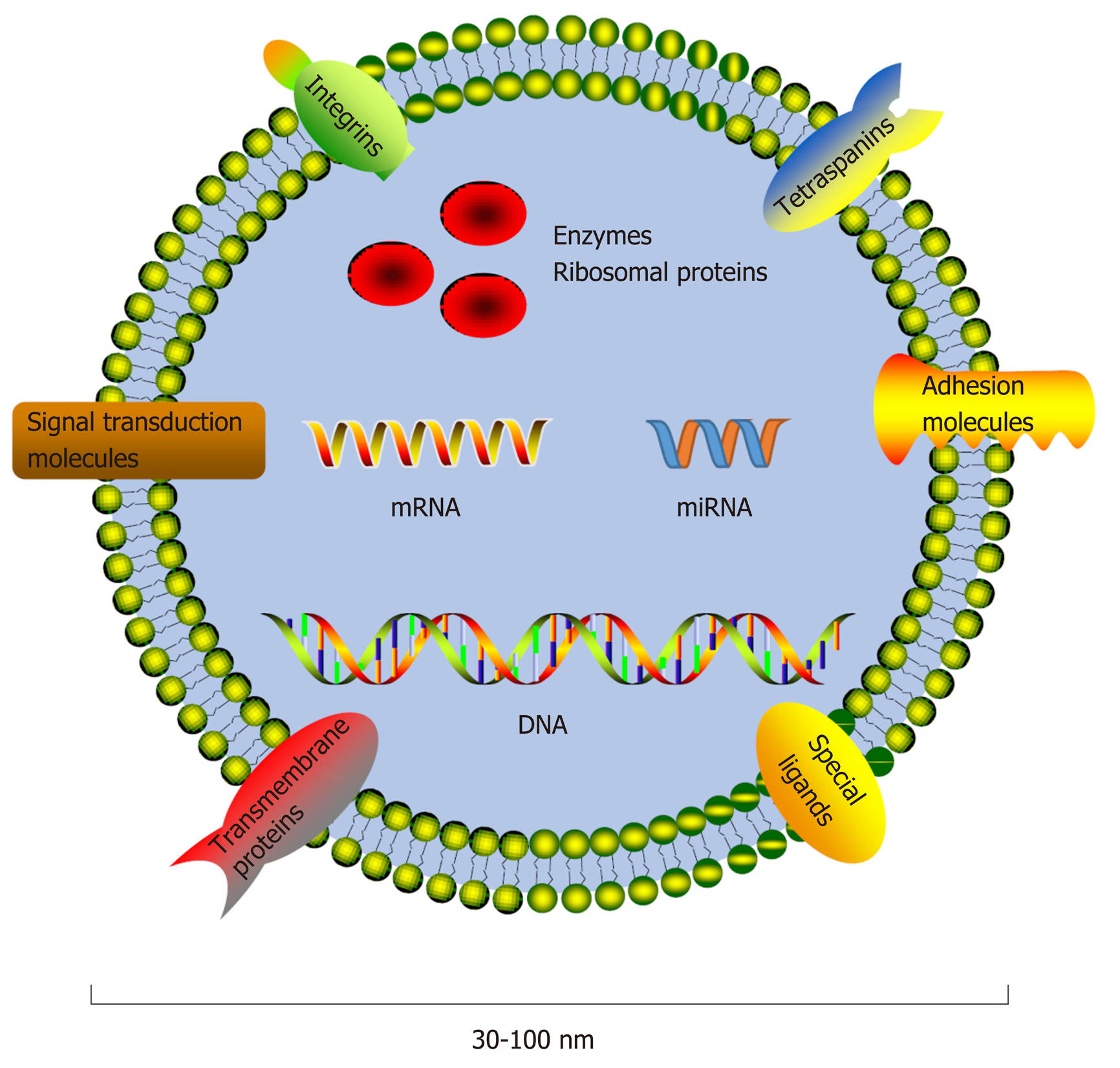
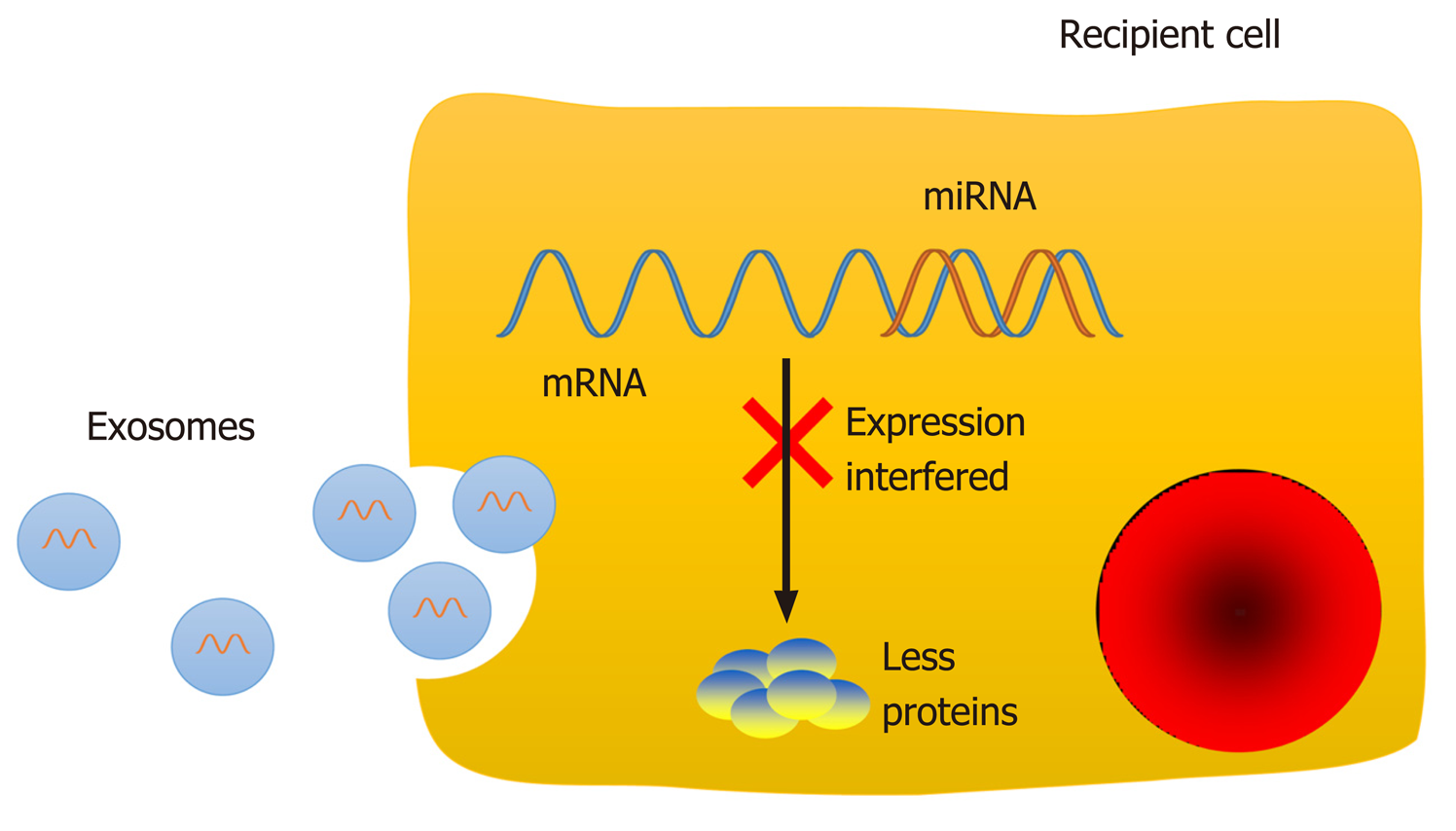
Molecules performing specific functions are frequently separated and compartmentalized into organelles, and the components of organelles are exchanged dynamically with the rest of the cell to perform their physiological functions[1]. The majority of organelles stay within the cell, while others, such as exosomes, are secreted into extracellular spaces. Exosomes, a class of membrane-bound extracellular vesicles, are released from their parental cell membrane by shedding or budding spontaneously[2]. Exosomes are approximately 30-100 nm in diameter and can only be revealed by electron microscopy by their "cup shaped" morphology[3]. Exosome secretion is a general phenomenon, and exosomes can be found in blood as well as other biological fluids[4-9], such as saliva, breast milk, urine, and semen[10-13]. Since differential ultracentrifugation enabled exosome isolation[14], studies about exosomal components have appeared, and exosomes have already become an attractive topic in medical research. Differences in exosomal content have greatly facilitate their characterizations, indicating heterogeneity within the exosome population[2,15]. Exosomes contain a set of proteins, lipids, and nucleic acids, such as mRNAs and noncoding RNAs (ncRNAs) (Figure 1)[16]. Exosomal protein compositions vary depending on the cell origins and unique tissue types. However, a set of proteins were identified that were present or enriched in exosomes despite cell origins and tissue types, such as tetraspanins (CD9, CD63, and CD81), integrins, and adhesion molecules[17]. The protein analytic approaches include Western blot, flow cytometry, and global proteomic analysis[17]. Moreover, some metabolic enzymes, ribosomal proteins, and transmembrane and signal transduction molecules could also be detected in exosomes. Specific ligands present on the exosomal membrane could be recognized by the corresponding receptors[2]; thus, exosomes could fuse with the recipient cell membrane directly and release their inner contents into the cytoplasm. However, not all cells internalize exosomes but instead interact with surface molecules to deliver signals as another communication method[18]. The RNA molecules in exosomes are also cell-specific and tissue-specific. As a type of small ncRNA, microRNAs (miRNAs) can interact with the mRNA 3'-untranslated region via their seed sequence and then interfere with the expression of target genes post-transcriptionally to regulate physiological and pathological processes (Figure 2)[19-24]. Thus, exosomes could carry and transfer miRNAs into recipient cells, and the transferred miRNAs may play a role in cell biological activities, such as cell proliferation, differentiation, migration, and apoptosis[13,19-21]. Therefore, exosomes could regulate the functions of neighboring cells and distant cells by means of autocrine, paracrine, and endocrine mechanisms[25], as a noval communication method between cells[26].
