Published online Sep 21, 2019. doi: 10.3748/wjg.v25.i35.5266
Peer-review started: April 22, 2019
First decision: June 10, 2019
Revised: July 18, 2019
Accepted: August 7, 2019
Article in press: June 10, 2019
Published online: September 21, 2019
Processing time: 153 Days and 18.8 Hours
Hepatitis B virus (HBV) has been recognized as a leading cause of hepatocellular carcinoma (HCC). Numerous reports suggest that immune infiltration can predict the prognosis of HCC. Nonetheless, no creditable markers for prognosis of HBV-related HCC have been established by systematically assessing the immune-related markers based on tumor transcriptomes.
To establish an immune-related marker based on the cell compositions of immune infiltrate obtained based on tumor transcriptomes, so as to enhance the prediction accuracy of HBV-related HCC prognosis.
RNA expression patterns as well as the relevant clinical data of HCC patients were obtained from The Cancer Genome Atlas. Twenty-two immunocyte fraction types were estimated by cell type identification by estimating relative subsets of RNA transcripts. Subsequently, the least absolute shrinkage and selection operator (LASSO) Cox regression model was employed to construct an immunoscore based on the immunocyte fraction types. Afterwards, the receiver operating characteristic (ROC) curve, Kaplan-Meier, and multivariate Cox analyses were performed. Additionally, a nomogram for prognosis that integrated the immunoscore as well as the clinical features was established. Meanwhile, the correlation of immunoscore with immune genes was also detected, and gene set enrichment analysis (GSEA) of the immunoscore was conducted.
A total of 22 immunocyte fraction types were predicted and compared among the tumor as well as non-tumor samples. An immunoscore was constructed through adopting the LASSO model, which contained eight immunocyte fraction types. Meanwhile, the areas under the ROC curves for the immunoscore biomarker prognostic model were 0.971, 0.912, and 0.975 for 1-, 3-, and 5-year overall survival (OS), respectively. Difference in OS between the high-immunoscore group and the low-immunoscore group was statistically significant [hazard ratio (HR) = 66.007, 95% confidence interval (CI): 8.361-521.105; P < 0.0001]. Moreover, multivariable analysis showed that the immunoscore was an independent factor for predicting the prognosis (HR = 2.997, 95%CI: 1.737-5.170). A nomogram was established, and the C-index was 0.757 (95%CI: 0.648-0.866). The immunoscore showed a significant negative correlation with the expression of PD-1 (P = 0.024), PD-L1 (P = 0.026), PD-L2 (P = 0.029), and CD27 (P = 0.033). Eight pathways were confirmed by GSEA.
The established immunoscore can potentially serve as a candidate marker to estimate the OS for HBV-related HCC cases.
Core tip: Hepatitis B virus (HBV) infection is epidemiologically related to hepatocellular carcinoma (HCC) development. Immune cells have been recognized to play crucial roles in the prognosis of various types of cancers, including HCC. The immune signature for HBV-related HCC remains unknown. In this study, we explored the proportions of immunocytes, and constructed a robust immune signature to predict the prognosis. These results might contribute to risk stratification and facilitate individualized clinical approach for HBV-related HCC.
- Citation: Chen QF, Li W, Wu PH, Shen LJ, Huang ZL. Significance of tumor-infiltrating immunocytes for predicting prognosis of hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2019; 25(35): 5266-5282
- URL: https://www.wjgnet.com/1007-9327/full/v25/i35/5266.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i35.5266
Although the screening, diagnostic, and therapeutic techniques for hepatocellular carcinoma (HCC) have progressed, it remains a major cause of cancer-related death worldwide, with dismal clinical outcomes[1]. Hepatitis viruses have been epidemiologically related to HCC development[2,3], of which hepatitis B virus (HBV) has been recognized as a major factor for HCC in Asian countries[4]. Typically, HBV infection is more prevalent in Eastern Asia, such as China, where over two-thirds of patients are detected[5].
Although the etiological link between HBV and HCC has been well established, the effect of immunocytes on tumor-related microenvironment remains largely unclear. The tumor-infiltrating lymphocytes can serve as prognostic factors for HCC, and are a research hotspot[6,7]. Nonetheless, the non-lymphocyte immunocytes have also been recognized to play crucial roles in prognosis of various types of cancers[8]. Therefore, it is critical to comprehensively predict the immune-related prognosis.
The traditional approaches to measure immune infiltrate in tumors, like immunohistochemistry (IHC) and flow cytometry, cannot thoroughly evaluate the immune functions of various types of cells or effectively distinguish closely related cell populations, since few immune markers can be simultaneously detected using the existing technology. In recent years, the algorithm cell type identification by estimating relative subsets of RNA transcripts (CIBERSORT), known as the metagene means, is recognized as the method with the highest accuracy. Typically, CIBERSORT can accurately estimate 22 human immunocyte phenotypes[9].
This study explored the proportions of immunocytes by appropriately selecting HBV-related HCC cases, so as to identify candidate immune biomarkers using The Cancer Genome Atlas data. The least absolute shrinkage and selection operator (LASSO) algorithm was employed to determine key immunocytes. Thereafter, a HCC immunoscore system was formulated, aiming to establish an immune-related marker that could be used to predict the survival of HBV-related HCC patients.
