Published online Nov 14, 2019. doi: 10.3748/wjg.v25.i42.6299
Peer-review started: July 22, 2019
First decision: August 27, 2019
Revised: September 27, 2019
Accepted: November 1, 2019
Article in press: November 1, 2019
Published online: November 14, 2019
Processing time: 116 Days and 2.8 Hours
Studies have shown that long non-coding RNAs (lncRNAs) play a key role in almost all key physiological and pathological processes, including different types of malignant tumors. Our previous lncRNA microarray results have shown that lncRNA XLOC_001659 is upregulated in esophageal cancer (EC) tissues, with a fold change of 20.9 relative to normal esophageal tissues. But its effect and the molecular biological mechanisms on proliferation and invasion of EC cells remain unclear.
To investigate the effect of lncRNA XLOC_001659 on esophageal squamous cell carcinoma (ESCC) cells and explore the molecular biological mechanisms involved.
RT-qPCR assay was used to quantify the expression levels of lncRNAXLOC-001659 and miR-490-5p. The proliferative capacity of the cells was determined using CCK8 and colony formation assays, and the effect of lncRNAXLOC-001659 on the invasion of ESCC cells was determined by Transwell assay. Dual-luciferase reporter assay was used to detect the target genes of lncRNAXLOC-001659 and miR-490-5p.
The results of RT-qPCR showed that the expression of lncRNAXLOC_001659 was upregulated in ESCC cells. CCK-8 assay showed that knockdown of lncRNAXLOC_001659 significantly inhibited ESCC cell proliferation. Colony formation and Transwell invasion assays showed that knockdown of lncRNAXLOC_001659 or overexpression of miR-490-5p significantly inhibited ESCC cell growth and invasion. Furthermore, lncRNAXLOC_001659 acts as an endogenous sponge by competitively binding to miR-490-5p to downregulate miR-490-5p. Further results confirmed that miR-490-5p targeted PIK3CA, and the recovery of PIK3CA rescued lncRNAXLOC_001659 knockdown or miR-490-5p overexpression-mediated inhibition of cell proliferation and invasion, which suggested the presence of an lncRNAXLOC_001659/miR-490-5p/PIK3CA regulatory axis.
Knockdown of lncRNA XLOC_001659 inhibits proliferation and invasion of ESCC cells via regulation of miR-490-5p/PIK3CA, suggesting that it may play a role in ESCC tumorigenesis and progression.
Core tip: Studies have shown that long non-coding RNAs (lncRNAs) play a key role in almost all key physiological and pathological processes, including different types of malignant tumors. Our previous lncRNA microarray results have shown that lncRNA XLOC_001659 is upregulated in esophageal squamous cell carcinoma (ESCC) tissues, with a fold change of 20.9 relative to normal esophageal tissues. But its effect and the molecular biological mechanisms on proliferation and invasion of ESCC cells remain unclear. Here we showed that knockdown of lncRNA XLOC_001659 inhibited proliferation and invasion of ESCC cells via regulation of miR-490-5p/PIK3CA, suggesting that it may play a role in ESCC tumorigenesis and progression.
- Citation: Li FZ, Zang WQ. Knockdown of lncRNAXLOC_001659 inhibits proliferation and invasion of esophageal squamous cell carcinoma cells. World J Gastroenterol 2019; 25(42): 6299-6310
- URL: https://www.wjgnet.com/1007-9327/full/v25/i42/6299.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i42.6299
Esophageal cancer (EC) is a common malignant tumor, ranking eighth among all malignancies in the world[1]. It is the sixth most common cause of cancer death, with incidence varying geographically[2]. The incidence of EC is highest in China, with more than 90% of EC cases being esophageal squamous cell carcinoma (ESCC)[1]. Due to the lack of specific symptoms and effective methods for early diagnosis, ECSS tends to be diagnosed late. Only 15%-25% of ESCC patients survive five years after the initial diagnosis[1,2]. In addition, given the high incidence and mortality, un-derstanding the molecular mechanism of ESCC is urgently needed to enhance the survival of patients with ESCC[3].
Long-chain non-coding RNAs (lncRNAs) have been identified as a new class of evolutionarily conserved RNA molecules. They are more than 200 nucleotides in length and have no or limited protein-coding ability[4]. Studies over the past few decades have shown that lncRNAs play a key role in almost all key physiological and pathological processes[5], including different types of malignant tumors, such as lung cancer[6], thyroid cancer[7], colon cancer[8], and ESCC. Although the effects of lncRNAs on cancer progression have attracted considerable research attention, their abnormal expression and functional roles in ESCC development are not fully elucidated[9].
Our previous lncRNA microarray analysis has shown that lncRNA XLOC_001659 is upregulated in EC tissues, with a fold change of 20.9 relative to normal esophageal tissues distant from the tumor[10]. But its effect and the molecular biological mechanisms on proliferation and invasion of EC cells remain unclear. In this study, we investigated the expression of lncRNA XLOC_001659 in ESCC and its effect on proliferation and invasion of EC cells. We further explored the molecular and biological mechanisms underlying lncRNA XLOC_001659. To the best of our knowledge, this is the first study to report the expression and role of lncRNA XLOC_001659 in ESCC cells.
Human esophageal epithelial cell line, HET-1A, and ESCC cell lines, EC9706 and EC-1, were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and were sub-cultured and preserved in our laboratory. HET-1A cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, United States), 100 U/mL penicillin, and 100 μg/mL streptomycin. EC9706 and EC-1 cells were cultured in D6429-high glucose medium (Sigma-Aldrich, United Kingdom) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. All cell lines were kept at 37 °C in an incubator with a humidified atmosphere and 5% CO2.