The presence of exosomes in various biological fluids makes them easily accessible. Tumor cells have been found to release exosomes into the circulation and other biological fluids, and the levels of tumor-derived exosomes in cancer patients were higher than those in healthy controls[27,28]. Taverna et al[28] showed that exosomes play an important role in non-small cell lung cancer, especially exosomal miRNAs in lung cancer diagnosis and prognosis[29]. Moreover, cancer-specific exosomal miRNAs in ovarian cancer patients were detected, and the profiles varied between different disease stages[30]. In other words, exosomes may have potential effects on disease pathology and represent a new potent noninvasive tool for disease diagnosis[29,31-38], while their genetic content, such as mRNAs and miRNAs, allows easy screening for genetic markers of diseases. Therefore, more studies on the extensive characterizations of exosomal RNAs should be conducted in different disease settings, as well as in healthy human controls.
Esophageal cancer is a common malignancy worldwide, and esophageal adenocarcinoma (EAC) has experienced an incredibly rapidly increasing incidence in recent decades[39-41]. The five-year survival rate of EAC patients is less than 20% because of the advanced stages when patients are first diagnosed with the disease[42,43]. The incidence of EAC developing from Barrett's esophagus (BE) is much higher than that in the general population[44,45]. The only well-recognized precursor of EAC[46], BE, is defined as a metaplastic condition in which the stratified squamous epithelium lining of the lower esophagus is replaced by a columnar epithelium[47]. However, the underlying mechanisms of the pathogenesis and carcinogenesis for the progression from BE to EAC have not yet been clarified[48]. Thus, BE has become a focus of attention. Many researchers have shown that specific factors can contribute to the development of BE/EAC, and these identified risks include but are not limited to gastroesophageal reflux (GER), male gender, aging, obesity, tobacco consumption, and Helicobacter pylori eradication[42,49,50]. Of note, studies on BE pathogenesis could provide approaches to prevent EAC development, and even inhibit carcinogenesis[51]. However, preventive strategies are lacking. The roles of exosomes and exosomal miRNAs in BE/EAC progression cannot be ignored. Although studies of associations between miRNAs and BE/EAC are increasing, little is known about the relationships between circulating exosomes and the pathogenesis of BE/EAC. This review will focus on the features of circulating exosomal miRNAs in patients with BE/EAC, which could have the potential to promote disease development and be used as noninvasive biomarkers to help improve early diagnosis, therapeutic approaches, and outcome prediction in BE/EAC.