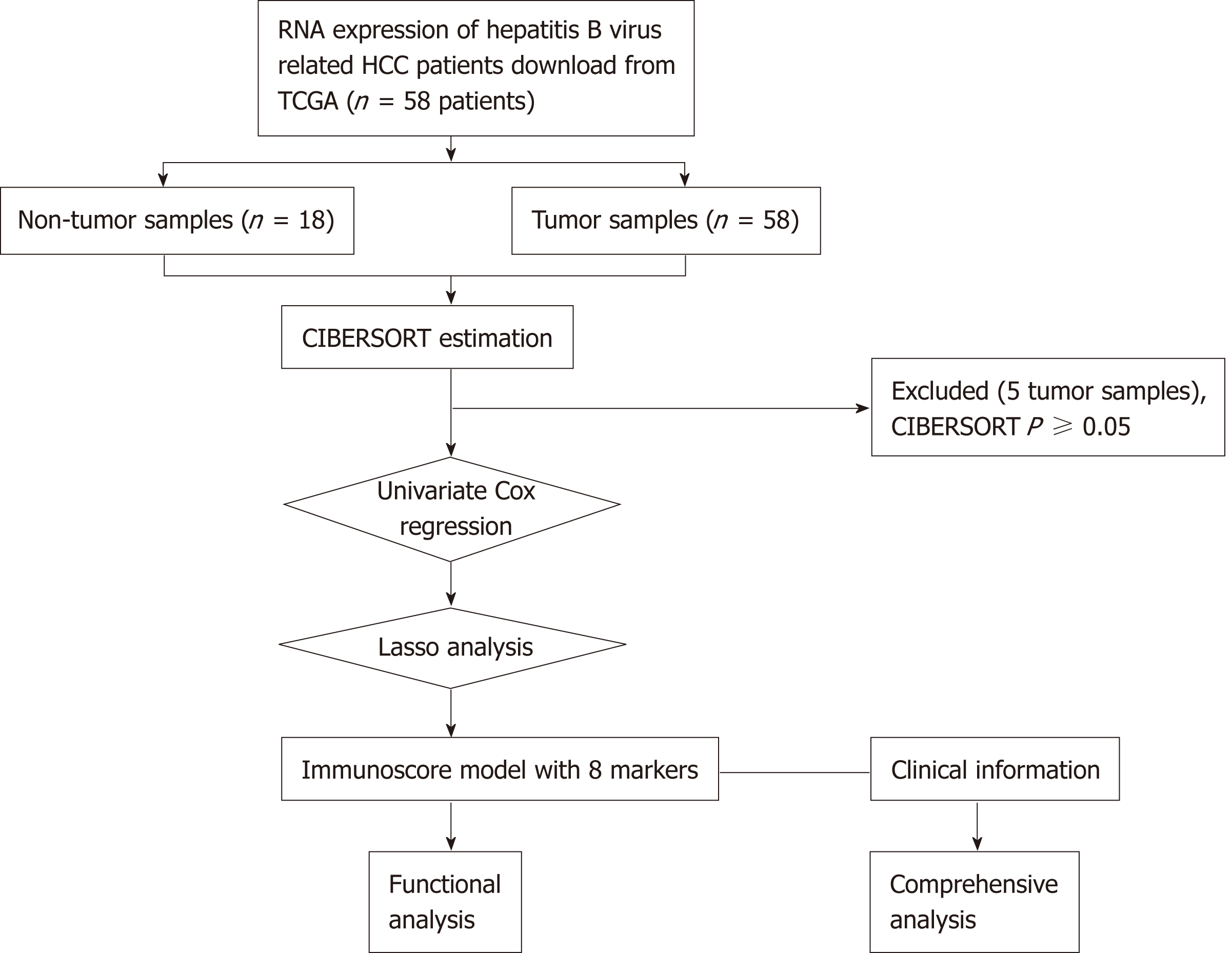
Data were downloaded from the TCGA database. The data transfer tool of the Genomic Data Commons applications was employed to download gene expression profiles as well as clinical information of HCC patients (https://tcga-data.nci.nih.gov/). HCC patients with seroprevalence of hepatitis B surface antigen (HBsAg) were included. Among them, 58 patients were HBV-related, but patients with CIBERSORT P ≥ 0.05 were excluded (n = 5), leaving 53 HCC cases in the final cohort for analysis. The flow chart for this analysis is shown in Figure 1. A total of 53 HBV-related HCC cases were enrolled for comprehensive integrated and functional analyses. The mRNA expression in HCC samples was derived from the Illumina HiSeq RNASeq platform and normalized based on TCGA. All data were publicly available and open access. Hence, approval from the ethics committee was not required. All data were processed in accordance with the guidelines from data access policy as well as the NIH TCGA human subject protection (http://cancergenome.nih. gov/publications/publicationguidelines).
The CIBERSORT method was used to quantify immunocyte fractions[9], which could sensitively and specifically distinguish 22 phenotypes of human hematopoietic cells. Pornpimol et al[10,11] had used the deconvolution method through the CIBERSORT tool, with the threshold set at P < 0.05; their results suggested that CIBERSORT could accurately predict the immunocyte population fractions. Hence, only those possessing the CIBERSORT P < 0.05 were deemed as eligible in subsequent analysis. Moreover, the immunocyte proportions were separately estimated for 76 samples. For all samples, each estimate of immunocyte type fractions added up to be 1.
The optimal threshold for each immunocyte fraction was generated using the survminer package. Then, the value of immunocyte fraction was assigned as 0 or 1; specifically, the value 0 was given to the cell type fraction below the corresponding threshold, otherwise the value 1 was assigned. The correlation of each immunocyte expression with the overall survival (OS) of patients was calculated using the univariate Cox model. Then, the immunocytes were screened, followed by verification through LASSO regression using “glmnet” package in R project. Finally, an immunoscore was constructed based on immunocytes by linearly combining the multiplication of regression model (β) by expression level as follows: Immunoscore = β1 × immunocyte 1 expression level + β2 × immunocyte 2 expression level + ····· + βn × immunocyte n expression level. In addition, the time dependent receiver operating characteristic (ROC) curves were employed to evaluate the accuracy of prognosis prediction in five years, which was achieved by the comparisons of sensitivity as well as specificity in predicting the survival using the immunoscore.
The patients were distributed based on the immunoscore and their survival information. Furthermore, all cases were classified into a high- or low-risk group according to the immunoscore threshold. In addition, related Kaplan-Meier survival curves for patients with high or low predicted risk were plotted. Thereafter, univariate as well as multivariate Cox proportional hazard regression analyses were performed. We developed a nomogram with “rms” package in R project. The prognosis prediction accuracy of the nomogram was verified for discrimination and calibration. Each test was two-sided, and a difference of P < 0.05 was deemed statistically significant. The R (version 3.4.1; R Foundation) software was adopted for all analyses.
Inhibitory checkpoint molecules can be potentially used in numerous cancer types, which are thereby recognized as the immunotherapy targets for cancer. The correlations of the immunoscore with corresponding genes were further investigated. In addition, the correlation of the immunoscore with the determined immune markers was analyzed through Pearson’s correlation analysis based on their expression levels. The mechanisms underlying “Molecular Signatures Database” for the c2.cp.kegg.v6.2.symbols were investigated according to gene set enrichment analysis (GSEA)[12] with JAVA procedure (http://software.broadinstitute.org/gsea/index.jsp). The random sample permutation number was deemed at 1000 and a significant level was P < 0.05.
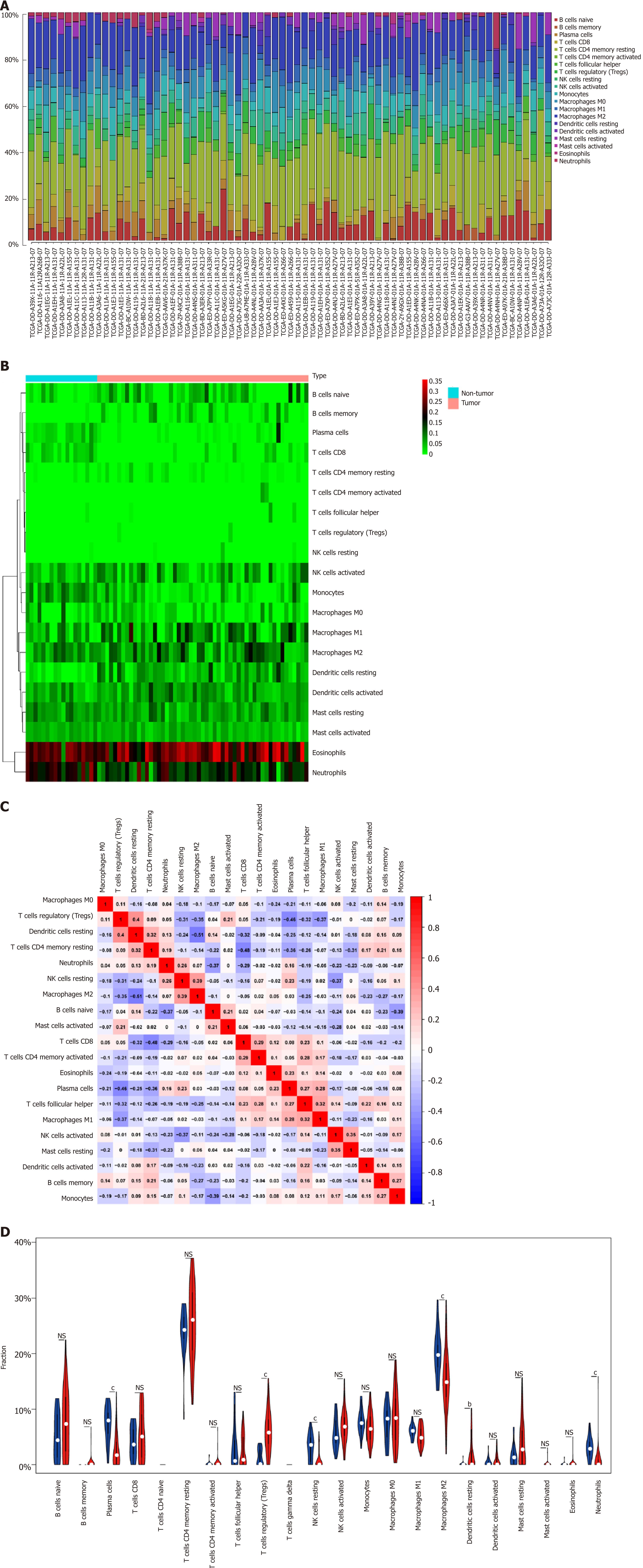
Detailed patient characteristics are listed in Table 1. The immunocyte compositions of cancer and normal tissues were analyzed. Figure 2A summarizes immunocyte composition in the entire cohort. Resting CD4 memory T cells ranked the first, accounting for 25% on average; while CD4 naive T cells and gamma delta T cells were not detected. Heatmap summarizing the distribution of immunocytes was also plotted (Figure 2B). Various immunocyte subpopulation fractions showed a low to moderate correlation (Figure 2C). Relative to para-carcinoma tissues, higher proportions of regulatory T cells as well as resting dendritic cells could be detected in cancer tissues, along with lower proportions of plasma cells, resting NK cells, M2 macrophages, and neutrophils (Figure 2D, P < 0.05).
| Variable | Number (n) | Percent (%) | |
| Age | 60.1 ± 15.4 | 53 | 100 |
| Gender | |||
| Male | 31 | 58.5 | |
| Female | 22 | 41.5 | |
| Race | |||
| White | 33 | 62.3 | |
| Others | 20 | 37.7 | |
| Grade | |||
| I | 18 | 34.0 | |
| II | 16 | 30.2 | |
| III | 14 | 26.4 | |
| IV | 5 | 9.4 | |
| Stage | |||
| I | 5 | 9.4 | |
| II | 25 | 47.2 | |
| III | 22 | 41.5 | |
| IV | 1 | 1.9 |
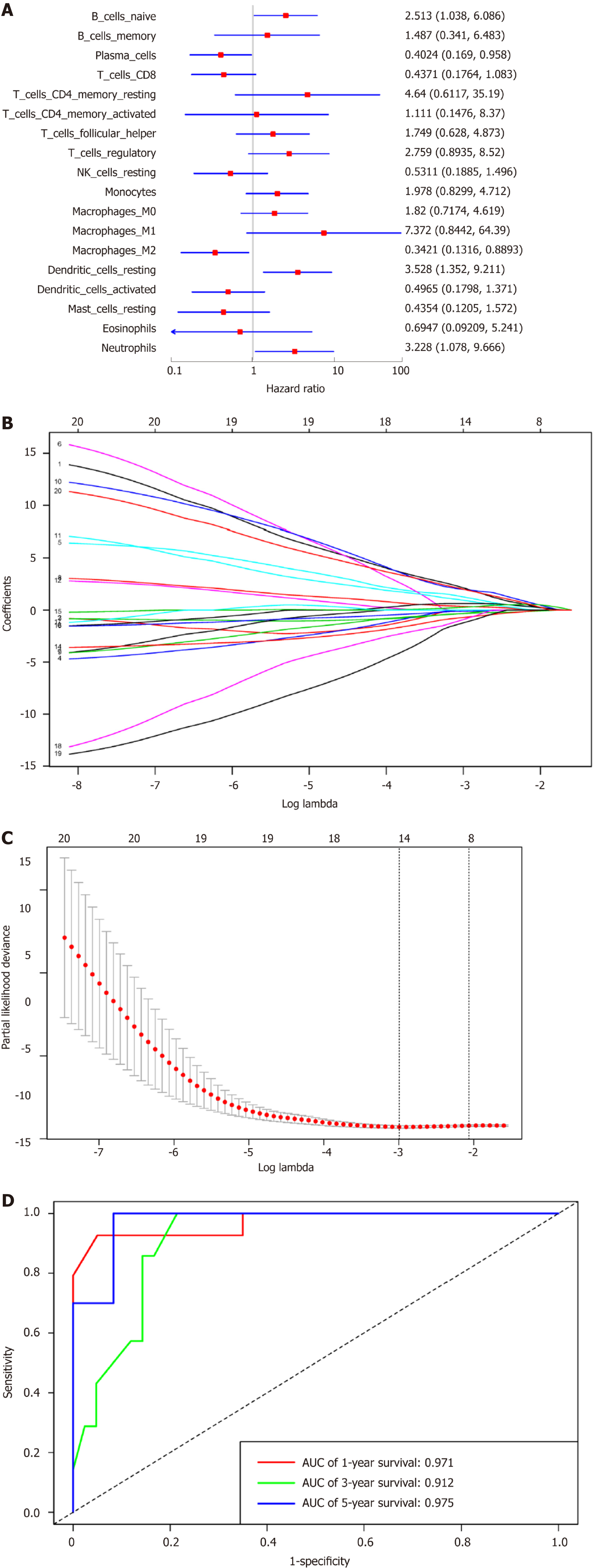
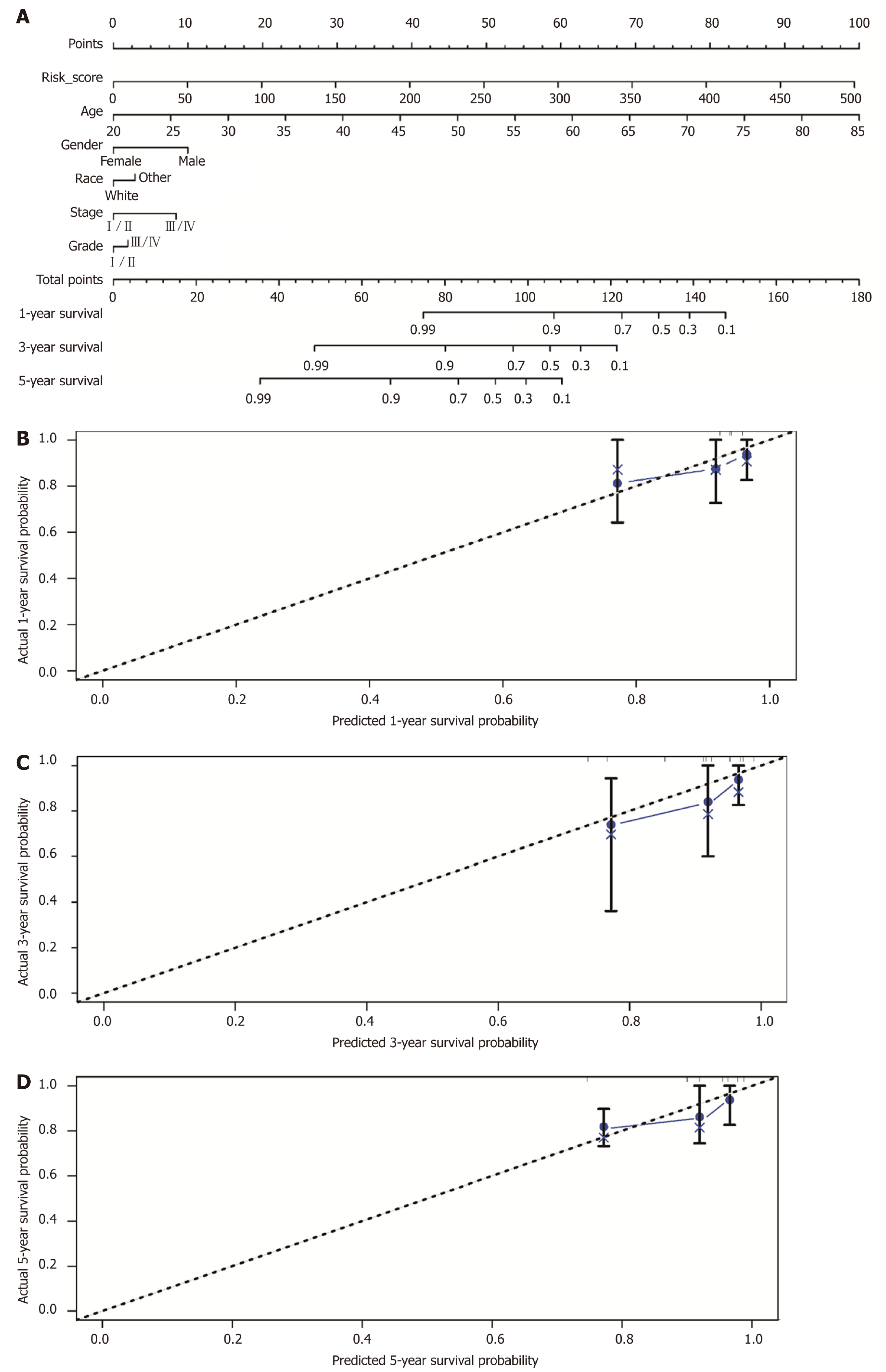
Table 2 displays the thresholds of all cell types. Figure 3A presents the forest plot of the correlation of 18 immunocyte subsets with OS. Through the LASSO algorithm (Figure 3B and C ), eight types of immunocytes (including plasma cells, naive B cells, CD4 memory resting T cells, M1 macrophages, activated NK cells, resting dendritic cells, M2 macrophages, and neutrophils) were selected to build the prognostic immunoscore model. The immunoscore was calculated according to the formula: (2.764 × fraction level of naive B cells) - (0.371 × fraction level of plasma cells) + (1.694 × fraction level of resting CD4 memory T cells) + (21.45 × fraction level of activated NK cells) + (1.669 × fraction level of M1 macrophages) - (1.649 × fraction level of M2 macrophages) + (0.504 × fraction level of resting dendritic cells) + (3.647 × fraction level of neutrophils). Moreover, the immunoscore prognosis performance, as the continuous variable, was also studied within the cohort through time-dependent ROC analysis. The areas under the curves of the immunoscore biomarker prognostic model were 0.971, 0.912, and 0.975 for 1-, 3-, and 5-year OS, respectively (Figure 3D).
| Cell type | Cut-off value |
| B cells naive | 0.107 |
| B cells memory | 0 |
| Plasma cells | 0.010 |
| T cells CD8 | 0.020 |
| T cells CD4 memory resting | 0.157 |
| T cells CD4 memory activated | 0 |
| T cells follicular helper | 0.054 |
| T cells regulatory | 0.108 |
| NK cells resting | 0.002 |
| NK cells activated | 0.035 |
| Monocytes | 0.082 |
| Macrophages M0 | 0.067 |
| Macrophages M1 | 0.030 |
| Macrophages M2 | 0.131 |
| Dendritic cells resting | 0.039 |
| Dendritic cells activated | 0.002 |
| Mast cells resting | 0.074 |
| Mast cells activated | 0 |
| Eosinophils | 0 |
| Neutrophils | 0.013 |
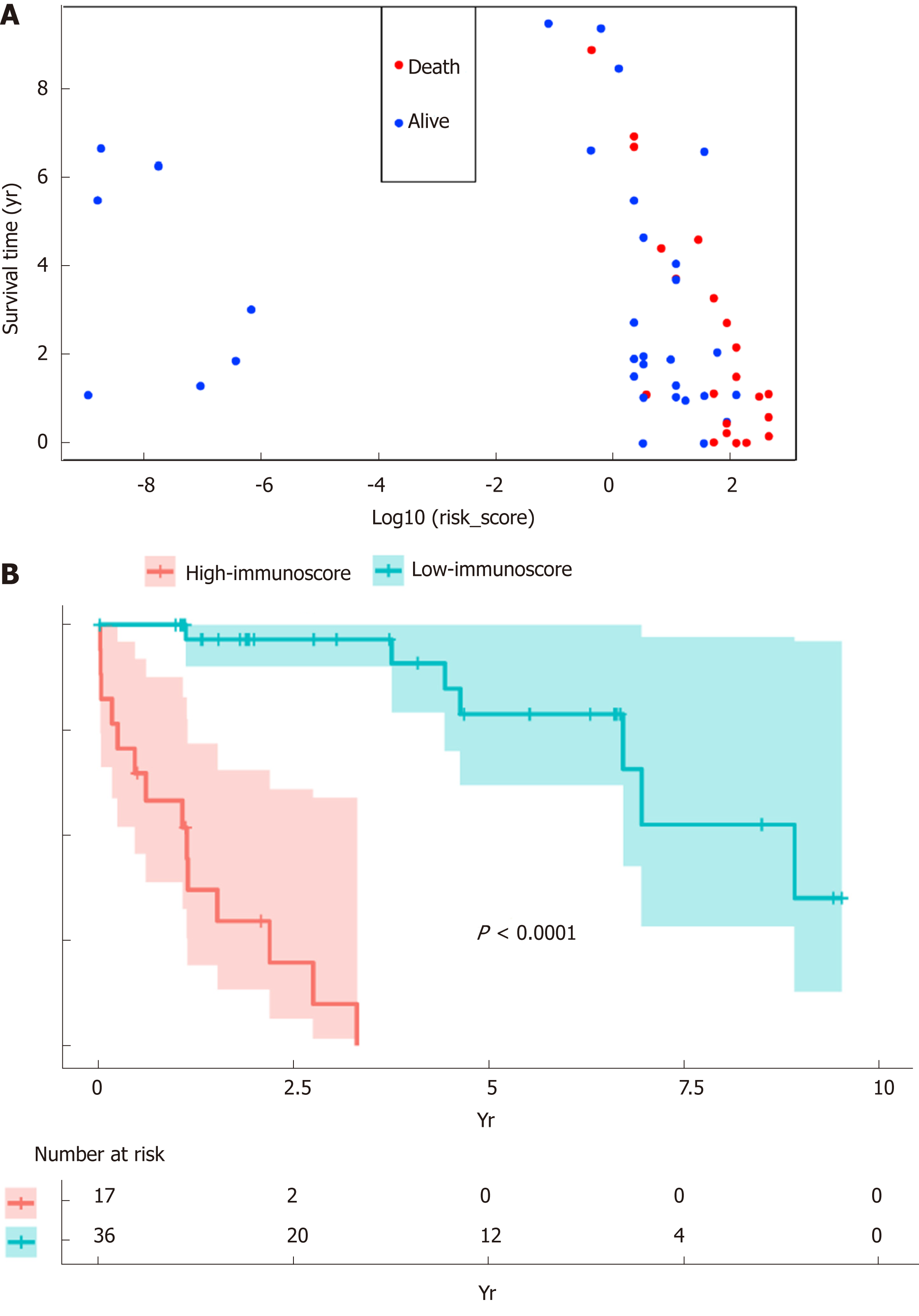
The patients were distributed based on the immunoscore and their survival information (Figure 4A). Subsequently, the patients were classified into high vs low immunoscore groups according to the threshold (36.47) acquired using survminer package. The Kaplan-Meier curves for both groups are presented in Figure 4B. The patients with a high immunoscore were associated with worse OS than those with a low immunoscore [hazard ratio (HR) = 66.007, 95% confidence interval (CI): 8.361-521.105; P < 0.0001]. The HR of the immunoscore was 2.010 (95%CI: 1.487-2.716) based on the univariate Cox proportional hazard regression model, and similar results were obtained from the multivariate Cox proportional hazard regression model (HR = 2.997, 95%CI: 1.737-5.170) after adjustment for clinical covariables (Table 3).
| Parameter | Univariate analysis | Multivariate analysis | ||
| HR (95%CI) | P-value | HR (95%CI) | P-value | |
| Risk_score | 2.010 (1.487-2.716) | < 0.001 | 2.997 (1.737-5.170) | < 0.001 |
| Age | 1.022 (0.985-1.060) | 0.255 | 1.091 (1.024-1.162) | 0.007 |
| Gender (vs male) | 1.001 (0.404-2.479) | 0.999 | 0.727 (0.215-2.459) | 0.608 |
| Race (vs white) | 1.369 (0.470-3.992) | 0.565 | 0.905 (0.195-4.208) | 0.899 |
| Grade (vs I/II) | 1.800 (0.743-4.359) | 0.193 | 0.987 (0.327-2.975) | 0.981 |
| Stage (vs I/II) | 2.161 (0.865-5.400) | 0.099 | 10.182 (2.251-46.058) | 0.003 |
Additionally, a prognostic nomogram incorporating the above prognostic factors for visualization and prediction was built to identify a composite factor to predict the prognosis for HCC cases (Table 3). Notably, the nomogram was constituted by six factors (Figure 5A), with a C-index of 0.757 (95%CI: 0.648-0.866). Meanwhile, the 1-, 3-, and 5-year calibration curves of the recurrence probability were consistent between predicted values by the nomogram and measured values (Figure 5B-D).
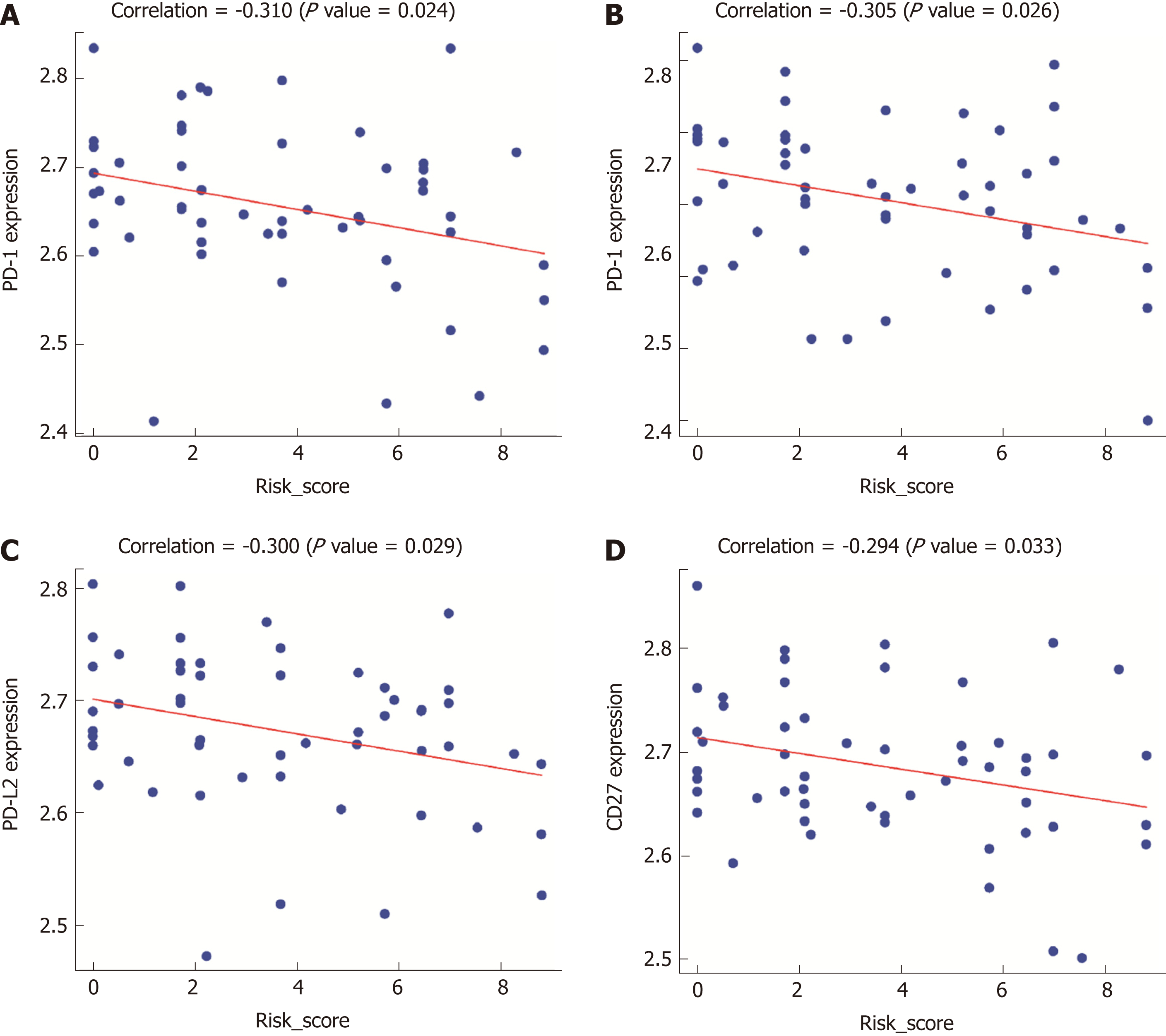
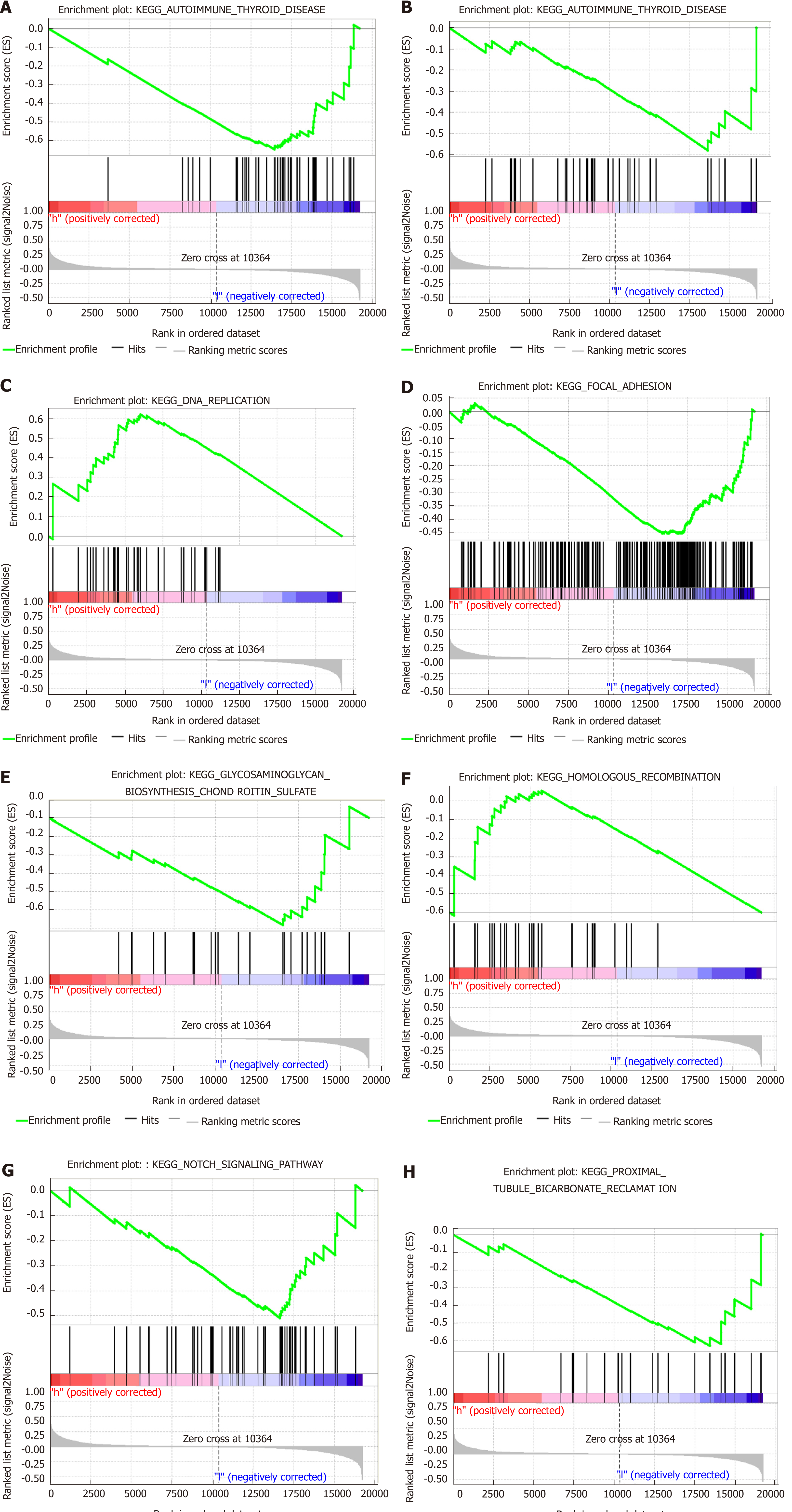
Gene expression profiles were examined for exploring the potential immunoscore model-related biological phenotypes. First, the correlation of the immunoscore with the screened immune-related gene expression was particularly emphasized. Figure 6 displays the significant negative correlations of the immunoscore with programmed death (PD)-1 (P = 0.024), PD-L1 (P = 0.026), PD-L2 (P = 0.029), and CD27 (P = 0.033) expression. Second, GSEA was performed for illustrating the biological roles for the prepared immunoscore model (Supplemental Table 1). The highly expressed genes showed significant enrichment in pathways including glycosaminoglycan chondroitin sulfate biosynthesis, proximal tubule bicarbonate reclamation, focal adhesion, autoimmune thyroid disease, the Notch signaling pathway, basal transcription factors, DNA replication, and homologous recombination (Figure 7).
The CIBERSORT was utilized for the quantification of composition of 22 immunocytes for HBV-related HCC. Furthermore, the immunoscore was developed, which was based on eight immunocyte fractions and served as a new tool to predict the prognosis of HBV-related HCC survival. The prognostic significance of the immunoscore signature was confirmed by the time-dependent ROC curve, Kaplan-Meier, and Cox regression analyses. The results suggested that the established immunoscore system displayed favorable accuracy for estimating OS of HBV-related HCC by nomogram analysis. Further functional analysis uncovered several immune checkpoints and pathways that played crucial roles in the immunoscore system during the progression of HCC.
There are few available immunoscore-based models for the prognosis of HCC. In 2015, Sun et al[13] confirmed that center tumor CD8+ T cells could predict the prognosis for HCC patients, and were a valuable marker for predicting patient survival and tumor recurrence. However, only CD3+ and CD8+ T cells were measured by IHC in their research. Similarly, in 2016, Gabrielson et al[14] concluded that immunoscore as well as PD-L1 expression could be used as valuable markers to predict the prognosis for HCC patients undergoing resection of primary tumor. However, regarding etiology of HCC, only 16.9% (11/65) of cases suffered from HBV infection in their study. In 2016, Petrizzo demonstrated that immunoscore was a meaningful marker to predict the prognosis for HCC patients undergoing resection for primary tumor[15]. However, 65 HCC patients with unknown virus condition were reviewed in that study, and IHC was only performed with anti-CD3, CD8, and PD-L1 monoclonal antibodies. In 2017, Yao et al[16] conducted a cohort study by recruiting 92 consecutive HBV-HCC patients, and concluded that the immunoscore classification showed close relation to patient outcome. However, only CD3+, CD8+, and CD45RO+ T cells were measured by IHC in that study.
The current study was superior to previous studies examining the prognosis value of immunoscore for HCC in several aspects. First, assessing CD3+ and CD8+ lymphocytes alone could not comprehensively represent local immune status. Prior research was limited by its narrow view of immunological reaction due to technical limitations, and immunocytes were evaluated through IHC based on a single surface marker, which was ineffective for distinguishing the tightly correlated cell types, since numerous marker proteins would be expressed among various types of cells. Technically, IHC is limited to existing phenotypic markers, and may lead to inconsistent findings in clinical studies. However, using transcriptomic data for computationally describing tumor microenvironment can serve as a promising tool to overcome the technical limitations related to IHC, and can be more readily employed for the further characterization of 22 different immune populations than IHC. Second, homogeneity in the case cohort was largely enhanced through pre-analysis case selection. Viral and non-viral HCC were suggested to be associated with different prognoses, and different treatment strategies were recommended; therefore, they must be distinctively analyzed to develop the models for prognosis[17-20]. In contrast to previous studies that combined viral and non-viral related HCC, the current study was conducted in HBV-positive patients only. Third, in terms of the methodology, LASSO penalized regression could increase the accuracy of bioinformatic analysis, and allowed for simultaneous analysis of each independent variable to select the most valuable parameters[21]. Therefore, such approach would show much higher accuracy than stepwise regression of the multivariate Cox model[22]. Fourth, an easy-to-use nomogram model based on Cox analysis was constructed in our meticulously chosen patient cohort with clinicopathological characteristics that might facilitate individualized prediction of patients.
Immune checkpoint inhibitors have shown significant therapeutic responses to tumors. Clinical trials of antibodies targeting the immune checkpoint inhibitors to treat advanced HCC are ongoing[23]. Our results showed that the immunoscore was negatively correlated with PD-1, PD-L1, PD-L2, and CD27 (Figure 6). There are two ligands for PD-1 receptor, namely, PD-L1 and PD-L2[24,25]. Our previous studies found that excessive PD-1-positive intratumoral lymphocytes could predict the cytokine-induced killer cell survival benefits for HCC patients[26], and serum soluble PD-1 and PD-L1 can independently predict the prognosis for HCC patients in distinct ways[27]. CD27 is a cytokine that facilitates naive T cell expansion specific to antigen, which is of vital importance to generate T cell memory[28]. Moreover, pathway enrichment analysis suggested that such immunocytes could affect HCC genesis as well as progression by eight pathways whose biological functions have been previously reported in HCC. Most of these pathways are canonical and crucial, which participate in HCC genesis as well as progression.
This study had several limitations. First, information, such as acute infection, immune system diseases, as well as anti-inflammatory drugs, could not be obtained since this study was conducted based on the publicly available datasets, and this could potentially result in error or bias. Second, each case was retrospectively chosen, so an underlying bias may exist related to the imbalanced clinicopathological characteristics with treatment heterogeneity. Third, all data were downloaded from Western countries to establish the models, so the findings should be cautiously applied to the Asian population. Finally, further large-sample research is needed for external validation of our results.
In summary, this study uncovered the immunocyte signature in HBV-related HCC. The proposed immunoscore model may comprehensively provide the required clinical data to improve the personalized treatment for HBV-related HCC cases.
Hepatocellular carcinoma (HCC) is a common subtype of liver cancer, which has become a serious health threat worldwide. Hepatitis B virus (HBV) has been identified as a leading cause of HCC. Increasing evidence indicates that cancer immune infiltration is associated with clinical outcomes. However, no credible prognosis signature for HBV-associated HCC has been constructed through systemically assessing bulk tumor transcriptomic immune landscape.
Tumor-infiltrating immunocytes represent a vital prognosis clue in HBV-associated HCC patients. In this study, a new immune model for HBV-associated HCC cases was developed through systematically assessing the bulk tumor transcriptomic immune landscape.
The current research aimed to establish a prognosis-related immune signature based on bulk tumor transcriptome-derived immune infiltrate compositions, thus improving the prognosis prediction accuracy for HBV-associated HCC.
The fractions of 22 immunocyte types extracted based on public datasets were predicated using the cell type identification by estimating relative subsets of RNA transcripts (CIBERSORT) algorithm. Furthermore, an immunoscore was calculated based on immunocyte type fractions by the least absolute shrinkage and selection operator (LASSO) model.
The immunoscore constituted by eight immunocyte type fractions was constructed by adopting the LASSO model, which displayed a high sensitivity and specificity for overall survival (OS), and the areas under the curves for the 1-year, 3-year, and 5-year OS were 0.971, 0.912, and 0.975, respectively. Difference in OS between the low-immunoscore and high-immunoscore groups was statistically significant. Additionally, a nomogram was established to expand the applied range of the model, which included the immunoscore as well as other clinical characteristics. The related pathways were enriched through gene set enrichment analysis.
The established immunoscore showed high prognosis prediction accuracy for patients with HBV-associated HCC, which may facilitate the risk stratification and provide valuable clinical suggestions for individual cases.
Findings of this study suggested that the tumor-infiltrating immunocytes could be used as promising biomarkers to predict the prognosis for HBV-associated HCC patients. This forms the framework for identification of efficient prognosis predictors and molecular biomarkers for HCC patients in the future.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kai K, El-Bendary M S-Editor: Ma RY L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Chen QF, Jia ZY, Yang ZQ, Fan WL, Shi HB. Transarterial Chemoembolization Monotherapy vs Combined Transarterial Chemoembolization-Microwave Ablation Therapy for Hepatocellular Carcinoma Tumors ≤ 5 cm: A Propensity Analysis at a Single Center. Cardiovasc Intervent Radiol. 2017;40:1748-1755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 2. | Zhou HY, Luo Y, Chen WD, Gong GZ. Hepatitis B virus mutation may play a role in hepatocellular carcinoma recurrence: A systematic review and meta-regression analysis. J Gastroenterol Hepatol. 2015;30:977-983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol (NY). 2018;43:13-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 302] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 4. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2501] [Article Influence: 192.4] [Reference Citation Analysis (2)] |
| 5. | de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62:1190-1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 348] [Cited by in RCA: 381] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 6. | Jang JE, Hajdu CH, Liot C, Miller G, Dustin ML, Bar-Sagi D. Crosstalk between Regulatory T Cells and Tumor-Associated Dendritic Cells Negates Anti-tumor Immunity in Pancreatic Cancer. Cell Rep. 2017;20:558-571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 284] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 7. | Wang TT, Zhao YL, Peng LS, Chen N, Chen W, Lv YP, Mao FY, Zhang JY, Cheng P, Teng YS, Fu XL, Yu PW, Guo G, Luo P, Zhuang Y, Zou QM. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut. 2017;66:1900-1911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 364] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 8. | Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3108] [Cited by in RCA: 3601] [Article Influence: 277.0] [Reference Citation Analysis (0)] |
| 9. | Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4763] [Cited by in RCA: 8853] [Article Influence: 885.3] [Reference Citation Analysis (0)] |
| 10. | Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017;18:248-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1446] [Cited by in RCA: 3167] [Article Influence: 395.9] [Reference Citation Analysis (0)] |
| 11. | Ali HR, Chlon L, Pharoah PD, Markowetz F, Caldas C. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS Med. 2016;13:e1002194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 446] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 12. | Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545-15550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27252] [Cited by in RCA: 37273] [Article Influence: 1863.7] [Reference Citation Analysis (0)] |
| 13. | Sun C, Xu J, Song J, Liu C, Wang J, Weng C, Sun H, Wei H, Xiao W, Sun R, Tian Z. The predictive value of centre tumour CD8⁺ T cells in patients with hepatocellular carcinoma: comparison with Immunoscore. Oncotarget. 2015;6:35602-35615. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Gabrielson A, Wu Y, Wang H, Jiang J, Kallakury B, Gatalica Z, Reddy S, Kleiner D, Fishbein T, Johnson L, Island E, Satoskar R, Banovac F, Jha R, Kachhela J, Feng P, Zhang T, Tesfaye A, Prins P, Loffredo C, Marshall J, Weiner L, Atkins M, He AR. Intratumoral CD3 and CD8 T-cell Densities Associated with Relapse-Free Survival in HCC. Cancer Immunol Res. 2016;4:419-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 243] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 15. | Petrizzo A, Buonaguro L. Application of the Immunoscore as prognostic tool for hepatocellular carcinoma. J Immunother Cancer. 2016;4:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Yao Q, Bao X, Xue R, Liu H, Liu H, Li J, Dong J, Duan Z, Ren M, Zhao J, Song Q, Yu H, Zhu Y, Lu J, Meng Q. Prognostic value of immunoscore to identify mortality outcomes in adults with HBV-related primary hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e6735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Hosaka T, Suzuki F, Kobayashi M, Seko Y, Kawamura Y, Sezaki H, Akuta N, Suzuki Y, Saitoh S, Arase Y, Ikeda K, Kobayashi M, Kumada H. Long-term entecavir treatment reduces hepatocellular carcinoma incidence in patients with hepatitis B virus infection. Hepatology. 2013;58:98-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 540] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 18. | Ogawa E, Furusyo N, Nomura H, Dohmen K, Higashi N, Takahashi K, Kawano A, Azuma K, Satoh T, Nakamuta M, Koyanagi T, Kato M, Shimoda S, Kajiwara E, Hayashi J; Kyushu University Liver Disease Study (KULDS) Group. Short-term risk of hepatocellular carcinoma after hepatitis C virus eradication following direct-acting anti-viral treatment. Aliment Pharmacol Ther. 2018;47:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 19. | Liu GM, Huang XY, Shen SL, Hu WJ, Peng BG. Adjuvant antiviral therapy for hepatitis B virus-related hepatocellular carcinoma after curative treatment: A systematic review and meta-analysis. Hepatol Res. 2016;46:100-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Shinn BJ, Martin A, Coben RM, Conn MI, Prieto J, Kroop H, DiMarino AJ, Hann HW. Persistent risk for new, subsequent new and recurrent hepatocellular carcinoma despite successful anti-hepatitis B virus therapy and tumor ablation: The need for hepatitis B virus cure. World J Hepatol. 2019;11:65-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7447] [Cited by in RCA: 8373] [Article Influence: 558.2] [Reference Citation Analysis (0)] |
| 22. | McNeish DM. Using Lasso for Predictor Selection and to Assuage Overfitting: A Method Long Overlooked in Behavioral Sciences. Multivariate Behav Res. 2015;50:471-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 202] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 23. | Kudo M. Immune Checkpoint Inhibition in Hepatocellular Carcinoma: Basics and Ongoing Clinical Trials. Oncology. 2017;92:50-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 140] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 24. | Syn NL, Teng MWL, Mok TSK, Soo RA. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18:e731-e741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 536] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 25. | Philips GK, Atkins M. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int Immunol. 2015;27:39-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 264] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 26. | Chang B, Shen L, Wang K, Jin J, Huang T, Chen Q, Li W, Wu P. High number of PD-1 positive intratumoural lymphocytes predicts survival benefit of cytokine-induced killer cells for hepatocellular carcinoma patients. Liver Int. 2018;38:1449-1458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 27. | Chang B, Huang T, Wei H, Shen L, Zhu D, He W, Chen Q, Zhang H, Li Y, Huang R, Li W, Wu P. The correlation and prognostic value of serum levels of soluble programmed death protein 1 (sPD-1) and soluble programmed death-ligand 1 (sPD-L1) in patients with hepatocellular carcinoma. Cancer Immunol Immunother. 2019;68:353-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 28. | Hendriks J, Gravestein LA, Tesselaar K, van Lier RA, Schumacher TN, Borst J. CD27 is required for generation and long-term maintenance of T cell immunity. Nat Immunol. 2000;1:433-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 561] [Cited by in RCA: 604] [Article Influence: 24.2] [Reference Citation Analysis (0)] |