The full-length PIK3CA cDNA was inserted into pcDNA3.1 vector (Sangon Biotech, Shanghai, China) to construct a vector overexpressing PIK3CA. EC9706 and EC-1 cells in the logarithmic growth phase were collected, seeded into six-well plates, and cultured overnight. Specific siRNA and non-target (i.e., negative control, NC) sequences of XLOC_001659 were synthesized by Sangon Biotech, followed by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions to transfect siRNA and NC into the EC cell lines and continuously culturing for 24-48 h for subsequent experiments. The sequences of XLOC_001659 siRNA and NC are 5′-AAGCCUCGUCACCACUUACUCGAGUAAGUGGUGACGAGGCUU-3′ and 5′-UUCUCCGAACGUGUCACGUTTACGUGACAGGUUCGGAGAATT-3′, respectively. The NCs and miR-490-5p mimics were purchased from RiboBio Co. Ltd (Guangzhou, China), transfected into EC cell lines using Lipofectamine 2000, and continuously cultured for 24-48h for subsequent experiments.
Total RNA was extracted from ESCC cell lines using an RNA extraction kit (Biomiga, San Diego, CA, United States). This was followed by using a NanoDrop 2000C Spectrophotometer (Thermo Scientific, United States) to determine the quality and quantity of the total RNA. The total RNA was reverse-transcribed into cDNA according to the instructions included in the cDNA synthesis kit (Thermo Fisher Scientific, Vilnius, Lithuania). Reverse transcription of miRNA cDNA was performed using a TaqMan miRNA reverse transcription kit. GAPDH and U6 were used as an internal reference gene for XLOC_001659 and miR-490-5p, respectively. RT-qPCR was performed on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) to detect the relative expression of XLOC_001659 and miR-490-5p.
The proliferation ability of ESCC cell lines was detected by CCK-8 assay (Dojindo, Japan). The transfected EC9760 and EC-1 cells were adjusted to a concentration of 5000 cells/mL per culture well, followed by inoculating 100 μL of diluted cells into each well of a new 96-well plate, with three identical wells per treatment group and per time point. In the 96-well plates, 10 μL of CCK-8 solution was added to each well of ECSS cells cultured for 0, 24, 48, and 72 h, followed by continuous incubation at 37 °C for 2 h and then absorbance at a wavelength of 450 nm was measured using a fully automated microplate reader (Bio-Rad Laboratory, Irvine, CA, United States).
The proliferation ability of EC9706 and EC-1 cells was examined by colony formation assay. Six-centimeter culture dishes were inoculated with 300 cells from different transfection groups. The cells were cultured at 37 °C in an incubator containing 5% CO2 for 10-14 d, and 500 μL of culture medium was added to each culture dish every three days. The incubation was discontinued once visible clones appeared in the culture dishes. Then the culture medium was discarded, and cells were washed twice in phosphate-buffered saline (PBS), fixed in methanol for 30 min, and stained with 0.05% crystal violet dye for 15 min. After the dye was washed off, the clones were air-dried and counted under a microscope. Only clones containing more than 50 cells were counted.
The invasion ability of the ESCC cells was measured using Transwell invasion assay in 24-well plates. First, the Transwell chambers were placed in each well of the 24-well plates, followed by addition of 700 μL of serum-free culture medium to soak the chamber at 37 °C for 2 h. Cells from each group after transfection were adjusted to 50000 cells/well using serum-free medium. The cell suspensions were collected and 100 μL of diluted cell suspension was added to the filter membrane of a Transwell insert (the upper chamber). Culture medium containing 20% FBS was added to the lower chamber of each well of the 24-well plates. The Transwell invasion chamber system was continuously incubated at 37 °C for 12-48 h, followed by collecting each of the Transwell inserts and discarding the culture medium. After washing twice with PBS, the cells were fixed in methanol for 30 min, air-dried, and stained with 0.05% crystal violet for 15 min. The un-migrated cells from the top of Transwell membrane were gently removed with a cotton-tipped applicator, followed by two rounds of washing with PBS. Transwell membranes were kept wet, and the numbers of cells in five different fields of view were counted to get an average sum of cells under a microscope (100× magnification).
The EC cell lines were inoculated in 24-well plates at a density of 1 × 105 cells per well. Once the confluency reached 60%-70%, miRNA mimics or miRNA NC and wide-type (LncRNA XLOC_001659WT) and recombinant reporter (LncRNA XLOC_001659MUT) vectors were co-transfected into the EC cells using Lipofectamine 2000; similarly, miRNA mimics or miRNA NC and wide-type (PIK3CA-WT) or mutant (PIK3CA-MUT) recombinant vectors were co-transfected into the EC cells for 48 h, after which the cells were harvested and fluorescein signals were assessed using a dual-luciferase reporter assay system (Promega).
After transfection, the cells of different treatment groups were trypsinized, centrifuged, and collected into new Eppendorf tubes. This was followed by addition of RIPA lysis buffer (Solarbio Co., Ltd) and phenylmethylsulfonyl fluoride (Solarbio Co., Ltd) at a ratio of 100:1, and quantification with a BCA Protein Assay Kit (Solarbio Co., Ltd). The extracted protein samples were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, and then transferred to polyvinylidene fluoride membranes (Solarbio Co., Ltd). The protein blots were blocked with 5% skim milk solution at room temperature for 2 h and separately incubated with the corresponding primary antibodies (1:1000 anti-β-actin antibody purchased from Bioass Antibodies, Beijing, China; 1:2000 anti-PIK3CA antibody purchased from Bioass Antibodies) at 4 °C overnight. The protein blots were incubated with 1:3000 goat anti-rabbit IgG antibody conjugated to horseradish peroxidase at room temperature for 2 h. After eight washes in TBST solution (5 min each), the signals of the protein blots were detected using enhanced chemiluminescence substrate.
Each experiment above was performed three times, and data obtained in the three independent experiments are presented as the mean ± standard deviation. GraphPad Prism7 software was used for graph preparation and statistical analyses. The t-test was used to analyze the differences in the expression of lncRNA XLOC_001659 in different groups of tissues and cells. One-way ANOVA was used to detect differences in the effects of lncRNA XLOC_001659 on cell growth and invasion in the transfected groups. P < 0.05 was considered statistically significant.