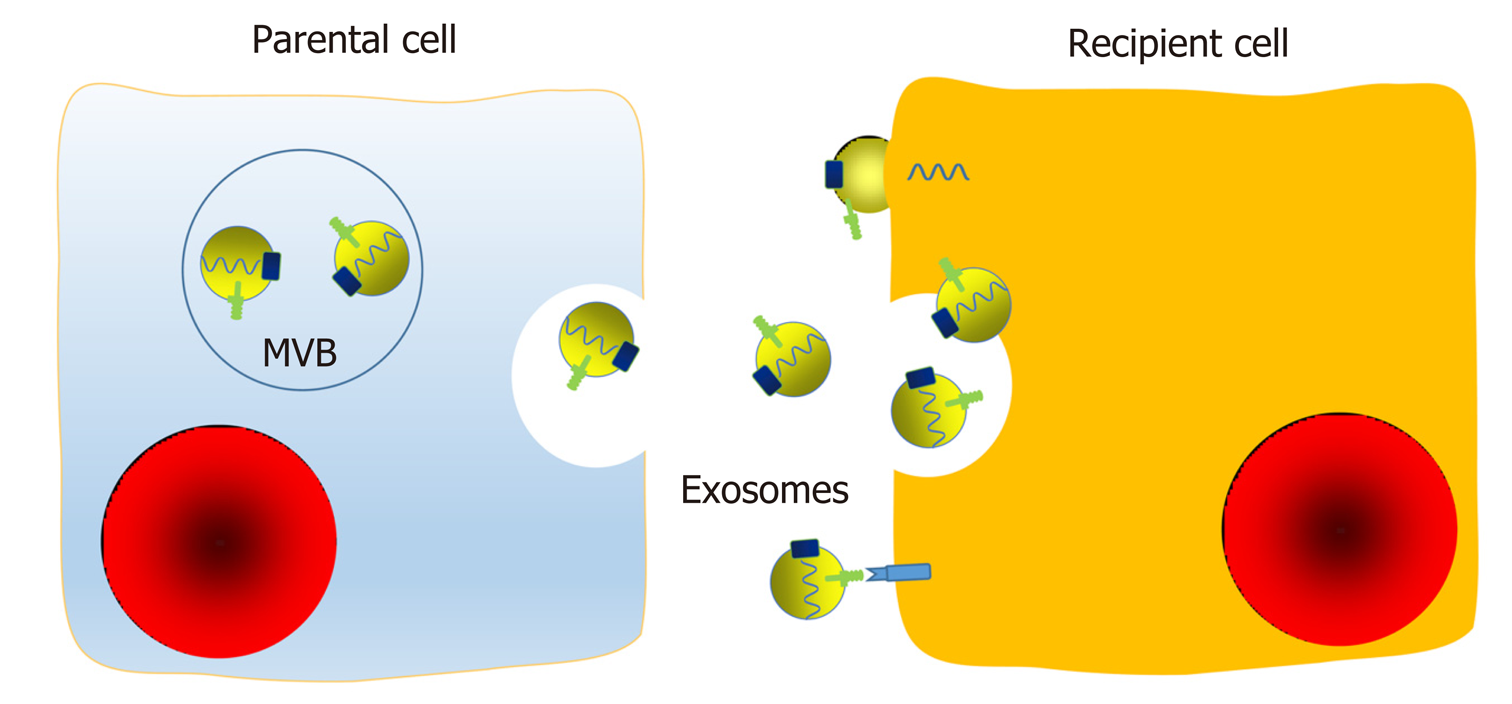
Foremost among all exosomal functions is the information transfer from parental cells to recipient cells[26]. Cellular communication can occur in several ways, including chemical receptor-mediated interaction, direct cell-cell contact, and synapses. Communication between cells is pivotal, and interactions between tumor cells are important for progression, angiogenesis, and invasiveness. Exosomes may represent a novel and essential way of intercellular communication and interaction, as exosomes not only participate in the local environment but also function at a distance by endocrine means[25,52]. After fusion with the cellular membrane, exosomes are released into the extracellular microenvironment and then circulate to serve as communication agents. Their surface proteins, lipids, and inner contents, including soluble factors, enzymes, DNA and RNA, are biologically active and capable of influencing the functions of recipient cells[2,53,54]. Exosomes can interact with surface membrane molecules, fuse with the recipient cell membrane directly, or be endocytosed in a nonselective way to transfer their inner components (Figure 3). In addition, exosomes can release small aggregates or be cleaved by proteases and then be taken up via a phagocytic mechanism by neighboring cells or act as ligands.
In addition to protein transfer, exosomes also contribute to the intercellular transmission of genetic information in the form of miRNAs[3,55], which are protected from degradation by ribonucleases (RNase)[31]. Some exosomal miRNAs can be detected in parental cells, while some can only be detected in either exosomes or parental cells, which may suggest programmed sorting of miRNAs into exosomes[31,56]. A number of studies have proposed several potential mechanisms by which exosomal miRNAs participate in human carcinogenesis through complicated interactions with the human host immune system and signaling pathways[2,10,15]. Tumor-derived exosomes can carry immunosuppressive molecules and factors that interfere with immune cell functions and then affect the development and maturation of immune cells[2]. Moreover, exosomes may reprogram the receipt cell functions to promote tumor progression after delivering genomic miRNAs[2]. Circulating exosomes definitely have the potential to be noninvasive biomarkers of disease progression and exert certain effects on disease pathophysiology. Therefore, the interactions and underlying mechanisms between exosomes and various diseases should be determined.
The diagnosis of BE and EAC is mainly based on gastrointestinal endoscopic procedures and histopathological examinations. To investigate tissue miRNAs for BE diagnosis, Li et al[57] analyzed the miRNA expression profiles of two independent sets of patients with BE and without BE. After microarray and Nanostring nCounter assays and target prediction using the related database and microarray datasets, the increased expression of miR-192 and miR-194 attracted their attention. The authors indicated that miR-194 negatively regulated GRHL3 expression, which can result in the development of squamous cell carcinoma and is known to activate PTEN transcription[58]. Thus, the miR-194-GRHL3-PTEN network may regulate normal esophageal cell growth and be involved in BE pathogenesis[57]. Luzna et al[59] indicated that the combination of upregulated miR-192 and miR-196a and downregulated miR-203 in esophageal tissues could be the biomarker to distinguish BE patients from the general population and independently diagnose patients with BE in spite of the fact that the histological examinations were unclear. Their data also showed that miR-196a could be considered a BE molecular biomarker, demonstrating that increased expression of miR-196a correlated with the progression from intestinal metaplasia to EAC[59]. MiR-196a was also proposed to be a potential marker of progression from BE to EAC by Maru et al[60], and they suggested that miR-196a could suppress the protein translation of KRTS, SPRRZC, and S100Ag to promote BE proliferation. Moreover, miR-145 could alter BMP4 signaling by inhibiting the translation of the transcription factor GATA6 to promote columnar epithelial differentiation and proliferation, which may be important for BE formation, development, and malignant transformation[61,62]. More studies have been conducted, and current data indicates that the expression levels of miR-21, miR-25, miR-143, miR-145, miR-192, miR-194, miR-196a, miR-215, and miR-223 are increased in BE/EAC tissues, while miR-136, miR-203, and miR-205 have been reported to be downregulated[57,59-72]. Moreover, the upregulated levels of miR-194-5p and miR-215-5p were shown to be specific to BE epithelium and have a high accuracy in BE diagnosis[57,73]. Furthermore, the miRNAs were differentially expressed among tissues from controls, BE, low-grade dysplasia, high-grade dysplasia (HGD), and EAC, indicating altered expression in the metaplasia-dysplasia-adenocarcinoma sequence[64]. However, Garman et al[74] indicated that miRNAs differ between squamous esophageal epithelium and BE/EAC but could not differentiate between BE and EAC patients. Nevertheless, it is clear that miRNAs play an important role in BE and EAC pathogenesis.
The studies above were mainly focused on esophageal tissue samples, and the sampling procedures were not pleasant, convenient, or economical. Researchers and medical doctors have made great efforts to seek a potent diagnostic tool to identify patients with BE/EAC from patients with other esophageal diseases. Due to the nature of chronic inflammatory diseases with progressive stages, searching for economic and convenient miRNA biomarkers for BE/EAC has been a challenge.
Biological fluids have a complicated extracellular milieu and include several kinds of RNA transporters[17], such as serum and plasma. Extracellular RNAs are bound to different carriers, including protein complexes, lipoproteins, and exosomes[17,75]. Numerous studies on extracellular miRNAs have indicated that circulating miRNAs can be potential biomarkers and reflect different aspects of pathophysiological conditions in a vast array of diseases[9,65,75-77]. Moreover, studies identifying the correlations between circulating and tissue miRNAs could support the hypothesis that circulating miRNAs could serve as biomarkers for various diseases[66]. In 2016, Bus et al[65] performed miRNA expression profiling using plasma from 8 EAC patients, 8 BE patients, and 6 controls, and validation of the array data (6 selected miRNAs) was conducted in 115 plasma samples of another set of 59 EAC patients, 41 BE patients, and 15 controls. They proposed that circulating miRNAs were differentially expressed in EAC and BE. Specifically, their data showed that the expression levels of miR-194-5p and miR-451a were significantly increased while miR-136-5p was significantly decreased in BE. In addition, miRNA-382-5p was significantly upregulated and miR-133a-3p was significantly downregulated in EAC[65]. After comparison with their previous data using tissue samples[61], the authors indicated that miR-194-5p and miR-451a were upregulated in both tissue and plasma samples from BE patients compared to controls, and miR-194-5p was more highly expressed in both tissue and plasma samples from EAC patients compared to controls[65]. Moreover, miR-451a was shown to be downregulated in both tissue and plasma samples from EAC patients compared to BE patients[65]. Fassan et al[78] collected a series of 280 gastroesophageal biopsies and 80 plasma samples from patients with a spectrum of phenotypic changes involved in the gastroesophageal carcinogenetic process, and the results indicated that miR-223 was overexpressed in tissue samples at an early stage in BE patients and that its level in plasma samples was also upregulated in EAC patients. Another study conducted by Cabibi et al[66] showed that the expression levels of miR-143, miR-145, miR-194, and miR-215 were increased in BE tissues compared to controls, and indicated that circulating serum miR-143, miR-194, and miR-215 were also upregulated in BE patients. Compared to the current routine surveillance and management endoscopy program with histopathological examination of tissue biopsies, a blood-derived test could be considerably more convenient, less invasive, and more cost-effective. Several studies[79,80] analyzed the miRNA expression profiles in both plasma and serum samples and found little differences in the expression of circulating miRNAs; however, further validation is required.
Circulating miRNAs are present in various forms, and circulating exosomal miRNAs could be detected more stably and accurately because of the membranous envelope and protection from RNase degradation[8,9,81,82]. Researchers have investigated whether exosomal miRNAs could be more related to disease states than total miRNAs in the circulation. Ashby et al[75] separated different miRNA carriers in serum using asymmetrical flow field flow fractionation and demonstrated that accurate quantification yielded different distribution profiles in each fraction. Distribution profiling could reveal more distinct differences in miRNA expression between healthy controls and cancer patients[75]. Zhao et al[83] compared the miRNA expression profiles in bovine serum and exosomes and showed that the profiles of total miRNAs in serum and exosomes were quite different, such as miRNA numbers, types, and expression levels. Exosomes had fewer miRNAs and represented about 78% of the total miRNAs detected in serum. In the same year, Castoldi et al[84] investigated the characteristics of vesicular and nonvesicular miRNAs in hepatectomized rats, and their results suggested that the altered expression profile of either vesicular or non-vesicular miRNAs may be more related to diseases and could be novel disease-associated biomarkers instead of total miRNAs isolated from serum/plasma.
Many existing studies have indicated that circulating exosomes in biological fluids could play a role in cellular communication and pathogenesis by delivering specific molecules from the parental cells to distant target cells. Research on miRNA profiling of disease-related circulating exosomes may help determine the potential use of exosomes as diagnostic biomarkers through noninvasive blood tests[1]. Moreover, the pathological state of cancer cells may be reflected by exosomal shuttle profiling[85]. Thus, circulating exosomal miRNAs have attracted great attention and could be a potential approach in biomarker exploration[8].
For the first time, Warnecke-Eberz et al[85] compared the exosomal miRNA profiles of EAC patients with those obtained from healthy controls in a study of matched primary tumor and normal tissues from EAC patients. They proposed that "exosomal onco-miRNAs", including miR-126-5p, miR-146a-5p, miR-192-5p, miR-196b-5p, miR-223-3p, miR-223-5p, miR-409-3p, and miR-483-5p, were overexpressed in exosomes and tumor tissues compared with the corresponding normal tissues. Moreover, miR-22-3p, miR-23b-5p, miR-27b-3p, miR-149-5p, miR-203-5p, miR-224-5p, miR-452-5p, miR-671-3p, miR-1201-5p, and miR-944-5p were significantly downregulated in EAC tissues or were undetectable in serum exosomes isolated from EAC patients. Hence, the authors suggested that the "exosomal onco-miRNAs" may play a major role in carcinogenesis and could be applied for diagnosis and therapy monitoring of EAC patients as a noninvasive tool. While Chiam et al[86] first compared the expression profiles of circulating exosomal miRNAs among 18 patients with locally advanced EAC, 10 patients with BE, and 19 healthy controls to investigate the value of circulating exosomal miRNAs to discriminate EAC patients from BE patients and healthy controls. The researchers mainly focused on the miRNA ratios and all possible permutations of miRNA ratios for each subject and indicated that a multi-biomarker panel, which included RNU6-1/miR-16-5p, let-7e-5p/miR-15b-5p, miR-17-5p/miR-194-5p, miR-25-3p/miR-320a, and miR-30a-5p/miR-324-5p, had a higher sensitivity and specificity over single miRNA ratios to identify EAC patients from BE patients and healthy controls[86]. The ratio method that they used can be potentially used to overcome the limitations associated with global normalization approaches, which is vital for accurate assessment of miRNA data[86]. Despite the exciting results, the number of studies on circulating exosomal miRNAs in BE/EAC is limited to date, and further studies on exploration and validation are needed to identify specific and unique exosomal miRNAs in BE/EAC. Furthermore, there remains a critical need to develop molecular biomarkers to stratify cancer risks in BE patients as well[87].
Among the studies mentioned above, some focused on tissue miRNAs, while others focused on total circulating miRNAs or isolated exosomal miRNAs, some of which used the miRNA expression panels for differentially expressed miRNAs. Circulating biomarkers could provide an exciting approach to overcome sampling issues and be more convenient, stable, and reliable to translate into a clinically useful test. The ideal biomarker for clinical application should become positive at the early stage of a disease process and remain positive throughout the rest of the disease stages for surveillance[87]. The diagnosis of BE/EAC at an early stage could improve prognosis, and circulating exosome analysis may be implicated in diagnosis and therapy monitoring. MiRNAs that are dysregulated in both tumor tissues and circulating exosomes may satisfy this diagnostic criterion[85]. The value of circulating exosomal miRNAs as biomarkers for early disease detection in the clinic has to be examined and verified in a prospective study. Since the exact origins and functions of circulating miRNAs and circulating exosomal miRNAs are unknown and the study subjects and methods for detecting miRNA data are diverse, we could not fully explain the obtained results or clearly elucidate the underlying mechanisms. Additionally, data normalization remains an important issue because of variations between studies, and miRNAs have not been implemented in the clinical setting because of conflicting results from various studies. The reproducibility of circulating miRNAs is quite important, and their changes in expression over time also need to be clarified[2,77].
Since exosomes carry cell-specific cargoes that could regulate recipient cell functions, is there any possibility that exosomes could be used as a therapeutic approach that can be applied in the clinic?
The findings of RNA-containing exosomes in serum/plasma suggest the possibility of delivering RNAs to distant cells at least theoretically. Valadi et al[31] indicated that exosomes isolated from cell supernatants of a mouse and a human mast cell line, as well as primary bone marrow-derived mouse mast cells, contain RNAs, many of which are not present in the cytoplasm of exosome-derived cells. These RNAs, including mRNAs and miRNAs, could be transferred into recipient cells and could function in these cells[31]. In addition, exosome-mediated transfer of RNA information from human liver stem cells could stimulate liver regeneration in a model of 75% hepatectomy[88]. Camussi et al[89] also suggested that exosomes could transfer selected proteins, mRNA, and miRNAs between stem cells and differentiated cells and act as paracrine signaling mediators. Exosomes derived from injured tissues may also reprogram the phenotype of bone marrow or resident stem cells by transferring genetic information[89]. Additionally, stem cell-derived exosomes may reprogram cells that survive injury and favor tissue regeneration[89]. The mRNA and miRNA cargoes may raise the possibility of genetic information transfer and functional alteration in recipient cells[90]. The ability of exosomes with different origins to modulate different functions suggests that exosomes may be used as a therapeutic method[89].
Although studies on how exosomal miRNAs alter target cell functions are rare in BE/EAC, there are still some exciting and promising clues that could encourage continuing research. For instance, miR-223-overexpressing EAC cells had significantly higher migratory and invasive potential than transfected control cells, and miR-223 may also modulate sensitivity to chemotherapy by targeting PARP1[63]. In addition, because miR-145 could alter BMP4 signaling by inhibiting the translation of the transcription factor GATA6 to promote columnar epithelial differentiation and proliferation, manipulation of BMP4 signaling by targeting miR-145 may be of great use to prevent BE development and even progression in BE patients at high risk[61,62]. Future validation work is needed to explore the possible therapeutic role of miR-145[61], and whether mRNAs and miRNAs entering target cells can activate translational control mechanisms remains to be further investigated. However, several clinical trials have tested the use of exosomes with limited success[91-93]. Despite these ambiguous data, targeting exosomes or generating exosomes containing requisite miRNAs may be a viable method to enhance therapeutic activity, and the reduction of exosome release or depletion of certain circulating exosomes could also be another method for therapy[94].
The dismal outcome of EAC patients highlights the need for novel prognostic biomarkers. In the interest of exploring the predictive value of miRNAs in the progression from BE to EAC, Revilla-Nuin et al[95] conducted the first long-term follow-up study to our knowledge. MiR-192, miR-194, miR-196a, and miR-196b were shown to have significantly higher expression in tissues from BE patients with progression to EAC compared to that from other BE patients without progression to EAC[95], which suggested that altered miRNA expression could identify BE patients with higher risks for progressing to EAC and indicated the potential use of miRNAs for surveillance and management of this malignancy. Leidner et al[87] compared the miRNA expression profile in esophageal tissues, including disease tissues and paired normal esophageal squamous tissues from patients with BE, HGD, and EAC using next-generation sequencing. They found that the majority of dysregulated miRNAs were commonly expressed in HGD and EAC lesions[87]. However, the authors indicated that the reduced expression of miR-31 and miR-375 may be candidate biomarkers of multistep malignant progression after comparison of BE, HGD, and EAC lesions and suggested that these two downregulated miRNAs may be associated with a poor prognosis in patients with EAC after analysis of survival data from EAC patients[87]. In addition, Mathe et al[68] indicated that low expression of miR-375 in cancerous tissues was strongly associated with a worse prognosis in EAC patients with BE. MiR-223 was also significantly upregulated along with the metaplasia-dysplasia-adenocarcinoma progression sequence[63]. Moreover, Saad et al[71] showed that the expression of miR-21, miR-31, miR-192, and miR-194 was upregulated in BE adjacent to HGD compared to isolated BE samples. In addition, downregulated miR-203 was significantly associated with tumor progression of EAC, while the expression of miR-192, miR-194, and miR-200a was higher in stage I than in advanced stages of EAC, which suggested that these miRNAs might serve as biomarkers for disease progression and outcome prediction.
These data suggested that the miRNAs could be used as biomarkers for BE/EAC to screen and monitor patients at high risk of progression to EAC. However, it is still uncertain whether these miRNAs have a role in the progression sequence or are merely biomarkers. Additionally, these results were based on tissue samples, not circulating exosomal miRNAs. Current studies have established the role of circulating exosomal miRNAs in BE and EAC, but associations between miRNA profiles and the outcome of BE/EAC patients are still under investigation.
A molecular biomarker that predicts the progression in the metaplasia-dysplasia-adenocarcinoma progression sequence of BE and EAC should be of high value in identifying individuals who have higher risks and need closer surveillance and would be of great use to formulate rational therapeutic interventions[87]. The most promising surveillance and prediction programs should contain risk stratifications with a variety of biomarkers. MiRNA expression profiling could expand the current knowledge on molecular pathology and BE/EAC carcinogenesis, and enable the identification of molecular biomarkers for the early detection and stratification of BE and prediction of progression to EAC[64]. A large-scale multi-center study is important for clinical application to rationalize these strategies in BE/EAC patients.
Almost all cells release exosomes, and exosomal miRNAs can either repress the translation of or induce the degradation of multiple target mRNAs, and thus regulate the expression of certain genes in recipient cells[96,97]. Circulating exosomes from patients with different cancers carry cancer-specific miRNAs that might be correlated with cancer progression and therapeutic reactions[98,99]. BE provides a good mechanism to study carcinogenesis of EAC[48], and the number of studies on how circulating exosomes relate to BE/EAC is increasing. However, only a few studies have focused on the effects of exosomes from specific cell types in vivo[100], and little attention has been paid to the etiology and molecular mechanisms. Exploring and understanding how circulating exosomes derived from specific cells function to influence pathophysiology and progression could improve the early diagnosis, prevention, and therapeutic approaches of BE and EAC.
Despite the progress made by studies on the feasibility and safety of using circulating exosomal miRNAs of BE and EAC in the clinic, their application in diagnosis, therapeutic utility, and outcome prediction remains a difficult problem to solve. Moreover, there are some limitations of exosomal analysis based on biological fluids. First, the techniques for extraction and isolation of circulating exosomes are not widespread, and they are mainly used in scientific research. Moreover, the experimental conditions should be standardized in such studies, and the results should be reproducible. Second, the cellular origins of exosomes are important, so further investigations are needed to determine the cells that secrete exosomes containing functional RNAs. Although exosomes are mainly derived from platelets in the circulation of healthy controls, exosomes can originate from a wide array of cells under pathological conditions[89], and normal cells could also secrete exosomes into biological fluids, which may affect the purity of biological fluid-extracted exosomes. To understand the molecular specialty of cell-derived exosomes in diseases, exosomal preparation free of normal cell-derived exosomes is needed. Therefore, the cell-specific and tissue-specific signatures should be verified, and the purification strategies should be well defined. A new valuable tool is ExoCarta[101,102], a database of previously conducted exosomal proteomic and transcriptomic studies. Tissue-specific proteins and RNAs in exosomes from certain diseases are identified and listed in ExoCarta[101,102]. Third, the functions of circulating exosomal miRNAs are largely unknown, and how the direct and indirect interactions between exosomal miRNAs and esophageal epithelium regulate local inflammation is still not completely understood. Thus, functional studies on potent miRNAs in the extracellular microenvironment and recipient cells should be conducted, which could provide important information on the mechanisms of the pathophysiology and progression of BE/EAC. Additionally, the timing of the changes in exosomal miRNAs in the circulation during disease progression should be elucidated. Since exosomes in biological fluids could be easily screened for cancer genetic markers[31], the removal of harmful plasma exosomes might be beneficial in certain pathological conditions[94]. Moreover, if engineering exosomes by specific nucleic acids could be achieved, the delivery of nucleic acids by exosomes would make exosomes ideal candidates as vectors for gene therapy[31]. Further investigation should also be helpful in defining the different regulatory activities of exosomes and the mechanisms of their targeting to recipient cells. Last, the sample sizes of related studies were not large enough, and current data are too limited to draw definitive conclusions. Study results are not completely reliable and consistent, and no consensus about the changes in circulating exosomal miRNAs has been researched. Thus, circulating exosomal miRNAs that could differentiate BE/EAC patients from other subjects have not been identified. Of note, miRNA panels have shown a better sensitivity and specificity than one single miRNA, and we still need to identify the distinct exosomal miRNAs that could be biomarkers for BE/EAC[90]. Without doubt, confirming the underlying mechanisms is complicated but also crucial, and the identification of a panel of miRNAs responsible for BE/EAC requires considerable effort.
Currently, endoscopic procedures and histopathological examinations are still the gold standard for BE/EAC diagnosis, and endoscopic therapy and surgery continue to be the optimal therapeutic approaches. The cost-effectiveness and feasibility of gastrointestinal endoscopy application urge us to seek a more effective and less invasive method to screen BE and EAC[103]. Since early detection of EAC will improve patient survival rates and quality of life[42], individuals with high risks for BE/EAC will benefit from screening and early diagnosis, and medical resources could be distributed more effectively. The associations between circulating exosomal miRNAs and BE/EAC have been indicated in the discussed studies, and the exciting roles of exosomal miRNAs give us an opportunity to explore carcinogenesis from another respective. Future prospective studies are needed to determine when circulating exosomal miRNAs change during disease onsets, identify potential targets for prevention, early diagnosis, and therapy, and improve our understanding of pathogenesis.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chiu KW, Dinç T, Shiryajev Y S-Editor: Wang JL L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1718] [Cited by in RCA: 1945] [Article Influence: 129.7] [Reference Citation Analysis (0)] |
| 2. | Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126:1216-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 442] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 3. | Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1410] [Cited by in RCA: 1727] [Article Influence: 157.0] [Reference Citation Analysis (0)] |
| 4. | Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809-3814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 205] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 5. | Poliakov A, Spilman M, Dokland T, Amling CL, Mobley JA. Structural heterogeneity and protein composition of exosome-like vesicles (prostasomes) in human semen. Prostate. 2009;69:159-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 254] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 6. | Palanisamy V, Sharma S, Deshpande A, Zhou H, Gimzewski J, Wong DT. Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS One. 2010;5:e8577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 269] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 7. | Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med. 2011;9:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 463] [Cited by in RCA: 589] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 8. | Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087-2092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1048] [Cited by in RCA: 1028] [Article Influence: 68.5] [Reference Citation Analysis (0)] |
| 9. | Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4900] [Cited by in RCA: 6147] [Article Influence: 512.3] [Reference Citation Analysis (0)] |
| 10. | Bobrie A, Théry C. Exosomes and communication between tumours and the immune system: are all exosomes equal? Biochem Soc Trans. 2013;41:263-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 11. | Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 12. | Boyiadzis M, Whiteside TL. Information transfer by exosomes: A new frontier in hematologic malignancies. Blood Rev. 2015;29:281-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 13. | Sato K, Meng F, Glaser S, Alpini G. Exosomes in liver pathology. J Hepatol. 2016;65:213-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 14. | Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161-1172. [PubMed] |
| 15. | Greening DW, Gopal SK, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015;40:72-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 483] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 16. | Camussi G, Deregibus MC, Cantaluppi V. Role of stem-cell-derived microvesicles in the paracrine action of stem cells. Biochem Soc Trans. 2013;41:283-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 166] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 17. | Lötvall J, Hill AF, Hochberg F, Buzás EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Théry C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1880] [Cited by in RCA: 2042] [Article Influence: 185.6] [Reference Citation Analysis (0)] |
| 18. | Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 461] [Cited by in RCA: 604] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 19. | Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442-17452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1586] [Article Influence: 105.7] [Reference Citation Analysis (0)] |
| 20. | Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107:6328-6333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1168] [Cited by in RCA: 1304] [Article Influence: 86.9] [Reference Citation Analysis (0)] |
| 21. | Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 963] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 22. | Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249-e258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 623] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 23. | Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1029] [Cited by in RCA: 1052] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 24. | Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1629] [Article Influence: 162.9] [Reference Citation Analysis (0)] |
| 25. | Sano S, Izumi Y, Yamaguchi T, Yamazaki T, Tanaka M, Shiota M, Osada-Oka M, Nakamura Y, Wei M, Wanibuchi H, Iwao H, Yoshiyama M. Lipid synthesis is promoted by hypoxic adipocyte-derived exosomes in 3T3-L1 cells. Biochem Biophys Res Commun. 2014;445:327-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 26. | Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, Blot B, Haase G, Goldberg Y, Sadoul R. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci. 2011;46:409-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 453] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 27. | Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-beta1. Haematologica. 2011;96:1302-1309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 360] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 28. | Taverna S, Giallombardo M, Gil-Bazo I, Carreca AP, Castiglia M, Chacártegui J, Araujo A, Alessandro R, Pauwels P, Peeters M, Rolfo C. Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice. Oncotarget. 2016;7:28748-28760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 29. | Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 901] [Cited by in RCA: 1010] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 30. | Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1730] [Cited by in RCA: 1858] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 31. | Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8246] [Cited by in RCA: 9767] [Article Influence: 542.6] [Reference Citation Analysis (0)] |
| 32. | Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4100] [Cited by in RCA: 3938] [Article Influence: 231.6] [Reference Citation Analysis (0)] |
| 33. | Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, Ohkuchi A, Matsubara S, Takeshita T, Takizawa T. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 389] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 34. | Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 558] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 35. | Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 424] [Cited by in RCA: 481] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 36. | Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, 't Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272-9285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 481] [Cited by in RCA: 582] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 37. | Huan J, Hornick NI, Shurtleff MJ, Skinner AM, Goloviznina NA, Roberts CT Jr, Kurre P. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2013;73:918-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 215] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 38. | Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126:1163-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 268] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 39. | Bird-Lieberman EL, Fitzgerald RC. Early diagnosis of oesophageal cancer. Br J Cancer. 2009;101:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 40. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11831] [Article Influence: 845.1] [Reference Citation Analysis (4)] |
| 41. | de Jonge PJ, van Blankenstein M, Grady WM, Kuipers EJ. Barrett's oesophagus: epidemiology, cancer risk and implications for management. Gut. 2014;63:191-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 42. | Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:390-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 380] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 43. | Hur C, Miller M, Kong CY, Dowling EC, Nattinger KJ, Dunn M, Feuer EJ. Trends in esophageal adenocarcinoma incidence and mortality. Cancer. 2013;119:1149-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 396] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 44. | Shaheen NJ, Crosby MA, Bozymski EM, Sandler RS. Is there publication bias in the reporting of cancer risk in Barrett's esophagus? Gastroenterology. 2000;119:333-338. [PubMed] |
| 45. | Solaymani-Dodaran M, Logan RF, West J, Card T, Coupland C. Risk of oesophageal cancer in Barrett's oesophagus and gastro-oesophageal reflux. Gut. 2004;53:1070-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 207] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 46. | Underwood JA, Williams JW, Keate RF. Clinical findings and risk factors for Candida esophagitis in outpatients. Dis Esophagus. 2003;16:66-69. [PubMed] |
| 47. | Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R; Global Consensus Group. The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus. Am J Gastroenterol. 2006;101:1900-20; quiz 1943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2368] [Cited by in RCA: 2449] [Article Influence: 128.9] [Reference Citation Analysis (2)] |
| 48. | Quante M, Graham TA, Jansen M. Insights Into the Pathophysiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:406-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 49. | Cook MB, Wild CP, Forman D. A systematic review and meta-analysis of the sex ratio for Barrett's esophagus, erosive reflux disease, and nonerosive reflux disease. Am J Epidemiol. 2005;162:1050-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 50. | Quante M, Abrams JA, Wang TC. The rapid rise in gastroesophageal junction tumors: is inflammation of the gastric cardia the underwater iceberg? Gastroenterology. 2013;145:708-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 51. | Rubenstein JH. Risk factors for Barrett's esophagus. Curr Opin Gastroenterol. 2014;30:408-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 52. | Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 594] [Cited by in RCA: 704] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 53. | Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211-34222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 1191] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 54. | Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, Milosevic J, Tkacheva OA, Divito SJ, Jordan R, Lyons-Weiler J, Watkins SC, Morelli AE. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1093] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 55. | EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1956] [Cited by in RCA: 2567] [Article Influence: 213.9] [Reference Citation Analysis (0)] |
| 56. | Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1233] [Cited by in RCA: 1432] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 57. | Li X, Kleeman S, Coburn SB, Fumagalli C, Perner J, Jammula S, Pfeiffer RM, Orzolek L, Hao H, Taylor PR, Miremadi A, Galeano-Dalmau N, Lao-Sirieix P, Tennyson M, MacRae S, Cook MB, Fitzgerald RC. Selection and Application of Tissue microRNAs for Nonendoscopic Diagnosis of Barrett's Esophagus. Gastroenterology. 2018;155:771-783.e3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 58. | Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT, Pandolfi PP, Pearson RB, Jane SM. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 185] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 59. | Luzna P, Gregar J, Uberall I, Radova L, Prochazka V, Ehrmann J Jr. Changes of microRNAs-192, 196a and 203 correlate with Barrett's esophagus diagnosis and its progression compared to normal healthy individuals. Diagn Pathol. 2011;6:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 60. | Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, Rashid A, Luthra R. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940-1948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (1)] |
| 61. | van Baal JW, Verbeek RE, Bus P, Fassan M, Souza RF, Rugge M, ten Kate FJ, Vleggaar FP, Siersema PD. microRNA-145 in Barrett's oesophagus: regulating BMP4 signalling via GATA6. Gut. 2013;62:664-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 62. | Pavlov K, Honing J, Meijer C, Boersma-van Ek W, Peters FT, van den Berg A, Karrenbeld A, Plukker JT, Kruyt FA, Kleibeuker JH. GATA6 expression in Barrett's oesophagus and oesophageal adenocarcinoma. Dig Liver Dis. 2015;47:73-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 63. | Streppel MM, Pai S, Campbell NR, Hu C, Yabuuchi S, Canto MI, Wang JS, Montgomery EA, Maitra A. MicroRNA 223 is upregulated in the multistep progression of Barrett's esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin Cancer Res. 2013;19:4067-4078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 64. | Slaby O, Srovnal J, Radova L, Gregar J, Juracek J, Luzna P, Svoboda M, Hajduch M, Ehrmann J. Dynamic changes in microRNA expression profiles reflect progression of Barrett's esophagus to esophageal adenocarcinoma. Carcinogenesis. 2015;36:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 65. | Bus P, Kestens C, Ten Kate FJ, Peters W, Drenth JP, Roodhart JM, Siersema PD, van Baal JW. Profiling of circulating microRNAs in patients with Barrett's esophagus and esophageal adenocarcinoma. J Gastroenterol. 2016;51:560-570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 66. | Cabibi D, Caruso S, Bazan V, Castiglia M, Bronte G, Ingrao S, Fanale D, Cangemi A, Calò V, Listì A, Incorvaia L, Galvano A, Pantuso G, Fiorentino E, Castorina S, Russo A. Analysis of tissue and circulating microRNA expression during metaplastic transformation of the esophagus. Oncotarget. 2016;7:47821-47830. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 67. | Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135:255-60; discussion 260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 304] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 68. | Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, Altorki NK, Casson AG, Liu CG, Wang XW, Yanaihara N, Hagiwara N, Dannenberg AJ, Miyashita M, Croce CM, Harris CC. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res. 2009;15:6192-6200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 306] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 69. | Wijnhoven BP, Hussey DJ, Watson DI, Tsykin A, Smith CM, Michael MZ; South Australian Oesophageal Research Group. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 70. | Fassan M, Volinia S, Palatini J, Pizzi M, Baffa R, De Bernard M, Battaglia G, Parente P, Croce CM, Zaninotto G, Ancona E, Rugge M. MicroRNA expression profiling in human Barrett's carcinogenesis. Int J Cancer. 2011;129:1661-1670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 71. | Saad R, Chen Z, Zhu S, Jia P, Zhao Z, Washington MK, Belkhiri A, El-Rifai W. Deciphering the unique microRNA signature in human esophageal adenocarcinoma. PLoS One. 2013;8:e64463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 72. | Kong X, Gong S, Su L, Li C, Kong Y. Expression signatures and roles of MicroRNAs in human oesophageal adenocarcinomas. J Cell Mol Med. 2018;22:123-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 73. | Bansal A, Hong X, Lee IH, Krishnadath KK, Mathur SC, Gunewardena S, Rastogi A, Sharma P, Christenson LK. MicroRNA Expression can be a Promising Strategy for the Detection of Barrett's Esophagus: A Pilot Study. Clin Transl Gastroenterol. 2014;5:e65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 74. | Garman KS, Owzar K, Hauser ER, Westfall K, Anderson BR, Souza RF, Diehl AM, Provenzale D, Shaheen NJ. MicroRNA expression differentiates squamous epithelium from Barrett's esophagus and esophageal cancer. Dig Dis Sci. 2013;58:3178-3188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 75. | Ashby J, Flack K, Jimenez LA, Duan Y, Khatib AK, Somlo G, Wang SE, Cui X, Zhong W. Distribution profiling of circulating microRNAs in serum. Anal Chem. 2014;86:9343-9349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 76. | Bansal A, Gupta V, Wang K. MicroRNA Expression Signatures During Malignant Progression From Barrett's Esophagus. J Cell Biochem. 2016;117:1288-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 77. | Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139:1443-1448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 147] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 78. | Fassan M, Saraggi D, Balsamo L, Realdon S, Scarpa M, Castoro C, Coati I, Salmaso R, Farinati F, Guzzardo V, Arcidiacono D, Munari G, Gasparini P, Veronese N, Luchini C, Valeri N, Rugge M. Early miR-223 Upregulation in Gastroesophageal Carcinogenesis. Am J Clin Pathol. 2017;147:301-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 79. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6301] [Article Influence: 370.6] [Reference Citation Analysis (0)] |
| 80. | Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the MicroRNA spectrum between serum and plasma. PLoS One. 2012;7:e41561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 441] [Cited by in RCA: 515] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 81. | Köberle V, Pleli T, Schmithals C, Augusto Alonso E, Haupenthal J, Bönig H, Peveling-Oberhag J, Biondi RM, Zeuzem S, Kronenberger B, Waidmann O, Piiper A. Differential stability of cell-free circulating microRNAs: implications for their utilization as biomarkers. PLoS One. 2013;8:e75184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 155] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 82. | Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3:23743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 465] [Cited by in RCA: 628] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 83. | Zhao K, Liang G, Sun X, Guan le L. Comparative miRNAome analysis revealed different miRNA expression profiles in bovine sera and exosomes. BMC Genomics. 2016;17:630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 84. | Castoldi M, Kordes C, Sawitza I, Häussinger D. Isolation and characterization of vesicular and non-vesicular microRNAs circulating in sera of partially hepatectomized rats. Sci Rep. 2016;6:31869. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 85. | Warnecke-Eberz U, Chon SH, Hölscher AH, Drebber U, Bollschweiler E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015;36:4643-4653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 86. | Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, Smith L, White IA, Bowen JM, Keefe D, Thompson SK, Jones ME, Hussey DJ. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J Gastrointest Surg. 2015;19:1208-1215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (1)] |
| 87. | Leidner RS, Ravi L, Leahy P, Chen Y, Bednarchik B, Streppel M, Canto M, Wang JS, Maitra A, Willis J, Markowitz SD, Barnholtz-Sloan J, Adams MD, Chak A, Guda K. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett's esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 88. | Herrera MB, Fonsato V, Gatti S, Deregibus MC, Sordi A, Cantarella D, Calogero R, Bussolati B, Tetta C, Camussi G. Human liver stem cell-derived microvesicles accelerate hepatic regeneration in hepatectomized rats. J Cell Mol Med. 2010;14:1605-1618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 232] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 89. | Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 808] [Cited by in RCA: 906] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 90. | Camussi G, Deregibus MC, Bruno S, Grange C, Fonsato V, Tetta C. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res. 2011;1:98-110. [PubMed] |
| 91. | Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 697] [Cited by in RCA: 752] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 92. | André F, Chaput N, Schartz NE, Flament C, Aubert N, Bernard J, Lemonnier F, Raposo G, Escudier B, Hsu DH, Tursz T, Amigorena S, Angevin E, Zitvogel L. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004;172:2126-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 382] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 93. | Escudier B, Dorval T, Chaput N, André F, Caby MP, Novault S, Flament C, Leboulaire C, Borg C, Amigorena S, Boccaccio C, Bonnerot C, Dhellin O, Movassagh M, Piperno S, Robert C, Serra V, Valente N, Le Pecq JB, Spatz A, Lantz O, Tursz T, Angevin E, Zitvogel L. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 763] [Cited by in RCA: 1005] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 94. | Robbins PD, Dorronsoro A, Booker CN. Regulation of chronic inflammatory and immune processes by extracellular vesicles. J Clin Invest. 2016;126:1173-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 220] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 95. | Revilla-Nuin B, Parrilla P, Lozano JJ, de Haro LF, Ortiz A, Martínez C, Munitiz V, de Angulo DR, Bermejo J, Molina J, Cayuela ML, Yélamos J. Predictive value of MicroRNAs in the progression of barrett esophagus to adenocarcinoma in a long-term follow-up study. Ann Surg. 2013;257:886-893. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 96. | Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163-179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 336] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 97. | Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5:5439-5452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 237] [Cited by in RCA: 277] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 98. | Brase JC, Johannes M, Schlomm T, Fälth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sültmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128:608-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 458] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 99. | Gracias DT, Katsikis PD. MicroRNAs: key components of immune regulation. Adv Exp Med Biol. 2011;780:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 100. | Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M, Théry C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920-4930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 503] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 101. | Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, Gangoda L, Mathivanan S. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J Mol Biol. 2016;428:688-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 722] [Cited by in RCA: 1062] [Article Influence: 106.2] [Reference Citation Analysis (0)] |
| 102. | Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241-D1244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 697] [Cited by in RCA: 836] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 103. | di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology. 2018;154:421-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |