Published online Nov 14, 2019. doi: 10.3748/wjg.v25.i42.6322
Peer-review started: September 3, 2019
First decision: October 14, 2019
Revised: October 29, 2019
Accepted: November 7, 2019
Article in press: November 7, 2019
Published online: November 14, 2019
Processing time: 72 Days and 6.4 Hours
Nonalcoholic steatohepatitis-related cirrhosis is one of the liver complications in type 2 diabetes mellitus (T2DM) and reported to be a risk factor for developing hepatocellular carcinoma (HCC). A reliable screening biomarker of liver cirrhosis (LC) and HCC among T2DM patients is important to reduce the morbidity and mortality of this disease. MicroRNA (miRNA) is considered a key player in HCC and T2DM, and it might be a hidden culprit in diabetes-associated HCC, making it a promising reliable prognostic tool.
To investigate the signature of serum miRNAs as early biomarkers for the screening of HCC among diabetic patients.
Expression profiles of miRNAs in serum samples of diabetic LC and diabetic HCC patients were assessed using Illumina sequencing; then, RT-qPCR was used to validate significantly altered miRNAs between the two groups. Candidate miRNAs were tested in serum samples of 200 T2DM patients, 270 LC patients, 200 HCC patients, and 225 healthy control subjects. Additionally, receiver operating characteristic (ROC) analysis, with area under the curve (AUC), was performed to assess the diagnostic performance of the screened miRNAs for discriminating HCC from LC and nonmalignant patients (LC + T2DM).
Expression of the sequenced miRNAs in serum was different in HCC vs LC-positive T2DM patients. Two miRNAs (miR-34a, miR-221) were significantly up-regulated and five miRNAs (miR-16, miR-23-3p, miR-122-5p, miR-198, miR-199a-3p) were significantly down-regulated in HCC compared to LC patients. Analysis of ROC curve demonstrated that the combination of these seven miRNAs can be used as a reliable biomarker for detection of HCC in diabetic patients, as it could identify HCC with high diagnostic accuracy in diabetic LC patients (AUC = 0.993) and in diabetic nonmalignant patients (AUC = 0.961).
This study validates a panel of serum miRNAs that can be used as a reliable noninvasive screening biomarker of HCC among T2DM cirrhotic and noncirrhotic patients. The study recommends further research to shed light on a possible role of c-Met in T2DM-associated HCC via the miRNA regulatory pathway.
Core tip: The identification of high-sensitivity biomarkers for detection of he-patocellular carcinoma (HCC) in high-risk individuals is essential. The present study was undertaken to identify and validate serum microRNAs as potential biomarkers for HCC among type 2 diabetes patients.
- Citation: Elemeery MN, Mohamed MA, Madkour MA, Shamseya MM, Issa NM, Badr AN, Ghareeb DA, Pan CH. MicroRNA signature in patients with hepatocellular carcinoma associated with type 2 diabetes. World J Gastroenterol 2019; 25(42): 6322-6341
- URL: https://www.wjgnet.com/1007-9327/full/v25/i42/6322.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i42.6322
The incidence of many progressive cancers among diabetic patients especially type 2 diabetes mellitus (T2DM) is rapidly growing worldwide, including that of hepatocellular carcinoma (HCC)[1]. The chief etiologies for liver cirrhosis (LC) and HCC in diabetic patients include chronic hepatitis B and C, nonalcoholic steatohepatitis (NASH), and alcoholic liver disease. Nonalcoholic fatty liver disease (NAFLD) is a condition commonly occurring among diabetic patients, due to insulin resistance. It has been demonstrated by many studies to be a cause of liver fibrosis and cirrhosis, thereby increasing the risk of HCC. Consequently, some researchers tend to advocate the presence of a relation between diabetes and the risk of developing HCC.
In a study by Huang et al[2], a significant relation was found between diabetes and HCC, even after optimized conditions for uncovering reverse causation and bias, which might indicate the presence of a clinically important and influential connection. Moreover, many observational researches have correlated T2DM with the decreased overall survival rate among patients with HCC[3]. Not only that, but some meta-analyses refer to a possible effect of certain treatment modalities of T2DM like metformin in lowering the risk of HCC and even positively affecting HCC prognosis[4]. On the other hand, treatments like insulin or sulphonylureas might be related to higher risk of HCC[3].
The fundamental biological mechanisms relating HCC and T2DM are intricate, reflecting a close interconnection between obesity, T2DM and NAFLD. These conditions are known to induce hepatic/systemic resistance to insulin, which consequently leads to release of pro-oxidant molecules and pro-inflammatory cytokines that have a potential role in HCC occurrence and progression[2].
While great efforts are made daily to control diabetes complications, the issue of HCC as a possible diabetes-associated disease with poor prognosis has not yet been sufficiently tackled. Early diagnosis of HCC in diabetic patients and timely treatment can greatly improve life expectancy and reduce mortality. However, not more than 40% of all HCC patients are responsive to the available treatment modalities, partially due to lack of biomarkers for early recognition of the disease with its clinical repercussions[5]. Thus, discovering new tumor biomarkers for HCC with high specificity and sensitivity is mandatory.
MicroRNAs (miRNAs) are noncoding, regulatory small RNA sequences that inversely modulate expression of their target genes[6,7]. Apart from being involved in several biological functions like cell apoptosis, differentiation and proliferation, miRNAs play a vital role in the occurrence and progression of various diseases, including diabetes and HCC[3,8]. Evidence has shown that miRNAs are differentially expressed by normal tissue compared to adjacent tumor tissue and have a possible dual function as either tumor suppressors or oncomirs, depending on the cellular needs[9,10]. MiRNAs are tissue specific and are stable in plasma or serum even after being subjected to unstable conditions like low pH or extreme temperature, which strengthens their role as possible reliable noninvasive biomarkers in disease diagnosis[8,11-15].
Considering the different alterations in miRNA expression occurring in diabetes, numerous miRNAs would theoretically qualify as reliable target biomarkers for better prognosis and treatment of diabetes[16]. For most researchers, it has been hard to specifically target a very small cell population like the pancreatic β-cells, so they have expanded their focus to include miRNAs’ alterations in other insulin-targeted organs, like the liver[17].
This study’s aim was to perform profiling of miRNAs by high-throughput small RNA sequencing in order to identify a panel of miRNAs that can act as efficient screening biomarkers for distinguishing T2DM patients who are at high risk of developing HCC.
A total of 200 diabetic patients with nonviral HCC (HCC group), 270 diabetic patients with nonviral LC (LC group), 200 T2DM patients (T2DM group) without liver disease (persistently normal liver profile parameters for at least 6 mo and no cirrhosis by ultrasound examination) and 225 healthy control subjects were enrolled. All were recruited from the Medical Research Institute (Alexandria, Egypt) and National Hepatology and Tropical Medicine Research Institute (Cairo, Egypt). HCC was diagnosed according to the practice guidelines of the American Association for the Study of Liver Diseases[18] and staged according to the Barcelona Clinic Liver Cancer (BCLC) system[19]. LC was diagnosed by abdominal ultrasonography examination combined with laboratory investigations of liver profile parameters, with or without liver biopsy (according to patient’s consent).
Only patients with F4-fibrosis stage, i.e., cirrhosis (by ultrasound-based transient elastography, with a cut-off value of 12.2 kPa for diagnosing F4-fibrosis stage)[20], were enrolled, for homogeneity of the study population. Active hepatitis C virus (HCV) and hepatitis B virus (HBV) infections were excluded by screening for serum HCV antibodies and hepatitis B surface antigen, respectively, and confirmed by real-time PCR whenever necessary (to exclude the potential carcinogenic effect of HBV and HCV). Diabetics were chosen with T2DM, while type 1, gestational, drug-induced and postorgan transplantation diabetics were excluded. Healthy volunteers were chosen according to normal ultrasound image of the liver, normal liver profile parameters, and no history of HCV, diabetes or cancer, to serve as healthy controls.
The study was approved by the ethics committees of the Medical Research Institute and National Hepatology and Tropical Medicine Research Institute, and all the patients were required to sign the informed consent form.
As a preliminary step, pools of serum samples from 10 LC patients and 10 HCC patients, respectively, were subjected to Illumina sequencing to detect miRNAs that were expressed differently among the two groups (as the control step). The samples were subsequently analyzed by RT-qPCR assay to confirm each miRNA as a biomarker. The analysis was performed in two stages. In the first stage, 50 HCC patients and 50 LC patients were set as the first round validation groups. Then, the miRNAs that were identified as significantly different between the two groups were tested by RT-qPCR in each of the serum samples from the whole study population (200 HCC patients, 270 LC patients, 200 T2DM and 225 healthy controls).
Blood samples were drawn from HCC patients. RNA extraction was performed by miRNeasy Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. The resultant product was quantified using the Nanoquant Infinite M200 Microplate Reader (Tecan, Männedorf, Switzerland).
Total miRNA was isolated through polyacrylamide gel electrophoresis purification after a pair of adaptors was ligated to their 3’ and 5’ ends. Amplification of the miRNA (21 cycles) was performed, followed by isolation and gel purification of the fragments around 90 bp to prepare the yield for Illumina HiSeq 2500 sequencing analysis, according to the manufacturer’s instructions. Finally, data were computationally analyzed.
RT-qPCR was used to validate the sequencing of uncovered related miRNAs. Reverse transcription for miRNA was carried out by a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, United States) according to the manufacturer’s protocol (16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min) to create a miRNA cDNA library. PCR was then performed by the Roche 480 Light Cycler Real Time PCR System (Basel, Switzerland) using a TaqMan MicroRNA assay (Applied Biosystems) under the following conditions: 95 °C/10 min, 40 cycles of 95 °C/15 s, and 60 °C/1 min. Cel-miRNA-39 served as an external control. Expression of miRNA was assessed through the 2(-ΔΔCt) equation, where cel-miRNA-39 Ct value above 30 was set as an exclusion criterion[20,21].
The quantitative data were presented as mean ± standard deviation. Kolmogorov-Smirnov normality test was used for checking all continuous variables to show their distributions. The unpaired Student’s t-test and analysis of variance (ANOVA) test were used to compare the continuous variables data with normal distribution, while those with abnormal distribution were compared using the Mann-Whitney U test and ANOVA. The chi-square test was used for categorical variables. Finally, to establish the correlation between the validated miRNAs, Pearson’s correlation coefficient was calculated. Receiver operating characteristic (ROC) analysis was performed and curves were constructed; area under the curve (AUC) values were assessed and studied to evaluate the accuracy of individual and combined miRNAs in discriminating between HCC and controls as described[22,23]. P values below 0.05 were considered statistically significant. Statistical calculations were performed using SPSS 16.0 (IBM Corp., Armonk, NY, United States).
The statistical methods of this study were reviewed by the statistical review boards of the Medical Research Institute (Alexandria, Egypt) and the National Research Centre (Cairo, Egypt).
Initially, samples from 10 LC and 10 HCC patients were subjected to Illumina sequencing to detect miRNAs differentially expressed among the two groups (as the control step). Data of these patients are shown in Table 1. The result was confirmed through first confirmation analysis in 50 LC and 50 HCC patients. Further information about these patients are shown in Table 2. Statistical analysis showed no significant differences in sex, age, liver profile parameters and complete blood picture between the two groups. On the other hand, alpha-fetoprotein (AFP) level was significantly higher in HCC compared to LC patients (P < 0.005). A second confirmation analysis was performed on samples from 270 LC, 200 HCC, 200 T2DM, and 225 healthy control subjects. Statistical analysis of the clinical and routine laboratory characteristics of these groups showed significant differences in AST, ALT, AFP and total bilirubin (P < 0.001, for each). With liver disease progression (beginning from normal liver, through LC, and up to HCC), levels of albumin, hemoglobin, and platelet count tending to decrease among the studied groups (P < 0.001, for each). The imaging characteristics, routine laboratory, pathological and clinical data of the studied groups are shown in Tables 3 and 4.
| Sex | Age in yr | AST in IU/L | ALT in IU/L | Total bilirubin in mg/dL | AFP in ng/mL | Albumin in g/dL | Hemoglobin in g/dL | BCLC score | |
| HCC patients | |||||||||
| 1 | Male | 65 | 50.51 | 33.7 | 5.18 | 50.8 | 3.12 | 11.3 | A |
| 2 | Male | 55 | 38.9 | 44.29 | 1.92 | 17.8 | 3.03 | 12.2 | A |
| 3 | Male | 57 | 126.8 | 194.3 | 1.91 | 42.1 | 2.41 | 10.4 | B |
| 4 | Female | 59 | 31.42 | 26.56 | 1.38 | 25.82 | 2.63 | 9.4 | B |
| 5 | Female | 57 | 103.62 | 39.82 | 1.95 | 27.47 | 2.94 | 9.6 | B |
| 6 | Male | 57 | 39.5 | 43.19 | 1.81 | 15.8 | 2.96 | 10.2 | A |
| 7 | Female | 53 | 102.32 | 37.52 | 1.94 | 26.98 | 2.23 | 11.1 | B |
| 8 | Female | 60 | 28.62 | 21.77 | 1.2 | 2365 | 2.76 | 9.1 | B |
| 9 | Male | 57 | 137.9 | 204.3 | 1.71 | 41.7 | 2.43 | 10.0 | B |
| 10 | Male | 61 | 51.38 | 35.2 | 5.07 | 52.1 | 3.11 | 111.3 | A |
| LC patients | |||||||||
| 1 | Male | 61 | 39.4 | 18.8 | 1.58 | 26 | 3.23 | 9.0 | |
| 2 | Female | 59 | 36.8 | 59.7 | 1.51 | 43 | 3.45 | 10.6 | |
| 3 | Female | 55 | 34.9 | 58.2 | 1.49 | 40.9 | 3.01 | 10.8 | |
| 4 | Female | 52 | 50.1 | 52.3 | 1.58 | 32.9 | 2.54 | 9.2 | |
| 5 | Male | 63 | 78.5 | 30.9 | 4.99 | 7.2 | 2.46 | 10.4 | |
| 6 | Female | 57 | 51.6 | 55.5 | 1.69 | 33 | 2.25 | 10.5 | |
| 7 | Male | 62 | 77.7 | 31.7 | 5.06 | 7.6 | 3.75 | 9.6 | |
| 8 | Male | 59 | 95.4 | 74.3 | 1.22 | 3.6 | 2.78 | 10.3 | |
| 9 | Male | 53 | 93.1 | 71.9 | 1.19 | 3.5 | 2.33 | 11.2 | |
| 10 | Male | 60 | 38.9 | 18.2 | 1.53 | 25.9 | 2.54 | 10.9 | |
| Parameter | LC, n = 50 | HCC, n = 50 | P value |
| Age in yr | 55.63 ± 7.69 | 58.65 ± 8.97 | NS |
| Male sex | 66% | 66% | NS |
| ALT in IU/L | 76.98 ± 65.83 | 78.53 ± 43.19 | NS |
| AST in IU/L | 69.17 ± 63.36 | 62.78 ± 43.32 | NS |
| Total bilirubin in mg/dL | 2.53 ± 1.94 | 2.79 ± 1.78 | NS |
| Albumin in g/dL | 2.59 ± 1.47 | 2.45 ± 1.62 | NS |
| ALP in IU/L | 134.6 ± 83.5 | 103.18 ± 37.5 | NS |
| Creatinine in mg/dL | 0.98 ± 0.3 | 1.38 ± 0.87 | NS |
| Hemoglobin in g/dL | 11.42 ± 1.23 | 11.35 ± 1.68 | NS |
| AFP in ng/mL | 27.56 ± 41.23 | 165.49 ± 214.56 | < 0.005 |
| Platelet count as × 103/µL | 96.7 ± 51.6 | 149.7 ± 129.3 | < 0.005 |
| Leukocyte count as × 103/µL | 6.98 ± 2.98 | 7.61 ± 4.3 | < 0.005 |
| BCLC stage, n (%) | |||
| A | 20 (40) | ||
| B | 26 (52) | ||
| C | 4 (8) |
| Parameter | Healthy, n = 225 | T2DM, n = 200 | LC, n = 270 | HCC, n = 200 | P value |
| Age in yr | 49.92 ± 7.14 | 53.68 ± 9.48 | 57.18 ± 7.30 | 56.23 ± 8.14 | 0.26 |
| ALP in IU/L | 64.2 ± 20.8 | 134.3 ± 36.9 | 103.18 ± 37.5 | 134.6 ± 83.5 | < 0.0001 |
| ALT in IU/L | 30.2 ± 15.6 | 58 ± 18 | 31.8 ± 21.08 | 206.8 ± 133.31 | 0.001 |
| AST in IU/L | 28.6 ± 10.3 | 52.1 ± 18.0 | 58.18 ± 41.6 | 447.07 ± 249.90 | < 0.0001 |
| Bilirubin, total in mg/dL | 0.6 ± 0.2 | 0.39 ± 0.19 | 4.64 ± 7.90 | 8.39 ± 8.83 | < 0.0009 |
| Bilirubin, direct in mg/dL | 0.2 ± 0.1 | 0.86 ± 0.28 | 2.98 ± 6.5 | 6.36 ± 7.8 | < 0.0001 |
| Albumin in g/dL | 4.3 ± 1.2 | 4.01 ± 0.57 | 2.53 ± 0.49 | 2.33 ± 0.39 | < 0.0001 |
| Hemoglobin in g/dL | 11.9 ± 2.1 | 11.81 ± 4.2 | 9.20 ± 2.41 | 10.15 ± 1.98 | < 0.0001 |
| AFP in ng/mL | 9.33 ± 1.82 | 3.72 ± 1.15 | 35.56 ± 41.23 | 267.8 ± 221.9 | < 0.0001 |
| Platelet count as × 103/µL | 240 ± 97.6 | 320 ± 68.2 | 147.9 ± 129.6 | 97.8 ± 53.6 | < 0.0001 |
| leukocyte count as × 103/µL | 6.8 ± 2.4 | 8.6 ± 2.42 | 6.17 ± 4.1 | 7.18 ± 3.18 | < 0.0001 |
| INR | 1.25 ± 0.23 | 1.43 ± 0.51 | 1.81 ± 0.52 | 1.95 ± 0.63 | < 0.0001 |
| Creatinine in mg/dL | 0.7 ± 0.2 | 0.98 ± 0.31 | 0.99 ± 0.25 | 1.41 ± 0.78 | < 0.0001 |
| GGT in U/L | 12.0 ± 3.0 | 33.0 ± 12.8 | 41.54 ± 25.6 | 51.38 ± 32.86 | < 0.0001 |
| BUN in mg/dL | 29.2 ± 3.4 | 70.0 ± 20.3 | 39.9 ± 28.6 | 71.54 ± 52.83 | < 0.0001 |
| Variable | n (%) | |
| Size in cm, mean ± SD (range) | 7.41 ± 3.137 (3.2-14) | |
| Portal vein tumor thrombosis | Yes | 35 (17.5) |
| No | 165 (82.5) | |
| Number of focal lesions | Single | 25 (12.5) |
| Multiple | 175 (87.5) | |
| Site of focal lesions | Right lobe | 112 (56.0) |
| Left lobe | 60 (30.0) | |
| Both lobes | 28 (14.0) | |
| BCLC stage | A | 25 (12.5) |
| B | 81 (40.5) | |
| C | 94 (47.0) |
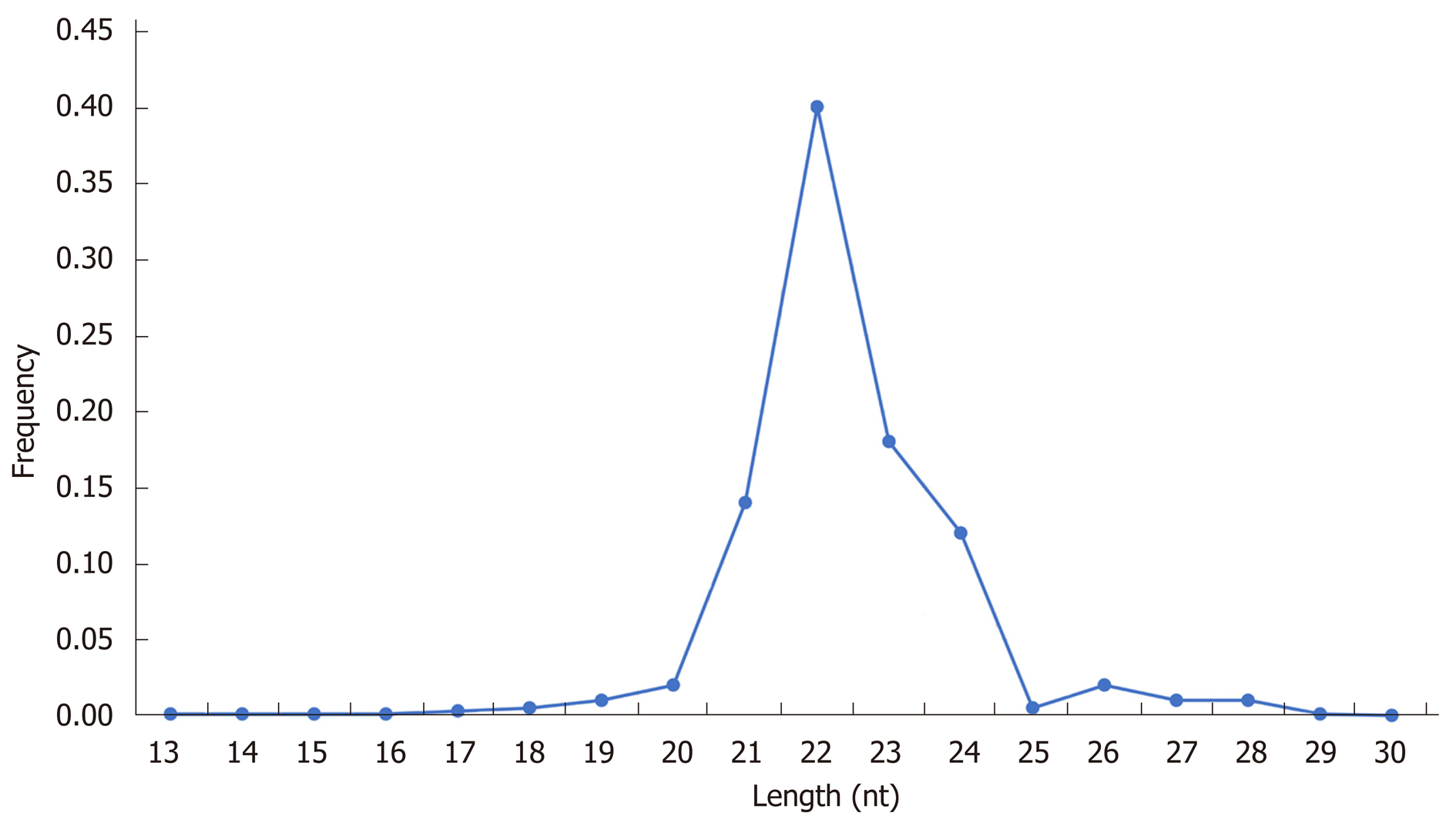
Comparison of the sum of 17784320 clean sequencing reads (60.8%) out of the 29233450 total reads in the diabetic-LC samples with the 21650404 clean sequencing reads (72.5%) out of the 29867922 total reads in the diabetic-HCC samples (Table 5) showed on Length Distribution Analysis that most reads were distributed around 22 nucleotides, as shown in Table 5 and Figure 1.
| Sample | LC | HCC |
| Total raw reads | 29233450 | 29867922 |
| Total raw bases | 1461672500 | 1493396100 |
| Total clean reads | 17784320 | 21650404 |
| Total clean bases | 432618636 | 498024958 |
| Clean reads rate, % | 60.8 | 72.5 |
| Low quality reads | 10854 | 1494 |
| Low quality rate, % | 0.04 | 0.01 |
| Without 3p adaptor reads | 2626766 | 1093718 |
| Without 3p adaptor rate, % | 8.99 | 3.7 |
| Without insert reads | 984336 | 14858 |
| Without insert rate, % | 3.4 | 0.05 |
| Poly A/T reads | 1335340 | 6929464 |
| Poly A/T rate, % | 4.6 | 23.2 |
| Ex-length reads | 6491834 | 177984 |
| Ex-length rate, % | 22.2 | 0.6 |
| Raw reads Q20, % | 97.3 | 96.7 |
| Raw reads Q30, % | 94.5 | 95.5 |
| Clean reads Q20, % | 97.5 | 97.4 |
| Clean reads Q30, % | 94.6 | 96.4 |
MiRBase database (release 22.1)[24] was used to identify known miRNAs, and 718 miRNAs were identified in LC patients while 1123 miRNAs were identified in HCC patients. Our criteria for considering differential expression among miRNAs were the following: Not less than 500 copies detected by sequencing in any recruited sample, and the miRNA showing ± 1.5 log2 fold difference between the compared groups of patients. Depending on these criteria, seven miRNAs were considered as possible miRNA candidates (Tables 6 and 7).
| MiRNA name | HCC | LC | Fold change | Log2 fold change | P value |
| hsa-miR-1 | 0.13 | 0.32 | 0.41 | -1.29 | 0.168 |
| hsa-miR-16 | 0.12 | 0.43 | 0.28 | -1.84 | 0.001a |
| hsa-miR-21 | 4.21 | 3.82 | 1.10 | 0.14 | 0.801 |
| hsa-miR-23b-3p | 0.27 | 0.78 | 0.35 | -1.51 | 0.001a |
| hsa-miR-26a | 0.33 | 0.51 | 0.64 | -0.64 | 0.009 |
| hsa-miR-29a | 0.54 | 0.42 | 1.29 | 0.37 | 0.001 |
| hsa-miR-34-a | 0.69 | 0.16 | 4.31 | 2.11 | 0.001a |
| hsa-miR-98 | 1.95 | 1.19 | 1.63 | 0.70 | 0.723 |
| hsa-miR-122-5p | 0.12 | 0.98 | 0.12 | -3.06 | 0.001a |
| hsa-miR-125b-5p | 0.01 | 0.05 | 0.20 | -2.32 | 0.823 |
| hsa-miR-127-3p | 0.29 | 0.78 | 0.37 | -1.43 | 0.059 |
| hsa-miR-150-5p | 2.16 | 0.78 | 2.78 | 1.48 | 0.071 |
| hsa-miR-185-5p | 0.21 | 0.88 | 0.24 | -2.06 | 0.083 |
| hsa-miR-198 | 0.09 | 0.29 | 0.31 | -1.69 | 0.001a |
| hsa-miR-199a-3p | 0.08 | 0.31 | 0.26 | -1.94 | 0.001a |
| hsa-miR-221 | 9.40 | 3.31 | 2.84 | 1.51 | 0.001a |
| hsa-miR-224-5p | 1.82 | 0.71 | 2.56 | 1.36 | 0.021 |
| hsa-miR-494-3p | 0.98 | 0.17 | 5.76 | 2.53 | 0.022 |
| hsa-miR-877-5p | 1.51 | 0.78 | 1.94 | 0.96 | 0.231 |
| hsa-miR-4433b-3 | 1.52 | 0.45 | 3.38 | 1.76 | 0.072 |
RT-qPCR assay was used to validate candidate miRNAs’ expression. The results of the first-round confirmation analysis (50 LC vs 50 HCC patients) demonstrating the fold change in expression quantities of candidate miRNAs displayed as (2-ΔΔCT) and considering cel-miRNA-39 as external control are shown in Table 7. A panel of seven miRNAs was found to be expressed differentially between the two groups (P ≤ 0.001, for all). On one hand, miR-16, miR-23-3p, miR-122-5p, miR-198, and miR-199a-3p were decreased significantly, while on the other hand miRNA-221 and miRNA-34 were increased significantly in the HCC group compared to the LC group.
In the second confirmation analysis round, these seven miRNAs were quantified in all three patient groups (T2DM, LC and HCC patients) as well as the normal control group. MiRNA-16, miRNA-23b-3p, miRNA-34a, miRNA-199a-3p, and miRNA-221 were found to be up-regulated significantly in T2DM patients vs controls; meanwhile, miRNA-122-5p and miRNA-198 were found to be significantly down-regulated in T2DM patients vs controls. On the other hand, miR-16, miRNA-23b-3p, miRNA-34a, miRNA-198, miRNA-122-5p and miRNA-199a-3p levels were found to be down-regulated significantly in LC patients compared to T2DM patients. The levels of miRNA-16, miRNA-23b-3p, miRNA-122-5p, miRNA-198 and miRNA-199a-3p were all found to be significantly decreased in HCC patients vs LC patients, T2DM patients, and healthy controls. The levels of miRNA-16, miRNA-199a-3p, miRNA-221, miRNA-23b-3p and miRNA-34a were found to be up-regulated significantly in diabetic patients and down-regulated in LC patients. Finally, in HCC patients, the levels of miRNA-221 and miRNA-34a were found to be up-regulated significantly, as compared to LC patients.
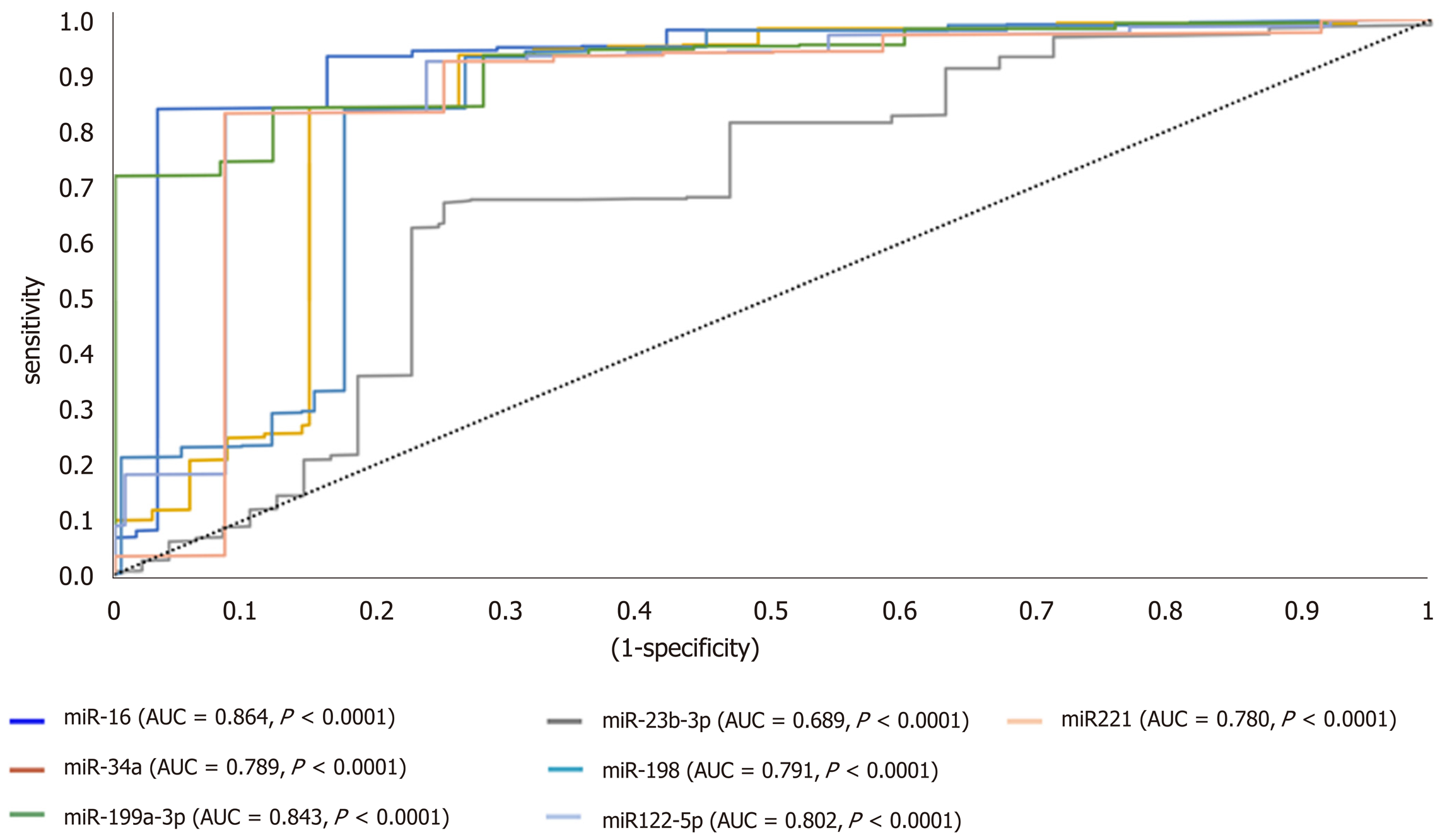
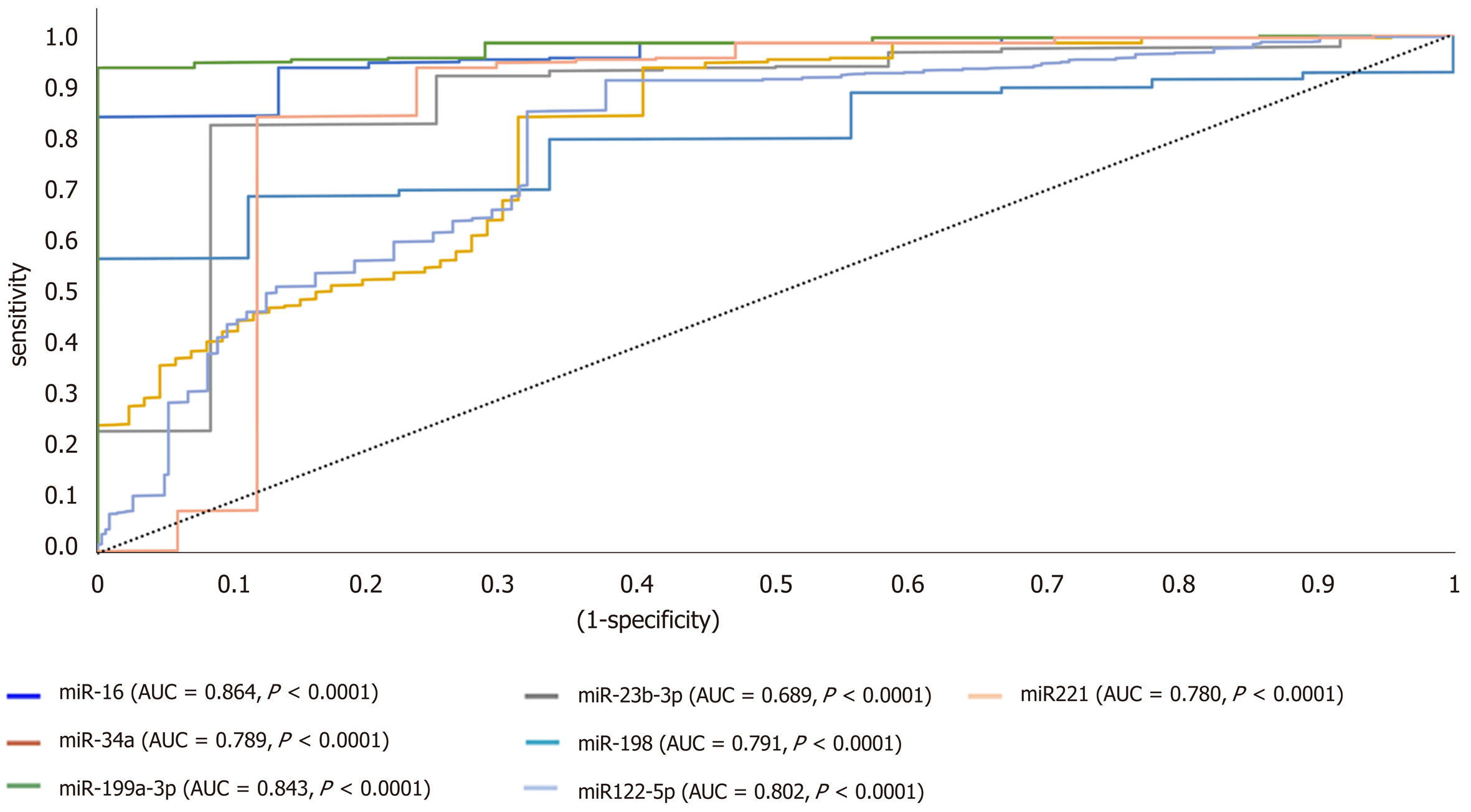
ROC analysis uncovered the reliability of the studied miRNAs for discriminating HCC from diabetic-LC with significant P value below 0.001, narrow boundaries of 95% confidence interval (CI), and AUC of 0.864, 0.689, 0.789, 0.791, 0.843, 0.802 and 0.780 for miRNA-16, miRNA-23b-3p, miRNA-34a, miRNA-198, miRNA-199a-3p, miRNA-122-5p and miRNA-221, respectively (Figure 2). For HCC discrimination from nonmalignant diabetic patients (T2DM and LC), the P values demonstrated a significant relation with narrow boundaries of 95%CI and AUCs of 0.897, 0.792, 0.761, 0.798, 0.901, 0.782 and 0.793 for miRNA-16, miRNA-23b-3p, miRNA-34a, miRNA-198, miRNA-199a-3p, miRNA-122-5p and miRNA-221, respectively (Figure 3).
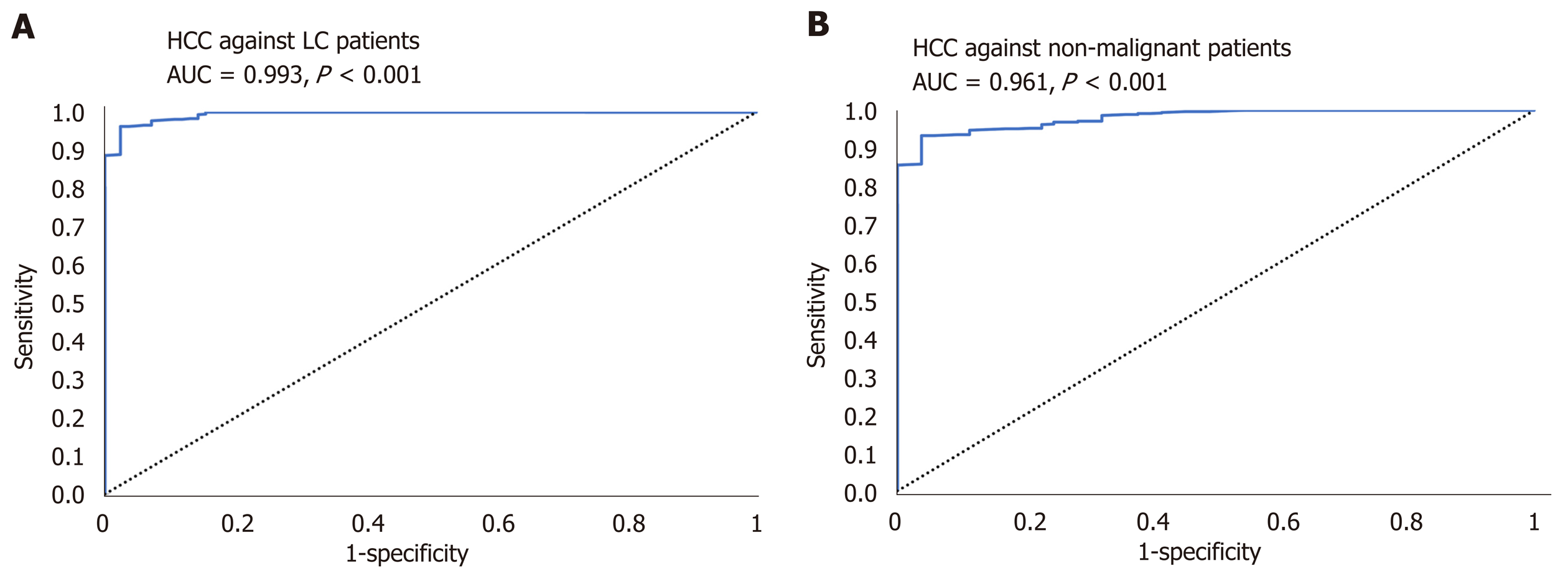
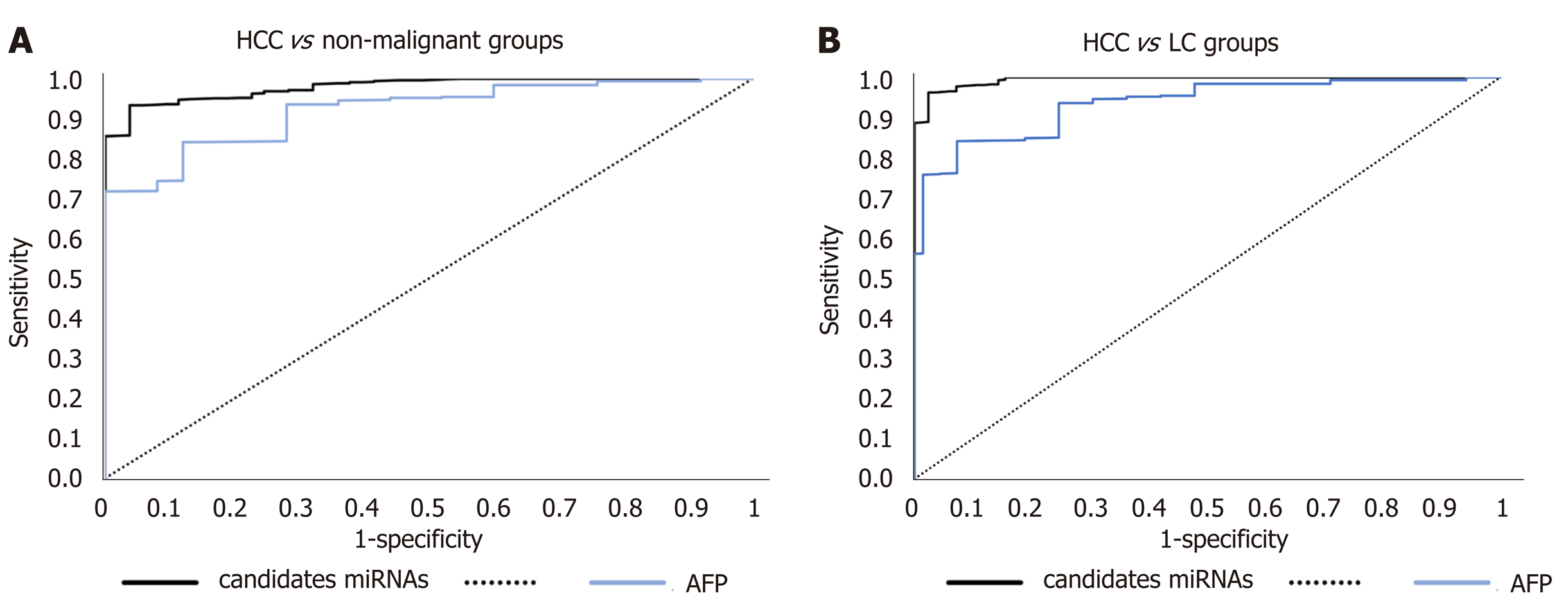
AUC of the combined panel of miRNAs for discriminating HCC patients from diabetic-LC patients reached 0.993 (95%CI, P < 0.001) and for discriminating HCC from nonmalignant diabetic patients (T2DM and LC) reached 0.961 (95%CI, P < 0.001), as shown in Figure 4.
ROC analysis for studied miRNAs proved to be useful in sorting out HCC patients from diabetic nonmalignant patients (T2DM and LC). All candidate miRNAs showed significant difference (P < 0.0001, 95%CI, AUC value of 0.897, 0.792, 0.761, 0.798, 0.901, 0.782 and 0.793 for miRNA-16, miRNA-23b-3p, miRNA-34a, miR-198, miRNA-199a-3p, miRNA-122-5p, and miRNA-221, respectively), as shown in Figure 3.
Furthermore, the candidate miRNAs demonstrated differential expression between HCC and LC groups, the latter being the group of patients at highest risk of developing HCC. ROC analysis of candidate miRNAs in LC vs HCC demonstrated AUC values as follows: 0.864, 0.689, 0.789, 0.791, 0.843, 0.802 and 0.780 for miRNA-16, miRNA-23b-3p, miRNA-34a, miR-198, miRNA-199a-3p, miRNA-122-5p and miRNA-221, respectively, as shown in Figure 2. The specificity, accuracy and sensitivity for each individual miRNA were examined for their ability to discriminate diagnostically, HCC from cirrhotic patients and/or normal healthy controls (Table 8).
| Serum miRNA | AUC value | Sensitivity | Specificity | PPV | NPV | Accuracy |
| miR-16 | ||||||
| HCC against ctrl | 0.897 | 81.0% | 92.0% | 90.6% | 86.2% | 90.1% |
| HCC against LC-T2D | 0.864 | 84.9% | 87.5% | 83.0% | 97.5% | 88.7% |
| miR-122-5p | ||||||
| HCC against ctrl | 0.793 | 79.8% | 76.2% | 80.0% | 81.0% | 81.3% |
| HCC against LC-T2D | 0.802 | 83.2% | 86.1% | 69.0% | 86.0% | 83.2% |
| miR-198 | ||||||
| HCC against ctrl | 0.798 | 81.4% | 79.4% | 77.1% | 82.0% | 80.3% |
| HCC against LC-T2D | 0.791 | 80.2% | 85.2% | 54.0% | 86.0% | 80.0% |
| miR-199a-3p | ||||||
| HCC against ctrl | 0.901 | 99.7% | 97.7% | 86.7% | 100% | 96.7% |
| HCC against LC-T2D | 0.843 | 82.6% | 85.3% | 75.7% | 96.6% | 83.6% |
| miR-221 | ||||||
| HCC against ctrl | 0.782 | 79.4% | 80.4% | 87.1% | 93.7% | 81.2% |
| HCC against LC-T2D | 0.780 | 78.6% | 81.8% | 34.9% | 91.0% | 60.8% |
| miR-23b-3p | ||||||
| HCC against ctrl | 0.792 | 81.0% | 83.2% | 76.1% | 82.0% | 79.8% |
| HCC against LC-T2D | 0.689 | 72.5% | 69.5% | 30.6% | 73.4% | 72.7% |
| miR-34a | ||||||
| HCC against ctrl | 0.761 | 96.4% | 59.5% | 83.4% | 90.7% | 75.4% |
| HCC against LC-T2D | 0.789 | 96.4% | 69.3% | 46.0% | 93.2% | 78.2% |
AFP as standard discriminator between nonmalignant and HCC individuals was assessed in the study groups, revealing an AUC value of 0.931 in discriminating HCC from LC and AUC of 0.952 in discriminating HCC from nonmalignant individuals, using a cut-off value of more than 200 ng/mL; the cut-off value for the validated panel was 40 ng/mL. A comparative analysis of AUC for miRNAs panel against AUC for AFP was done, finding no significant difference between them, as shown in Figure 5.
A strong correlation was found between miRNA-199a-3b and portal vein tumor thrombosis, which is a feature reflecting poor prognosis of HCC patients due to intra-hepatic metastasis via the portal vein route. In addition, focal lesion size significantly correlated with miRNA-23b-3p and 34a (P = 0.0008 and P ≤ 0.05, respectively). Finally, miRNA-23b-3p correlated significantly to the number of focal lesions and BCLC staging (P ≤ 0.0001 for both).
In diabetic HCC patients, miR-221 significantly correlated with miR-122-5p (r = 0.372, P = 0.003), miR-199a-3p (r = 0.381, P = 0.002), miR-198 (r = 0.365, P = 0.004), miR-34a (r = 0.392, P ≤ 0.001) and miR-23b-3p (r = 0.415, P = 0.008). MiR-34a demonstrated a correlation with miR-16 (r = 0.350, P = 0.003), miR-23b-3p (r = 0.429, P = 0.005), miR-198 (r = 0.450, P ≤ 0.001) and miR-122-5p (r = 0.421, P = 0.003). Additionally, miR-122-5p correlated with miR-23b-3p (r = 0.377, P ≤ 0.001) and with miR-199a-3p (r = 0.519, P = 0.001). Finally, miR-16 showed correlations with both miR-23b-3p and miR-198 (r = 0.682, P ≤ 0.001 and r = 0.587, P = 0.001, respectively) (Table 9).
| MiRNA name | miR-16 | miR-122-5p | miR-198 | miR-199a-3p | miR-221 | miR-23b-3p | miR-34a |
| miR-16 | NS | r = 0.587 | NS | NS | r = 0.682 | r = 0.350 | |
| P = 0.001 | P ≤ 0.001 | P = 0.003 | |||||
| miR-122-5p | NS | NS | r = 0.519 | r = 0.372 | r = 0.377 | r = 0.421 | |
| P = 0.001 | P = 0.003 | P ≤ 0.001 | P = 0.003 | ||||
| miR-198 | r = 0.587 | NS | NS | r = 0.365 | NS | r = 0.450 | |
| P = 0.001 | P = 0.004 | P ≤ 0.001 | |||||
| miR-199a-3p | NS | r = 0.519 | NS | r = 0.381 | r = 0.519 | NS | |
| P = 0.001 | P = 0.002 | P = 0.001 | |||||
| miR-221 | NS | r = 0.372 | r = 0.365 | r = 0.381 | r = 0.415 | r = 0.392 | |
| P = 0.003 | P = 0.004 | P = 0.002 | P = 0.008 | P ≤ 0.001 | |||
| miR-23b-3p | r = 0.682 | r = 0.377 | NS | r = 0.519 | r = 0.415 | r = 0.429 | |
| P ≤ 0.001 | P ≤ 0.001 | P = 0.001 | P = 0.008 | P = 0.005 | |||
| miR-34a | r = 0.350 | r = 0.421 | r = 0.450 | NS | r = 0.392 | r = 0.429 | |
| P = 0.003 | P = 0.003 | P ≤ 0.001 | P ≤ 0.001 | P = 0.005 |
A preliminary pilot study classifying patients according to type of diabetes treatment (i.e., treatment-naïve, metformin, insulin, or combined metformin plus insulin) was carried out. The result showed no significant effect of T2DM treatment regimen on the current miRNA panel expression compared to noncategorized assay (data not shown). We also compared groups according to T2DM-related parameters. The relationship between levels of candidate circulating miRNAs and the insulin sensitivity/resistance of liver determined by linear regression methods was adjusted by sex, age, triglycerides, high density lipoprotein, and body mass index. Results agreed that the hepatic insulin resistance index related significantly with all circulating miRNAs. Moreover, fasting glucose level was found to be correlated with miR-16, miR-122-5p, miR-199a-3p, miR-23b-3p and miR-34a level, while HbA1c seemed to be correlated with miR-16, miR-221, miR-23b-3p and miR-34 (Table 10). Also, miR-221 showed a tight correlation with triglycerides, while miR-16 showed correlation to both triglycerides and high density lipoprotein (r = -0.28 and r = 0.22, respectively).
| hsa-miR-16 | hsa-miR122-5p | hsa-miR-198 | hsa-miR199a-3p | hsa-miR221 | hsa-miR23b-3p | hsa-miR34a | ||
| HbA1c, % | r2 | 0.312 | 0.027 | 0.017 | 0.087 | 0.156 | 0.384 | 0.498 |
| P | < 0.001a | 0.758 | 0.369 | 0.08 | 0.003a | < 0.001a | < 0.001a | |
| GLU in mg/dL | r2 | 0.269 | 0.353 | 0.099 | 0.17 | 0.109 | 0.409 | 0.087 |
| P | 0.001a | < 0.001a | 0.026 | 0.003a | 0.016 | < 0.001a | 0.08 | |
| Insulin in mU/L | r2 | 0.091 | 0.412 | 0.041 | 0.47 | 0.031 | 0.497 | 0.063 |
| P | 0.003a | < 0.001a | 0.209 | < 0.001a | 0.003a | < 0.00a | 0.003a | |
| HOMA-IR | r2 | 0.096 | 0.419 | 0.013 | 0.098 | 0.49 | 0.492 | 0.027 |
| P | 0.03 | < 0.001a | 0.396 | 0.001a | < 0.001a | < 0.001a | 0.001a | |
| ISI | r2 | 0.095 | 0.421 | 0.102 | 0.019 | 0.499 | 0.545 | 0.0693 |
| P | 0.0074a | < 0.001a | 0.023 | 0.004a | < 0.001a | < 0.001a | 0.005a | |
| HIRI | r2 | 0.498 | 0.429 | 0.024 | 0.1807 | 0.427 | 0.672 | 0.049 |
| P | < 0.001a | < 0.001a | 0.005 | < 0.001a | < 0.001a | < 0.001a | 0.009a | |
NASH-related cirrhosis is a common liver complication in diabetics, and is reported to be a risk factor of developing HCC. Therefore, early reliable screening of LC and HCC among these patients is important to reduce the morbidity and possible mortality from this disease[25]. Advanced imaging modalities like triphasic computed tomography scan and/or liver biopsy are the current principal tools for HCC diagnosis. In spite of being relatively efficient tools, biopsy is known to increase the risk of tumor cell seeding along the needle track[26] and imaging techniques might be unable to discriminate malignant from benign tumors[27]. Thus, a reliable biomarker that can efficiently identify HCC is urgently needed.
In view of the intricate association between T2DM and HCC, some miRNAs could qualify as reliable HCC diagnostic molecules in diabetes, as several miRNAs have already been shown to be significantly dysregulated in the diabetic liver and in other insulin targeted cells. Indeed, this finding attracted the attention of our research group to validate a possible miRNA signature for T2DM-associated HCC.
MiRNAs, which are integral components of gene regulation networks controlling tumorigenesis are stably maintained in various biological fluids, including human serum/plasma[14,15]. The amounts of expressed miRNAs reflect the pathological and physiological conditions of the human body; as such, many circulating miRNAs have been approved as candidate blood biomarkers for prognosis and diagnosis of various cancers[11]. In the current study, small RNA sequencing was used as the initial screening step, followed by RT-qPCR-based validation steps to discover miRNAs specific for T2DM-associated HCC. The current miRNA profiling analysis of the sera of diabetic-LC patients and diabetic-HCC patients identified several miRNAs with variable expression patterns. The further validation analysis confirmed seven miRNAs (miRNA-16, miRNA-23b-3p, miRNA-34a, miRNA-122-5p, miRNA- 198, miRNA-199a-3p and miRNA-221) which were differentially expressed in the serum of diabetic-HCC vs diabetic-LC patients, verifying these miRNAs as reliable surrogate biomarkers of HCC.
The miRNA-122 family constitutes around 72% of total human liver-related miRNAs[28], making it one of the most disease-dysregulated miRNAs. This family also plays a critical role in regulating cholesterol and lipid metabolism, being tightly related to both risk of progression and susceptibility to metabolic syndrome and T2D in the general population[29]. Additionally, it plays a role in maintenance of liver cell integrity by serving as a tumor suppressor against HCC development via multiple target genes, such as Bcl-w (an anti-apoptotic gene)[30], metalloprotease 17 (ADAM17)[31], and cyclin G1 (CCGN1)[32]. It also regulates insulin resistance-related NAFLD[33]. However, some studies have demonstrated that miRNA-122 expression is down-regulated in HCC tumor tissues[32], while others have shown its serum level as up-regulated significantly in HCC patients compared to LC patients and healthy controls[34-37]. These contradictory findings have led some authors to hypothesize that the low level of miRNA-122 in tumor hepatic tissues might actually be due to its increased release in blood[37,38]. We found, in this study, that miRNA-122-5p was down-regulated significantly in the serum of our T2DM patients, as compared to controls, and that the serum level of miRNA-122-5p was significantly down-regulated in our diabetic HCC patients compared to nonmalignant diabetic patients or healthy controls. Thus, the current study’s findings confirm the signature/role of miRNA-122 as a diagnostic marker for detecting HCC in an LC group as well as in T2DM patients.
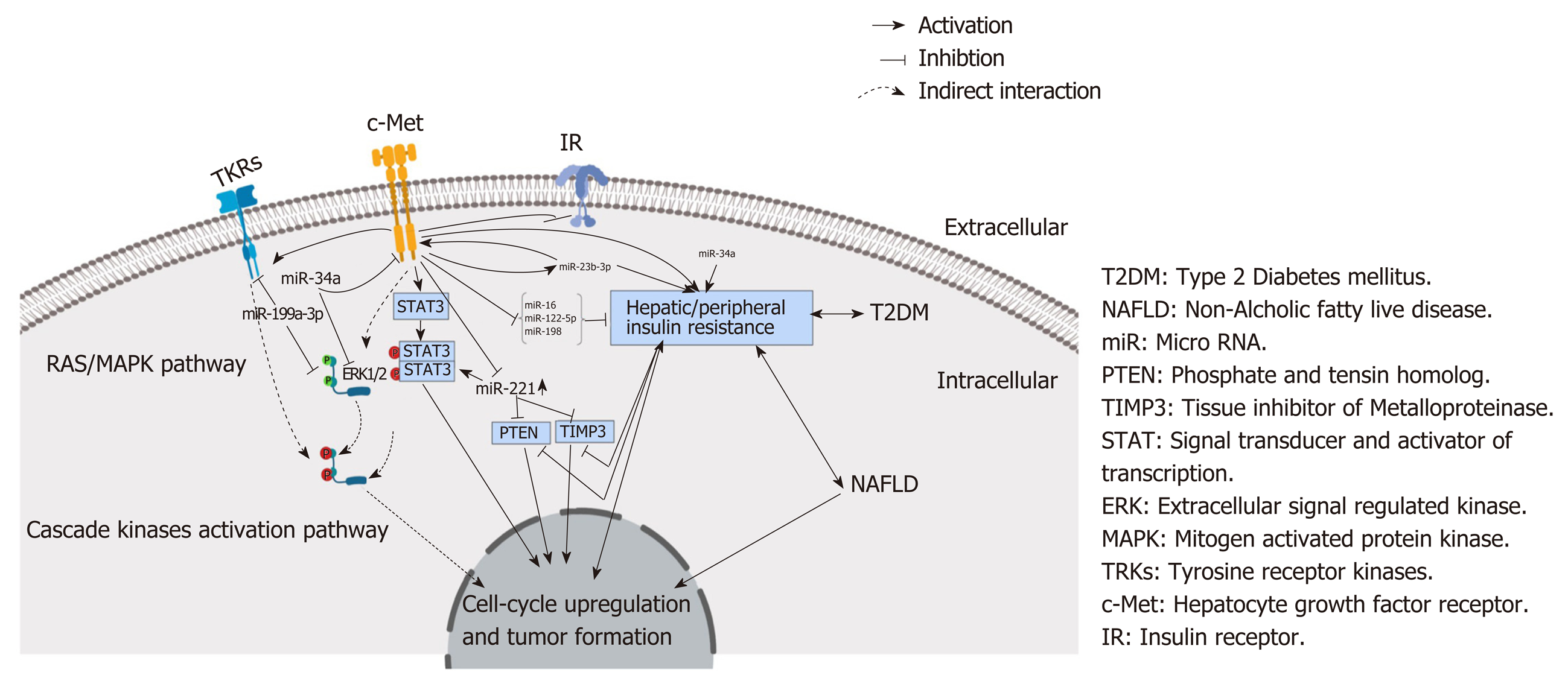
MiRNA-23b-3p has been reported by others to modulate neuropathic pain and cellular memory of high glucose in diabetic nephropathy patients[39]. Additionally, miRNA-23b-3p serves as a tumor suppressor of HCC migration and invasion by targeting Pyk2 by regulating epithelial-mesenchymal transition, which makes it a possible target for HCC treatment[40]. In addition, the plasma exosomal miR-23b-3p has been reported as being a promising noninvasive prognostic biomarker for nonsmall cell lung carcinoma in patients[41]. In HCC tissue, miRNA-23b-3p has been demonstrated to be abnormally down-regulated, in contrast to the adjacent noncancerous liver tissue[42]. In agreement with this reported down-regulation in tissue expression, our study found that serum levels of miRNA-23b-3p were also significantly down-regulated in our HCC patients compared to our LC, T2DM patients and healthy controls, suggesting that serum miRNA-23b-3p could be a novel reliable noninvasive biomarker for T2DM-associated HCC diagnosis and a possible therapeutic target due to its dual regulatory role for the receptor tyrosine kinase gene and c-Met (Figure 6).
In the present study, levels of serum miRNA-221 were found to be significantly up-regulated in HCC vs LC, T2DM patients and healthy controls. In HCC tissue, miRNA-221 has been shown by other research studies to be abnormally up-regulated and to possibly act as a tumor driver oncomir in HCC development via inhibition of caspase-3 mediated apoptosis[43] as well as by targeting multiple target genes, like nuclear factor-kB and CD44 (among others)[44], and promoting cell migration, invasion and proliferation by affecting the phosphatase and tensin homolog PTEN protein and the tissue inhibitor of metalloproteinases 3 TIMP3 protein[45]. Furthermore, kinases[46] and c-Met[45] have been reported to up-regulate miRNA-221 expression via Jun transcriptional activation in lung cancer.
The miRNA-34 family, including miRNA-34a, has been demonstrated to be commonly silenced in a diverse range of tumor types, emphasizing the role of these miRNAs in tumorigenesis, according to Giglio et al[47]. In fact, miRNA-34a has been shown to modulate cell invasion, proliferation and glucose metabolism, in an HCC cell line[16]. These results are in accordance with our finding that miRNA34a significantly correlated with tumor size and number of focal lesions in our diabetic HCC patients. The suggested mechanism behind this finding involves miR34a negatively controlling expression of lactate dehydrogenase A (referred to as LDHA), which consequently inhibits LDHAdependent glucose uptake in the liver, in addition to HCC invasion and proliferation[16]. Our findings are also in agreement with those from a recent study by Zhang et al[16], which suggested the negative regulatory role of miRNA34a on glucose metabolism and its ability to act as a novel biomarker for HCC prognosis. Another mechanism suggested by Li et al[48] in brain tumor and glioma involves miRNA-34a inhibition of cell proliferation, invasion and tumorigenesis, whereas c-Met up-regulation has been shown to partially reverse the cell cycle arrest and cell death induced by miRNA-34a, so miRNA-34a is up-regulated to modulate the phosphorylation signal cascade of kinases and c-Met[49]. This possible mechanism needs to be validated in cases of diabetic HCC. A third suggested molecular mechanism of miRNA-34a involves its ability to inhibit HCC cell growth by targeting the histone deacetylase 1 HDAC1 protein[50], the special AT-rich sequence-binding protein 2 SATB2 protein[51] and the cell cycle-dependent proteins (including p53), in addition to regulating protein output in a HepG2 cell line[52].
The miRNA-199 family is the third most abundant miRNA family in the liver tissue[53], with its member, miRNA-199a-3p, being known to serve as a tumor suppressor miRNA during hepatocarcinogenesis. In HCC patients, low-level miRNA-199a-3p has been reported to be present in both tissue and serum[53]. Consistent with these findings, the results of the current study demonstrated a significant down-regulation of serum miRNA-199a-3p levels in HCC patients compared to all other groups. Conversely, miRNA-199a-3p has been shown to be up-regulated in diabetic patients vs healthy subjects, being mostly correlated to diabetes-associated organ complications[54]. Likewise, our study demonstrated a significant up-regulation of serum miRNA-199a-3p in our T2DM patients compared to our healthy controls, whereas there was a sharp down-regulation observed in our LC and HCC patients compared to our diabetic group. In comparison, Kim et al[55] and Migloire et al[49] demonstrated that miRNA-199a-3p inhibits not only tumor proliferation but also its invasive capabilities and motility by down-regulating both c-Met and its downstream effector ERK2.
In the present study, we found serum levels of miR-198 to be significantly down-regulated in HCC patient samples compared to nonmalignant samples. In addition, down-regulation in T2DM was demonstrated when compared to the healthy group. Similar results were reported in nondiabetic patients by Huang et al[56], who demonstrated lower levels of miRNA-198 in HCC tissue compared to nonmalignant adjacent tissue. MiRNA-198 acts by suppression of motogenic and mitogenic pathways that diminish cell growth and migration[57]. As such, a possible mechanism that might modulate HCC progression in T2DM patient involves targeting of the c-Met/human growth factor signaling pathway, including the mTOR kinase, other kinases and STAT3, which play dual roles in both diabetes-related complications and HCC progression. MiRNA-198 is also known to directly target c-Met on its 3' untranslated region, leading to its overexpression, diminished human growth factor-induced phosphorylation of kinases, and, consequently, inhibition of HCC cell migration and invasion via a c-Met-dependent pathway[47].
MiRNA-16 is one of the tumor suppressive miRNAs that has been reported to act via several cell cycle tumorigeneses-incorporated pathways. For example, BCL-2 was found to be negatively regulated by miRNA-16, so that silencing of miRNA-16 leads to uncontrolled expression of BCL-2, leading to inhibition of apoptosis[58]. Our results demonstrated an elevated level of miRNA-16 in diabetic patients compared to controls, which is in accordance with findings by Bork-Jensen et al[59] of a significantly higher expression of miRNA-16 in T2DM vs type 1 diabetes mellitus, with a tight correlation to glucose/insulin homeostasis. The current study revealed a significant miRNA-16 elevation in T2DM compared to controls, followed by significant down-regulation in our LC and HCC patients. These findings suggest a significant role of this miRNA signature in diabetic HCC patients compared to nondiabetic patients. At this point, we might again suggest a potential role of c-Met and kinases as possible key targets of and regulators for miRNA-16.
Moreover, when we performed ROC analysis for miR-16, miR-122-5p, miR-198, miR-199a-3p, miR-221, miR-23b-3p, and miR-34a, the AUC values offered more solid proof that all seven miRNAs were able to significantly discriminate HCC from LC and nonmalignant groups. For discriminating HCC from LC, the AUC of miR-16 had the highest value (0.864), followed by miR-199a-3p (0.843). For discriminating HCC from nonmalignant patients, the AUC of miR-199a-3p was best (0.901), followed by miR-16 (0.897). Combining the seven miRNAs could efficiently discriminate HCC from LC (AUC of 0.993) and identify HCC from nonmalignant groups (AUC of 0.961).
To summarize, the current study demonstrated higher levels of serum miRNA-221 and miRNA-34a in HCC patients compared to LC patients, suggesting the roles of these miRNAs as potential biomarkers for HCC detection among patients at risk. MiRNA-122-5p and miR-198 were demonstrated to be down-regulated in serum of HCC and T2DM patients compared to the normal control group, with significant correlation between miRNA-122-5p and focal lesion size and number. Both miRNA198 and miRNA-122-5p were already reported to serve as tumor suppressors in HCC, working by targeting multiple genes and modulating NAFLD, which is a frequent finding in T2DM[31,57]. This suggests an important diagnostic role of both miRNAs in identifying HCC among high-risk diabetic LC patients, who mostly developed cirrhosis due to NAFLD.
Indeed, the relation between miRNAs and disease in humans, particularly cancers, has been demonstrated by many researchers shortly after the discovery of miRNA expression in human cells, in the early 2000s. MiRNA genes were shown to be located either in intergenic regions (30%) or protein-coding genes (70%)[60], but the majority of identified miRNAs were found to be located near or inside the cancer-related regions[61]. MiRNA expression signatures have the advantage of not only being both disease- and tissue-specific but also being relevant to the developmental stage of the disease[62]. Therefore, their future clinical application might extend to cover cancer diagnosis, grading, staging, treatment, prognosis, and monitoring of therapeutic response[63,64].
From our study, we conclude that the combined panel of seven miRNAs (miR-16, miR-122-5p, miR-198, miR-199a-3p, miR-221, miR-23b-3p, and miR-34a) might be used as a reliable screening biomarker for HCC among diabetic patients with or without NASH-induced cirrhosis. We recommend further studies to confirm the role of the current candidate miRNAs. Moreover, we suggest more investigation of the role of c-Met as an intermediate key player in diabetes-associated HCC.
Nonalcoholic steatohepatitis (NASH)-related cirrhosis is one of the liver complications in type 2 diabetes mellitus (T2DM) and is reported to be a risk factor for developing hepatocellular carcinoma (HCC). As such, a reliable screening biomarker of liver cirrhosis (LC) and HCC among T2DM patients will be important to reduce the morbidity and mortality of this disease. MicroRNA (miRNA) is considered a key player in HCC and T2DM, and it might be a hidden culprit in diabetes-associated HCC, which makes it a promising reliable prognostic tool.
The current study aimed to investigate the signature of serum miRNAs as early biomarkers for the screening of HCC among diabetic patients, to reduce the morbidity and mortality from HCC among diabetic patients.
The main study objective was the identification of high-sensitivity biomarkers for detection of HCC in high-risk individuals. The present study was undertaken to identify and then validate a panel (of seven) serum miRNAs as potential biomarkers for HCC among T2DM patients.
Expression profiles of miRNAs in serum samples of diabetic LC and diabetic HCC patients were assessed using Illumina sequencing, then RT-qPCR was used to validate the significantly altered miRNAs between the two groups. Candidate miRNAs were tested in serum samples of 200 T2DM patients, 270 LC patients, 200 HCC patients and 225 healthy control subjects. Additionally, receiver operating characteristic (ROC) analysis was performed to assess the diagnostic performance of the screened miRNAs for discriminating HCC from LC and nonmalignant patients (LC + T2DM).
Expression of the sequenced miRNAs in serum was different in HCC patients vs LC and nonmalignant patients. Two miRNAs (miR-34a, miR-221) were significantly up-regulated and five miRNAs (miR-16, miR-23-3p, miR-122-5p, miR-198, miR-199a-3p) were significantly down-regulated in HCC patients compared to LC patients. Analysis of the ROC curve demonstrated that the combination of these seven miRNAs can be used as a reliable biomarker panel for detection of HCC in diabetic patients, as it could identify, with high diagnostic accuracy, HCC in diabetic LC patients [area under the curve (AUC) of 0.993] and in diabetic nonmalignant patients (AUC of 0.961). The study findings support further research to shed light on a possible role of c-Met in T2DM-associated HCC via a miRNA regulatory pathway.
This study validates a panel of serum miRNAs that can be used as a reliable noninvasive screening biomarker of HCC among T2DM cirrhotic and noncirrhotic patients. Specifically, this study shed light on a new candidate panel of miRNAs that might serve in the diagnosis of HCC among diabetic patients. Furthermore, its findings from the miRNA screening suggest a possible role of c-Met in modulating HCC in T2DM patients. The current study aimed to investigate the signature of serum miRNAs as early biomarkers for the screening of HCC among diabetic patients, to reduce the morbidity and mortality from HCC among diabetic patients. It identified a new possible reliable panel that could be developed as a screening tool for patients. In addition, it provided evidence of a new correlation of miRNA with HCC susceptibility and hepatic disease progression in diabetic patients, especially the T2DM patients. Ultimately, however, it provided evidence to support the utility of screening miRNA in diabetes-associated HCC patients. Importantly, this study identified a miRNA-specific signature in diabetes-associated HCC patients. Our hypothesis of miRNAs comprising a signature through HCC development in diabetic patients was confirmed. The implications of our findings include that the identified miRNA panel may be useful for inclusion in the screening markers for T2DM patients with nonalcoholic fatty liver disease/NASH who are at risk of developing HCC.
Diabetic patients with dysregulated miRNA could be at high risk of developing HCC compared to nondiabetic patients. Future research should investigate the effect of c-Met dysregulation on the current panel in healthy vs disease-related patients. These investigations of the molecular mechanism of this miRNA interaction with c-Met should be carried out via a specific knock-out assay.
Technical support from the National Research Centre (Cairo, Egypt), Medical Research Institute (Alexandria, Egypt) and Korea Institute of Science and Technology (Republic of Korea, 2Z05620) is gratefully acknowledged.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kohla MAS, Gazouli M S-Editor: Tang JZ L-Editor: A E-Editor: Zhang YL
| 1. | American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care. 2019;42:S13-S28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1621] [Cited by in RCA: 1962] [Article Influence: 327.0] [Reference Citation Analysis (0)] |
| 2. | Huang TS, Lin CL, Lu MJ, Yeh CT, Liang KH, Sun CC, Shyu YC, Chien RN. Diabetes, hepatocellular carcinoma, and mortality in hepatitis C-infected patients: A population-based cohort study. J Gastroenterol Hepatol. 2017;32:1355-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 3. | Wang P, Kang D, Cao W, Wang Y, Liu Z. Diabetes mellitus and risk of hepatocellular carcinoma: a systematic review and meta-analysis. Diabetes Metab Res Rev. 2012;28:109-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 223] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 4. | Fujita K, Iwama H, Miyoshi H, Tani J, Oura K, Tadokoro T, Sakamoto T, Nomura T, Morishita A, Yoneyama H, Masaki T. Diabetes mellitus and metformin in hepatocellular carcinoma. World J Gastroenterol. 2016;22:6100-6113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 52] [Cited by in RCA: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 5. | Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 6. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27822] [Article Influence: 1324.9] [Reference Citation Analysis (0)] |
| 7. | Elemeery MN. Micro-RNA in Hepatocellular Carcinoma-Related Hepatitis C Virus Patients in Correlation to Disease Progression. Hepatitis C-From Infection to Cure: IntechOpen. 2018;. [DOI] [Full Text] |
| 8. | Elemeery MN, Badr AN, Mohamed MA, Ghareeb DA. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J Gastroenterol. 2017;23:3864-3875. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Gao B, Ning S, Li J, Liu H, Wei W, Wu F, Tang Y, Feng Y, Li K, Zhang L. Integrated analysis of differentially expressed mRNAs and miRNAs between hepatocellular carcinoma and their matched adjacent normal liver tissues. Oncol Rep. 2015;34:325-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5605] [Article Influence: 295.0] [Reference Citation Analysis (0)] |
| 11. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3218] [Cited by in RCA: 3550] [Article Influence: 208.8] [Reference Citation Analysis (0)] |
| 12. | Kelly BD, Miller N, Sweeney KJ, Durkan GC, Rogers E, Walsh K, Kerin MJ. A Circulating MicroRNA Signature as a Biomarker for Prostate Cancer in a High Risk Group. J Clin Med. 2015;4:1369-1379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 13. | Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G. A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110:2291-2299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 187] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 14. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5636] [Cited by in RCA: 6303] [Article Influence: 370.8] [Reference Citation Analysis (0)] |
| 15. | Li Y, Jiang Z, Xu L, Yao H, Guo J, Ding X. Stability analysis of liver cancer-related microRNAs. Acta Biochim Biophys Sin (Shanghai). 2011;43:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Zhang BH, Shen CA, Zhu BW, An HY, Zheng B, Xu SB, Sun JC, Sun PC, Zhang W, Wang J, Liu JY, Fan YQ. Insight into miRNAs related with glucometabolic disorder. Biomed Pharmacother. 2019;111:657-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Lorente-Cebrián S, González-Muniesa P, Milagro FI, Martínez JA. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: emerging roles as biomarkers and therapeutic targets. Clin Sci (Lond). 2019;133:23-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 18. | Bruix J, Sherman M; Practice Guidelines Committee, American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4333] [Cited by in RCA: 4506] [Article Influence: 225.3] [Reference Citation Analysis (0)] |
| 19. | Forner A, Reig ME, de Lope CR, Bruix J. Current strategy for staging and treatment: the BCLC update and future prospects. Semin Liver Dis. 2010;30:61-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 764] [Cited by in RCA: 865] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 20. | Wen Y, Han J, Chen J, Dong J, Xia Y, Liu J, Jiang Y, Dai J, Lu J, Jin G, Han J, Wei Q, Shen H, Sun B, Hu Z. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int J Cancer. 2015;137:1679-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 172] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 21. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133605] [Article Influence: 5566.9] [Reference Citation Analysis (1)] |
| 22. | Jiang L, Cheng Q, Zhang BH, Zhang MZ. Circulating microRNAs as biomarkers in hepatocellular carcinoma screening: a validation set from China. Medicine (Baltimore). 2015;94:e603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Ali HEA, Abdel Hameed R, Effat H, Ahmed EK, Atef AA, Sharawi SK, Ali M, Abd Elmageed ZY, Abdel Wahab AH. Circulating microRNAs panel as a diagnostic tool for discrimination of HCV-associated hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2017;41:e51-e62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T. A uniform system for microRNA annotation. RNA. 2003;9:277-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1324] [Cited by in RCA: 1329] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 25. | Mantovani A, Targher G. Type 2 diabetes mellitus and risk of hepatocellular carcinoma: spotlight on nonalcoholic fatty liver disease. Ann Transl Med. 2017;5:270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 108] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 26. | Stigliano R, Marelli L, Yu D, Davies N, Patch D, Burroughs AK. Seeding following percutaneous diagnostic and therapeutic approaches for hepatocellular carcinoma. What is the risk and the outcome? Seeding risk for percutaneous approach of HCC. Cancer Treat Rev. 2007;33:437-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 214] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 27. | Block T, Mehta AS, London WT. Hepatocellular carcinoma of the liver. Cancer Biomark. 2010;9:375-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Chang RM, Yang H, Fang F, Xu JF, Yang LY. MicroRNA-331-3p promotes proliferation and metastasis of hepatocellular carcinoma by targeting PH domain and leucine-rich repeat protein phosphatase. Hepatology. 2014;60:1251-1263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 29. | Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramírez CM, Goedeke L, Rotllan N, Bonora E, Hughes AD, Santer P, Fernández-Hernando C, Tilg H, Willeit J, Kiechl S, Mayr M. Circulating MicroRNA-122 Is Associated With the Risk of New-Onset Metabolic Syndrome and Type 2 Diabetes. Diabetes. 2017;66:347-357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 199] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 30. | Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 206] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 31. | Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, Hsu MT, Hsiao M, Huang HD, Tsou AP. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 477] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 32. | Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, Wu F. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int. 2012;32:752-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 170] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 33. | Jampoka K, Muangpaisarn P, Khongnomnan K, Treeprasertsuk S, Tangkijvanich P, Payungporn S. Serum miR-29a and miR-122 as Potential Biomarkers for Non-Alcoholic Fatty Liver Disease (NAFLD). Microrna. 2018;7:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 34. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 449] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 35. | Zekri AN, Youssef AS, El-Desouky ED, Ahmed OS, Lotfy MM, Nassar AA, Bahnassey AA. Serum microRNA panels as potential biomarkers for early detection of hepatocellular carcinoma on top of HCV infection. Tumour Biol. 2016;37:12273-12286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 36. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 37. | Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH, Wang JH, Huang CM, Lee CM, Lin CY, Yen YH, Chiu YC. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer. 2016;138:714-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 38. | Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 39. | Zhao S, Li T, Li J, Lu Q, Han C, Wang N, Qiu Q, Cao H, Xu X, Chen H, Zheng Z. miR-23b-3p induces the cellular metabolic memory of high glucose in diabetic retinopathy through a SIRT1-dependent signalling pathway. Diabetologia. 2016;59:644-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 40. | Cao J, Liu J, Long J, Fu J, Huang L, Li J, Liu C, Zhang X, Yan Y. microRNA-23b suppresses epithelial-mesenchymal transition (EMT) and metastasis in hepatocellular carcinoma via targeting Pyk2. Biomed Pharmacother. 2017;89:642-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 41. | Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z, Xiang Y, Wu N, Wu L, Bai L, Li Y. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget. 2017;8:13048-13058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 205] [Article Influence: 25.6] [Reference Citation Analysis (1)] |
| 42. | He RQ, Wu PR, Xiang XL, Yang X, Liang HW, Qiu XH, Yang LH, Peng ZG, Chen G. Downregulated miR-23b-3p expression acts as a predictor of hepatocellular carcinoma progression: A study based on public data and RT-qPCR verification. Int J Mol Med. 2018;41:2813-2831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 43. | Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 104] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 44. | Liu Z, Wang C, Jiao X, Zhao S, Liu X, Wang Y, Zhang J. miR-221 promotes growth and invasion of hepatocellular carcinoma cells by constitutive activation of NFκB. Am J Transl Res. 2016;8:4764-4777. [PubMed] |
| 45. | Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 650] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 46. | Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, Engelman JA, Ono M, Rho JK, Cascione L, Volinia S, Nephew KP, Croce CM. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat Med. 2011;18:74-82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 300] [Cited by in RCA: 314] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 47. | Giglio S, Vecchione A. c-Met and miRs in Cancer. Biomedicines. 2015;3:32-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 48. | Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569-7576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 476] [Article Influence: 29.8] [Reference Citation Analysis (1)] |
| 49. | Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, Comoglio PM, Giordano S. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128-10136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 50. | Sun TY, Xie HJ, Li Z, Kong LF, Gou XN, Li DJ, Shi YJ, Ding YZ. miR-34a regulates HDAC1 expression to affect the proliferation and apoptosis of hepatocellular carcinoma. Am J Transl Res. 2017;9:103-114. [PubMed] |
| 51. | Wu G, Li Z, Wang Y, Ju X, Huang R. miR-34a Inhibits Cell Proliferation by Targeting SATB2 in Hepatocellular Carcinoma. Biomed Res Int. 2018;2018:2863902. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 52. | Cheng J, Zhou L, Xie QF, Xie HY, Wei XY, Gao F, Xing CY, Xu X, Li LJ, Zheng SS. The impact of miR-34a on protein output in hepatocellular carcinoma HepG2 cells. Proteomics. 2010;10:1557-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 53. | Guan J, Liu Z, Xiao M, Hao F, Wang C, Chen Y, Lu Y, Liang J. MicroRNA-199a-3p inhibits tumorigenesis of hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am J Transl Res. 2017;9:2457-2465. [PubMed] |
| 54. | Li YB, Wu Q, Liu J, Fan YZ, Yu KF, Cai Y. miR199a3p is involved in the pathogenesis and progression of diabetic neuropathy through downregulation of SerpinE2. Mol Med Rep. 2017;16:2417-2424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, Choung S, Kim YJ, Choi YC. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem. 2008;283:18158-18166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 172] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 56. | Huang WT, Wang HL, Yang H, Ren FH, Luo YH, Huang CQ, Liang YY, Liang HW, Chen G, Dang YW. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: a clinicopathological and in silico study. Onco Targets Ther. 2016;9:5163-5180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 57. | Elfimova N, Sievers E, Eischeid H, Kwiecinski M, Noetel A, Hunt H, Becker D, Frommolt P, Quasdorff M, Steffen HM, Nürnberg P, Büttner R, Teufel A, Dienes HP, Drebber U, Odenthal M. Control of mitogenic and motogenic pathways by miR-198, diminishing hepatoma cell growth and migration. Biochim Biophys Acta. 2013;1833:1190-1198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166-5171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 590] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 59. | Bork-Jensen J, Scheele C, Christophersen DV, Nilsson E, Friedrichsen M, Fernandez-Twinn DS, Grunnet LG, Litman T, Holmstrøm K, Vind B, Højlund K, Beck-Nielsen H, Wojtaszewski J, Ozanne SE, Pedersen BK, Poulsen P, Vaag A. Glucose tolerance is associated with differential expression of microRNAs in skeletal muscle: results from studies of twins with and without type 2 diabetes. Diabetologia. 2015;58:363-373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 60. | Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902-1910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1501] [Cited by in RCA: 1454] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 61. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 62. | Chen M, Calin GA, Meng QH. Circulating microRNAs as Promising Tumor Biomarkers. Adv Clin Chem. 2014;67:189-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 63. | Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 346] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 64. | Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1354] [Cited by in RCA: 1476] [Article Influence: 92.3] [Reference Citation Analysis (0)] |