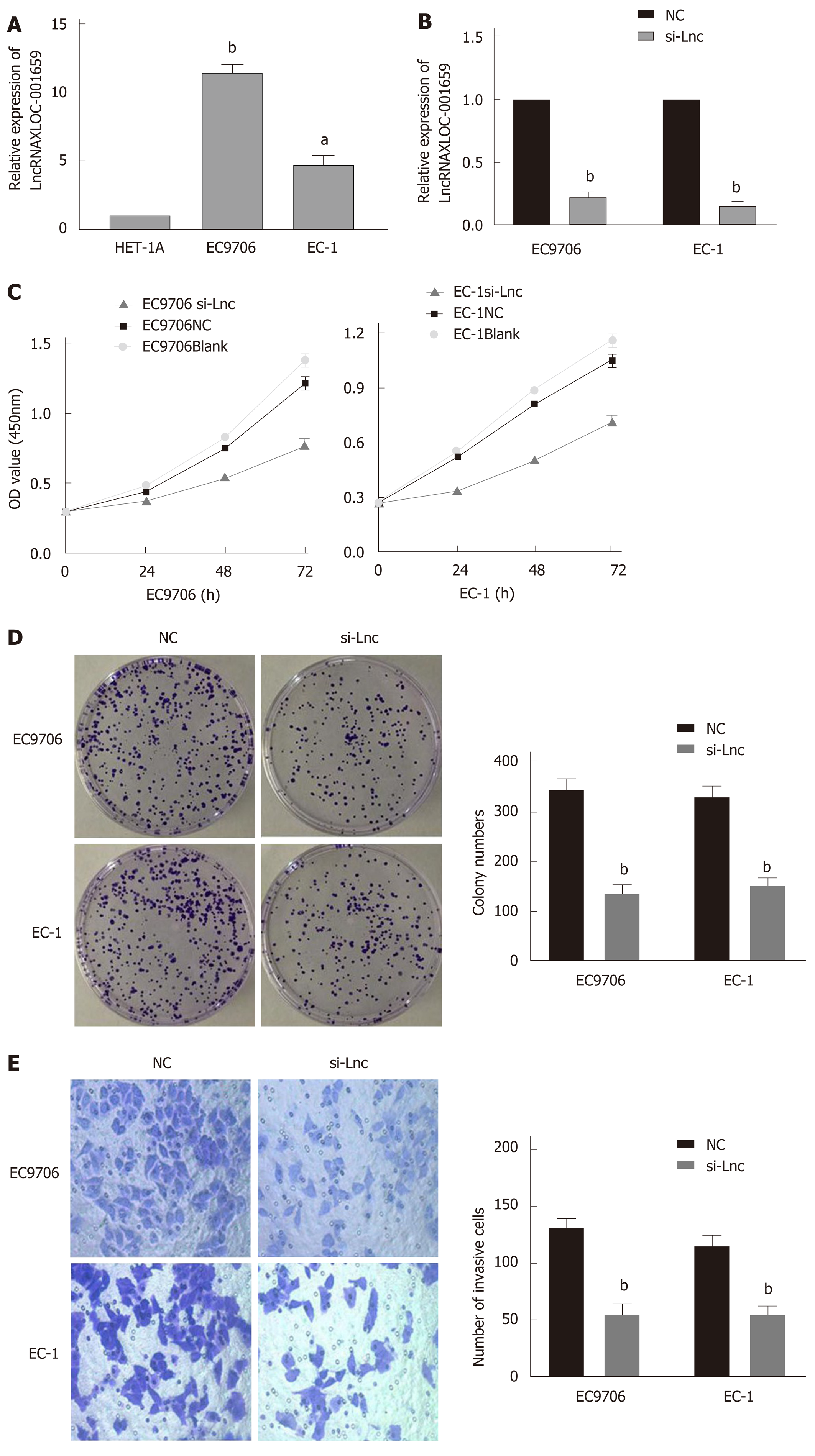
RT-qPCR was conducted to verify the lncRNA XLOC_001659 expression in normal esophageal epithelial cells (HET-1A) and ESCC cells (EC9706 and EC-1). RT-qPCR results showed that the lncRNA XLOC_001659 expression in the ESCC cell lines was significantly upregulated (Figure 1A). To further investigate the biological function of lncRNA XLOC_001659, we knocked down lncRNA XLOC_001659 in ESCC cell lines and performed RT-qPCR. Compared with the NC group, lncRNA XLOC_001659 expression was significantly reduced in ESCC cells transfected with siRNA-lncXLOC_001659 (Figure 1B). The cell proliferation was assessed using CCK-8 and colony formation assays. CCK-8 assay showed that ESCC cell lines transfected with siRNA-lncXLOC_001659 had a significantly lower proliferation rate than the ESCC cell lines in the NC group and the blank control group (Figure 1C). Colony formation assay showed that siRNA-lncXLOC_001659 significantly inhibited colony formation (Figure 2D). In addition, the cell migration measured by Transwell invasion assay showed that knockdown of lncXLOC_001659 significantly reduced the invasion of ESCC cell lines (Figure 1E). All of these results revealed that knockdown of lncRNA XLOC_001659 inhibited the growth and invasion of ESCC cells.
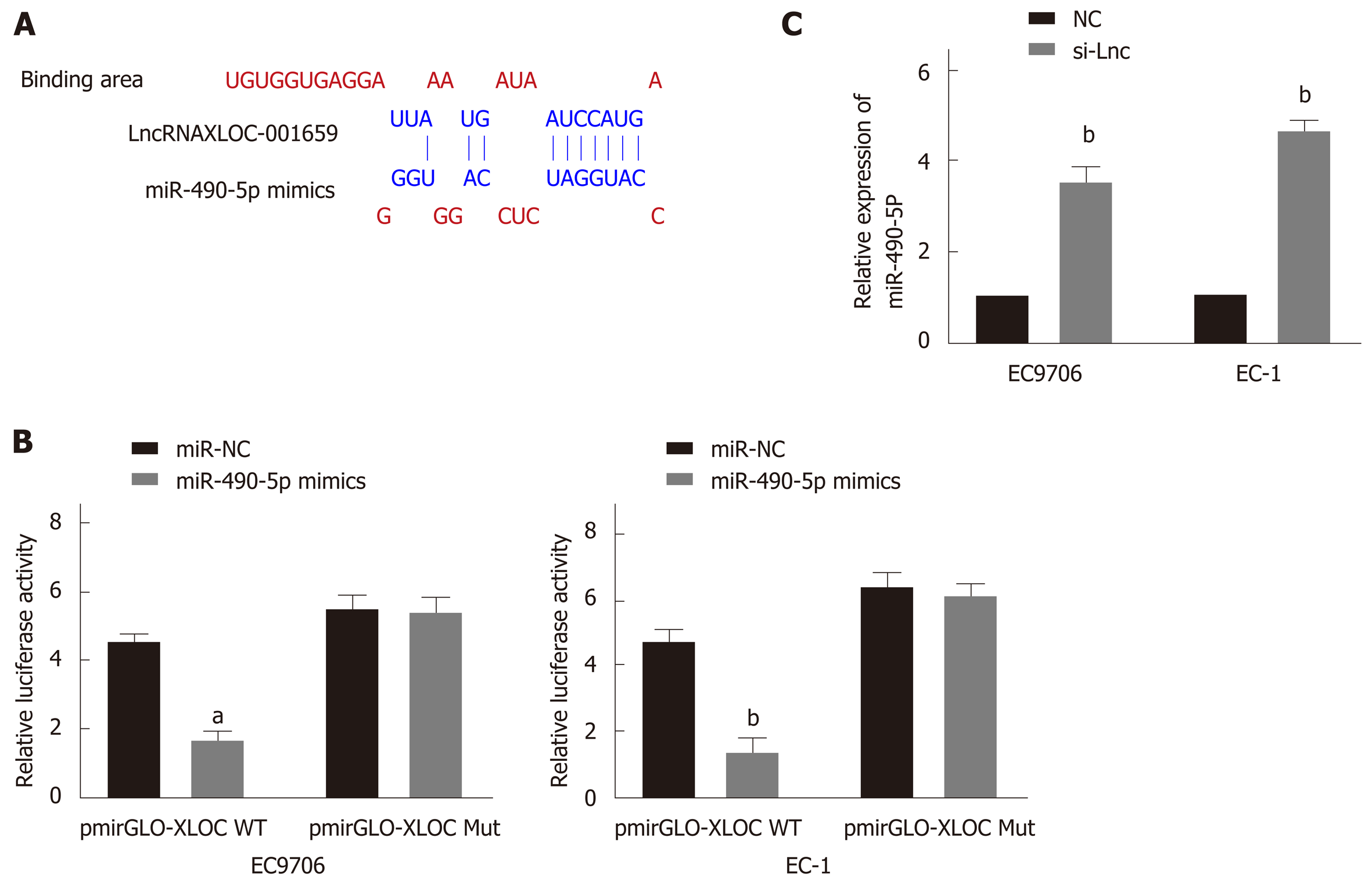
This study also performed extensive bioinformatics analysis on lncRNA XLOC_001659 to analyze miRNAs that may interact with lncRNA XLOC_001659. Predictive analysis revealed a possible interaction between lncRNA XLOC_001659 and miR-490-5p (Figure 2A). To further confirm the interaction between lncRNA XLOC_001659 and miR-490-5p, we constructed a recombinant reporter vector containing wild type (WT) or mutant (MUT) lncRNA XLOC_001659 to perform dual-luciferase reporter assay. The results showed that ESCC cell lines co-transfected with miR-490-5p mimics and WT lncRNA XLOC_001659 recombinant reporter vector had significantly lower luciferase activity than untransfected ESCC cell lines, while ESCC cell lines co-transfected with miR-490-5p mimics and MUT lncRNA XLOC_001659 recombinant reporter vector had similar levels of luciferase activity (Figure 2B). Subsequently, RT-qPCR was used to verify the effect of lncRNAXLOC_001659 knockdown on the miR-490-5p expression in ESCC cell lines. After transfecting siRNA-lncXLOC_001659 into the ESCC cell lines, there was significantly more miR-490-5p expression (Figure 2C). It was confirmed that lncRNA XLOC_001659 downregulated the expression of miR-490-5p by binding to the miR-490-5p seed region.
To determine whether miR-490-5p and lncRNA XLOC_001659 have similar effects on the proliferation and invasion of the ESCC cell lines, we overexpressed miR-490-5p and knocked down lncRNA XLOC_001659 in the EC9706 and EC-1 cells. The RT-qPCR results showed that miR-490-5p expression was significantly higher than in the NC group after transfecting EC9706 and EC1 cells with miR-490-5p mimics (Figure 3A). The results of colony formation assay showed that there was significantly fewer clones formed in EC9706 and EC-1 cell lines transfected with siRNA -lncXLOC_001659 and miR-490-5p mimics than in the corresponding NC groups (Figure 3B). The results of Transwell invasion assay showed that knockdown of lncXLOC_001659 and overexpression of miR-490-5p rendered the degree of invasion in ESCC cell lines far lower than in the corresponding NC groups (Figure 3C). These results confirmed that overexpression of miR-490-5p and knockdown of lncXLOC_001659 inhibited ESCC cell proliferation and migration, with consistent trends.
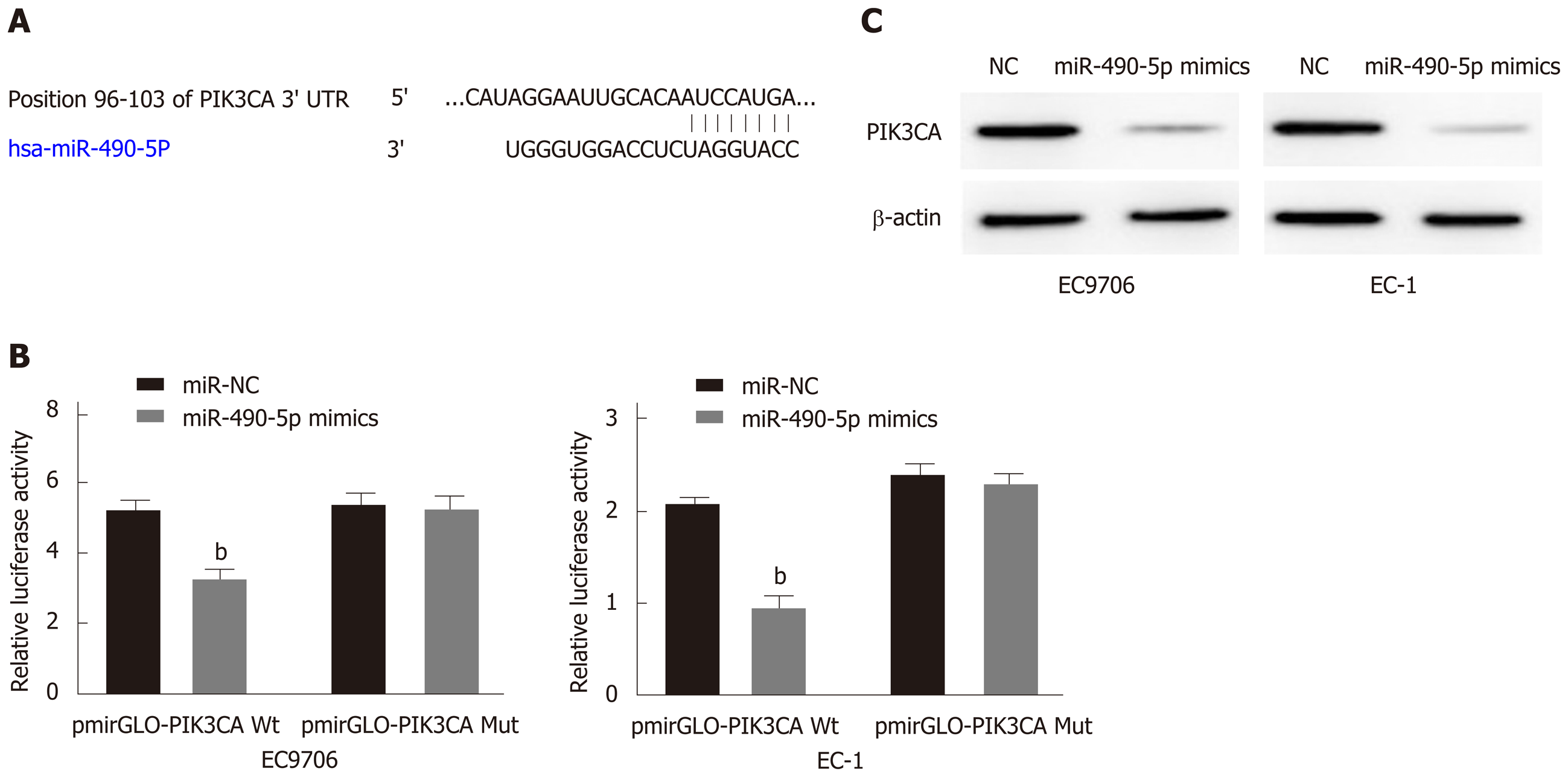
To further characterize the molecular mechanism by which miR-490-5p exerts its regulatory role in ESCC, predictive bioinformatics analysis was performed. It was revealed that miR-490-5p and PIK3CA had interacting targets (Figure 4A). We then transfected the WT and MUT PIK3CA 3’UTR recombinant reporter vector and miR-490-5p mimics or NC vector into the ESCC cell lines and performed dual-luciferase reporter assay. The results showed that co-transfection of miR-490-5p mimics and WT PIK3CA recombinant reporter vector into the ESCC cell lines reduced luciferase activity significantly; co-transfection of miR-490-5p mimics and MUT PIK3CA recombinant reporter vector produced no significant changes in the luciferase activity (Figure 4B). Western blot analysis showed that transfection of miR-490-5p mimics effectively reduced PIK3CA protein expression in the ESCC cell lines than in the control group (Figure 4C). These results indicated that PIK3CA is a target gene of miR-490-5p.
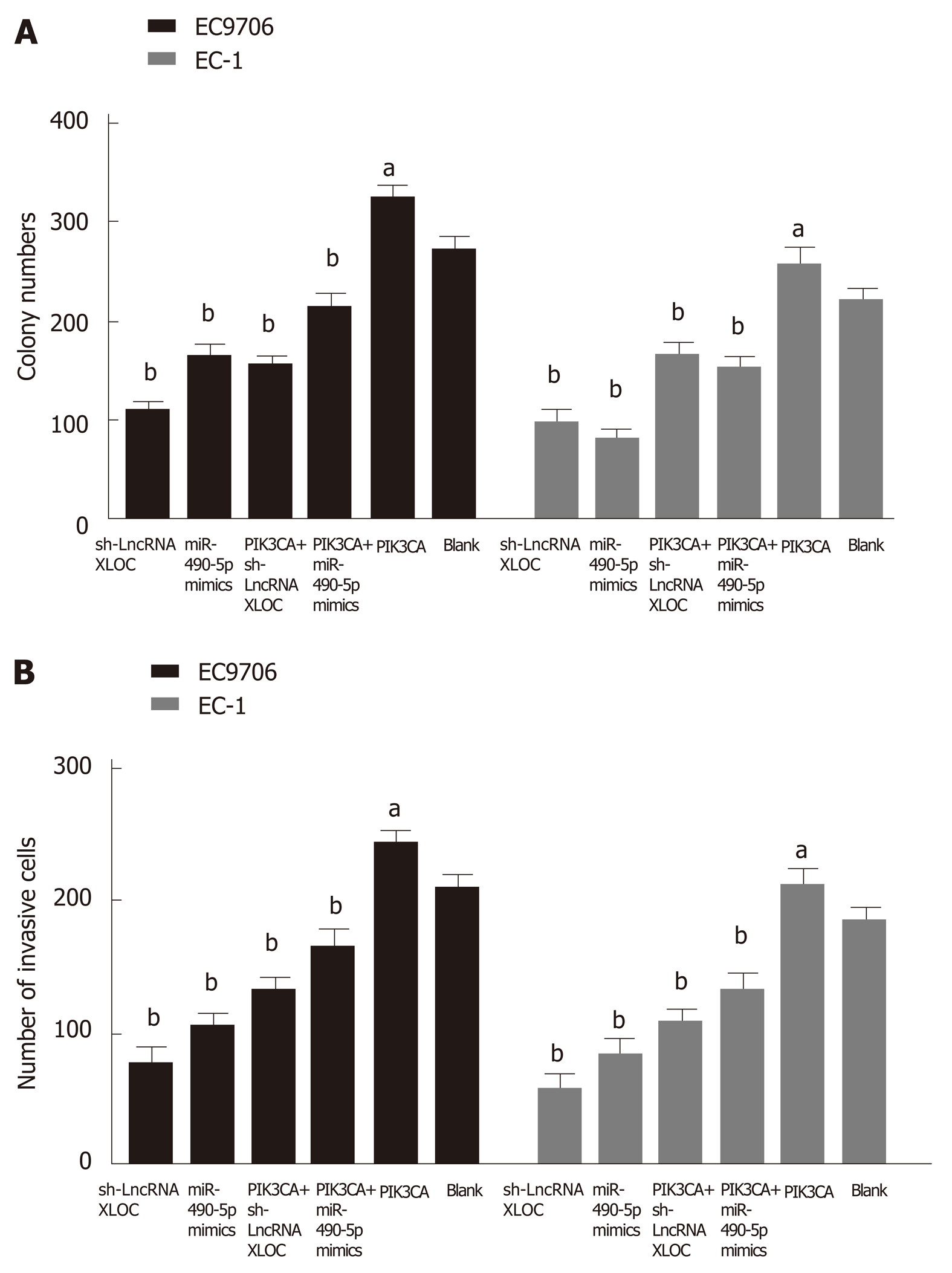
Based on the above findings, we concluded that an lncRNA XLOC_001659-miR-490-5p-PIK3CA regulatory axis might be involved in the development of ESCC. To validate this hypothesis, siRNA-lncXLOC_001659 or miR-490-5p mimics and pcDNA-PIK3CA were co-transfected into ESCC cell lines, followed by performing colony formation assay and Transwell invasion assay. Colony formation assay showed that ESCC cell lines transfected with either siRNA-lncXLOC_001659 or miR-490-5p mimics showed significantly less growth ability than the blank group. However, siRNA-lncXLOC_001659 or miR-490-5p mimics co-transfected with pcDNA-PIK3CA partially rescued the cell proliferation rate (Figure 5A). Transwell invasion assay showed that knockdown of lncXLOC_001659 or overexpression of miR-490-5p significantly inhibited the cell invasion ability, whereas co-transfection with pcDNA-PIK3CA partially eliminated the inhibitory effect (Figure 5B).
With the in-depth research performed in recent years, non-coding RNAs (ncRNAs) have ceased to be seen as the “noise” of genomic transcription[11]. NcRNAs are a class of RNA molecules that do not encode proteins. They mainly include lncRNAs, ribosomal RNAs (rRNAs), microRNAs (miRNAs), transfer RNAs (tRNAs), small nucleolar RNAs (snoRNAs), and small nuclear RNAs (snRNAs)[12]. LncRNAs are non-coding RNAs over 200 nucleotides in length. Its differential expression has been shown to be closely related to tumorigenesis and tumor progression. Several types of lncRNAs, such as lncRNA XIST[13], lncRNA XLOC[14,15], and lncRNA MALAT1[16], have been identified in ESCC.
MiRNAs are non-coding RNAs with 20-25 nucleotides and regulatory function[17]. They recognize target mRNAs by complementary base pairing and guide the silencing complexes to degrade the target mRNAs or to repress the translation of mRNAs based on the degree of complementarity[18]. Recent studies have shown that miRNA expression is associated with a variety of cancer, suggesting that miRNAs play a crucial role in tumorigenesis. These miRNAs play a role similar to the functions of tumor suppressor genes and oncogenes[19,20]. Many recent studies have shown that lncRNAs, together with miRNAs, act as ceRNAs to regulate target mRNAs, thereby playing an important role in tumorigenesis and tumor progression[21,22].
In this study, we analyzed the biological roles of lncRNA XLOC_001659, miR-490-5p, and PIK3CA in ESCC cells and confirmed the presence of an lnc-RNAXLOC_001659-miR-490-5p-PIK3CA regulatory axis. The lncRNA XLOC_001659 was upregulated in ESCC cells, and silencing the expression of lncRNA XLOC_001659 attenuated the proliferation and invasion of ESCC cells. It has been reported that miR-490-5p is downregulated in various cancers and cell lines, including liver cancer[23], kidney cancer[12], and bladder cancer[24,25]. For this reason, it is considered a potential tumor suppressor. In this study, we found that miR-490-5p was abnormally downregulated in ESCC cells, whereas miR-490-5p overexpression significantly inhibited the proliferation and invasion abilities of ESCC cells. LncRNA XLOC_001659 is a ceRNA that inhibits miR-490-5p. The results of dual-luciferase reporter assay and RT-qPCR confirmed that lncRNA XLOC_001659 targets miR-490-5p. Dual-luciferase reporter assay indicated that the activity of lncRNA XLOC_001659 luciferase-based reporter gene in the miR-490-5ptransfection group was significantly reduced. RT-qPCR showed that miR-490-5p was negatively correlated with lncRNA XLOC_001659 expression. Over the past few years, increasing numbers of studies have focused on the lncRNA-miRNA-mRNA regulatory mechanisms, and some lncRNAs negatively regulate miRNAs by acting as competing endogenous RNAs (ceRNAs), i.e., miRNA sponges or RNA antagonists[4,26]. MiR-490-5p has also been reported as a tumor suppressor in another study[27]. These results suggested that lncRNA XLOC_001659 acts as a ceRNA and regulates the expression of miR-490-5p.
Previous studies have shown that PIK3CA promotes the development of cancers, such as non-small cell lung cancer[28], breast cancer[29], colorectal cancer[30], and cervical cancer[31], suggesting that PIK3CA may be a cancer-promoting gene. The results of this study confirmed that miR-490-5p targets PIK3CA, thereby affecting the growth and invasion of ESCC cells.
In conclusion, we here investigated the biological roles of lncRNA XLOC_001659 in ESCC cells and showed that lncRNA XLOC_001659 was overexpressed in ESCC cells. Silencing lncRNA XLOC_001659 expression inhibited the proliferation and invasion of ESCC cells. Our results showed that lncRNA XLOC_001659 negatively regulated miR-490-5p expression, and miR-490-5p targets PIK3CA, suggesting the presence of an lncRNAXLOC_001659-miR-490-5p-PIK3CA regulatory axis. The lncRNAXLOC_001659-miR-490-5p-PIK3CA regulatory axis affected the growth and invasion of ESCC cells, suggesting that it may play a role in ESCC tumorigenesis and progression. For this reason, we speculate that lncRNA XLOC_001659 may be a cancer-promoting gene and a new target for the diagnosis and treatment of ESCC.
More and more studies have shown that long non-coding RNAs (lncRNAs) play a key role in almost all key physiological and pathological processes, including different types of malignant tumors. Although the effects of lncRNAs on cancer progression have drawn considerable research attention, their abnormal expression and functional roles in esophageal squamous cell carcinoma (ESCC) development are not fully elucidated. Our previous lncRNA microarray analysis has shown that lncRNA XLOC_001659 is upregulated in EC tissues. But its effect and the molecular biological mechanisms on proliferation and invasion of EC cells remain unclear.
Esophageal cancer is a common malignant tumor, ranking eighth among all malignancies in the world. Although the effects of lncRNAs on cancer progression have drawn considerable research attention, their abnormal expression and functional roles in ESCC development are not fully elucidated. In this study, we investigated the expression of lncRNAXLOC_001659 in ESCC and its effect on proliferation and invasion of EC cells. We further explored the molecular and biological mechanisms underlying lncRNAXLOC_001659. Our study is the first to report the expression and role of lncRNAXLOC_001659 in ESCC.
In this study, we analyzed the expression of lncRNAXLOC_001659 in ESCC and its effect on the proliferation and invasion of EC cells. We further explored the molecular and biological mechanisms underlying lncRNA XLOC_001659. The purpose of this study was to explore the role of lncRNA XLOC_001659 in ESCC tumorigenesis and progression.
The expression levels of LncRNAXLOC-001659 and miR-490-5p were quantified by RT-qPCR assay. CCK8 and colony formation assays were used to determine the proliferative capacity of the cells. Transwell assay were used to determine the invasion capacity of the cells. Dual-luciferase reporter assay was used to detect the target genes of lncRNAXLOC-001659 and miR-490-5p.
The expression of lncRNAXLOC_001659 was upregulated in ESCC. Knockdown of lncRNAXLOC_001659 significantly inhibited ESCC cell proliferation and invasion. Furthermore, lncRNAXLOC_001659 acts as an endogenous sponge by competitively binding to miR-490-5p to downregulate miR-490-5p. Further results confirmed that miR-490-5p targeted PIK3CA, and the recovery of PIK3CA rescued lncRNAXLOC_001659 knockdown or miR-490-5p overexpression-mediated inhibition of cell proliferation and invasion.
Knockdown of lncRNA XLOC_001659 inhibited proliferation and invasion of ESCC cells via regulation of miR-490-5p/PIK3CA, suggesting that it may play a role in ESCC tumorigenesis and progression. LncRNA XLOC_001659 may be a cancer-promoting gene and a new target for the diagnosis and treatment of ESCC.
LncRNA XLOC_001659 may plays an important role in ESCC tumorigenesis and progression. Exploring the role and mechanism of lncRNA XLOC_001659 in ESCC can provide an important reference for the diagnosis and treatment of ESCC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Que JY, Zou CB S-Editor: Wang J L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Deng HY, Wang YC, Ni PZ, Lin YD, Chen LQ. Long noncoding RNAs are novel potential prognostic biomarkers for esophageal squamous cell carcinoma: an overview. J Thorac Dis. 2016;8:E653-E659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 2. | Wu Y, Hu L, Liang Y, Li J, Wang K, Chen X, Meng H, Guan X, Yang K, Bai Y. Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2. Mol Cancer. 2017;16:150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 130] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 3. | Liu B, Pan CF, Yao GL, Wei K, Xia Y, Chen YJ. The long non-coding RNA AK001796 contributes to tumor growth via regulating expression of p53 in esophageal squamous cell carcinoma. Cancer Cell Int. 2018;18:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 4. | Li Y, Chen D, Gao X, Li X, Shi G. LncRNA NEAT1 Regulates Cell Viability and Invasion in Esophageal Squamous Cell Carcinoma through the miR-129/CTBP2 Axis. Dis Markers. 2017;2017:5314649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 587] [Cited by in RCA: 752] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 6. | Jing H, Qu X, Liu L, Xia H. A Novel Long Noncoding RNA (lncRNA), LL22NC03-N64E9.1, Promotes the Proliferation of Lung Cancer Cells and is a Potential Prognostic Molecular Biomarker for Lung Cancer. Med Sci Monit. 2018;24:4317-4323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 7. | Wang P, Liu G, Xu W, Liu H, Bu Q, Sun D. Long Noncoding RNA H19 Inhibits Cell Viability, Migration, and Invasion Via Downregulation of IRS-1 in Thyroid Cancer Cells. Technol Cancer Res Treat. 2017;16:1102-1112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 8. | Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, Xu RH. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017;8:e3011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 162] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 9. | Schmitt AM, Chang HY. Long Noncoding RNAs: At the Intersection of Cancer and Chromatin Biology. Cold Spring Harb Perspect Med. 2017;7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 10. | Zang W, Wang T, Wang Y, Chen X, Du Y, Sun Q, Li M, Dong Z, Zhao G. Knockdown of long non-coding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget. 2016;7:19960-19974. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 11. | Qu Z, Adelson DL. Bovine ncRNAs are abundant, primarily intergenic, conserved and associated with regulatory genes. PLoS One. 2012;7:e42638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Yao J, Shen X, Li H, Xu J, Shao S, Huang JX, Lin M. LncRNA-ECM is overexpressed in esophageal squamous cell carcinoma and promotes tumor metastasis. Oncol Lett. 2018;16:3935-3942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Wu X, Dinglin X, Wang X, Luo W, Shen Q, Li Y, Gu L, Zhou Q, Zhu H, Li Y, Tan C, Yang X, Zhang Z. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget. 2017;8:76015-76028. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 14. | Li Y, Zhao L, Zhang Y, Guan L, Zhang H, Zhou H, Gao T, Miao P, Sun M. Downregulation of the long non-coding RNA XLOC_010588 inhibits the invasion and migration of colorectal cancer. Oncol Rep. 2018;39:1619-1630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 15. | Liu YY, Chen ZH, Peng JJ, Wu JL, Yuan YJ, Zhai ET, Cai SR, He YL, Song W. Up-regulation of long non-coding RNA XLOC_010235 regulates epithelial-to-mesenchymal transition to promote metastasis by associating with Snail1 in gastric cancer. Sci Rep. 2017;7:2461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Wang X, Li M, Wang Z, Han S, Tang X, Ge Y, Zhou L, Zhou C, Yuan Q, Yang M. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290:3925-3935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 259] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 17. | Adlakha YK, Saini N. Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer. 2014;13:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 168] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 18. | Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V, Stopka T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer. 2014;14:448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 19. | Weiss CN, Ito K. A Macro View of MicroRNAs: The Discovery of MicroRNAs and Their Role in Hematopoiesis and Hematologic Disease. Int Rev Cell Mol Biol. 2017;334:99-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311-336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 406] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 21. | Liu D, Yu X, Wang S, Dai E, Jiang L, Wang J, Yang Q, Yang F, Zhou S, Jiang W. The gain and loss of long noncoding RNA associated-competing endogenous RNAs in prostate cancer. Oncotarget. 2016;7:57228-57238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Zhang Y, Xu Y, Feng L, Li F, Sun Z, Wu T, Shi X, Li J, Li X. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget. 2016;7:64148-64167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 162] [Article Influence: 23.1] [Reference Citation Analysis (1)] |
| 23. | Cai H, Hu B, Ji L, Ruan X, Zheng Z. Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling pathway. Am J Transl Res. 2018;10:1690-1702. [PubMed] |
| 24. | Xiang M, Yuan W, Zhang W, Huang J. Expression of miR-490-5p, miR-148a-3p and miR-608 in bladder cancer and their effects on the biological characteristics of bladder cancer cells. Oncol Lett. 2019;17:4437-4442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Lan G, Yang L, Xie X, Peng L, Wang Y. MicroRNA-490-5p is a novel tumor suppressor targeting c-FOS in human bladder cancer. Arch Med Sci. 2015;11:561-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 331] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 27. | Chen K, Zeng J, Tang K, Xiao H, Hu J, Huang C, Yao W, Yu G, Xiao W, Guan W, Guo X, Xu H, Ye Z. miR-490-5p suppresses tumour growth in renal cell carcinoma through targeting PIK3CA. Biol Cell. 2016;108:41-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Wang L, Hu H, Pan Y, Wang R, Li Y, Shen L, Yu Y, Li H, Cai D, Sun Y, Chen H. PIK3CA mutations frequently coexist with EGFR/KRAS mutations in non-small cell lung cancer and suggest poor prognosis in EGFR/KRAS wildtype subgroup. PLoS One. 2014;9:e88291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Merino VF, Cho S, Liang X, Park S, Jin K, Chen Q, Pan D, Zahnow CA, Rein AR, Sukumar S. Inhibitors of STAT3, β-catenin, and IGF-1R sensitize mouse PIK3CA-mutant breast cancer to PI3K inhibitors. Mol Oncol. 2017;11:552-566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 30. | Hao Y, Samuels Y, Li Q, Krokowski D, Guan BJ, Wang C, Jin Z, Dong B, Cao B, Feng X, Xiang M, Xu C, Fink S, Meropol NJ, Xu Y, Conlon RA, Markowitz S, Kinzler KW, Velculescu VE, Brunengraber H, Willis JE, LaFramboise T, Hatzoglou M, Zhang GF, Vogelstein B, Wang Z. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun. 2016;7:11971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 214] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 31. | Jiang W, He T, Liu S, Zheng Y, Xiang L, Pei X, Wang Z, Yang H. The PIK3CA E542K and E545K mutations promote glycolysis and proliferation via induction of the β-catenin/SIRT3 signaling pathway in cervical cancer. J Hematol Oncol. 2018;11:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |