Published online Jul 7, 2019. doi: 10.3748/wjg.v25.i25.3242
Peer-review started: April 15, 2019
First decision: May 16, 2019
Revised: June 2, 2019
Accepted: June 8, 2019
Article in press: June 9, 2019
Published online: July 7, 2019
Processing time: 84 Days and 22.7 Hours
Ulcerative colitis (UC) is considered to be closely associated with alteration of intestinal microorganisms. According to the traditional Chinese medicine (TCM) theory, UC can be divided into two disease syndromes called Pi-Xu-Shi-Yun (PXSY) and Da-Chang-Shi-Re (DCSR). The relationships among gut microbiota, TCM syndromes, and UC pathogenesis have not been well investigated.
To investigate the role of gut microbiota in UC and the distinction of microbiota dysbiosis between PXSY and DCSR syndromes.
From May 2015 to February 2016, UC patients presenting to LongHua Hospital who met the established inclusion and exclusion criteria were enrolled in this retrospective study. Fresh stool specimens of UC patients with PXSY or DCSR were collected. The feces of the control group came from the health examination population of Longhua Hospital. The composition of gut bacterial communities in stool samples was determined by the pyrosequencing of 16S ribosomal RNA. The high-throughput sequencing reads were processed with QIIME, and biological functions were predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
The composition of gut bacterial communities in 93 stool samples (30 healthy controls, 32 patients with PXSY syndrome, and 31 patients with DCSR syndrome) was determined by the pyrosequencing of 16S ribosomal RNA. Beta diversity showed that the composition of the microbiota was different among the three groups. At the family level, Porphyromonadaceae, Rikeneliaceae, and Lachnospiraceae significantly decreased while Enterococcus, Streptococcus, and other potential pathogens significantly increased in UC patients compared to healthy subjects. At the genus level, Parabacteroides, Dorea, and Ruminococcus decreased while Faeca-libacterium showed increased abundance in UC compared to healthy controls. Five differential taxa were identified between PXSY and DCSR syndromes. At the genus level, a significantly increased abundance of Streptococcus was observed in DCSR patients, while Lachnoclostridium increased in PXSY patients. The differential functional pathways of the gut microbiome between the PXSY and DCSR groups mainly included lipid metabolism, immunity, and the metabolism of polypeptides.
Our study suggests that the gut microbiota contributes to the distinction between the two TCM syndromes of UC.
Core tip: Ulcerative colitis (UC) is considered to be closely associated with alteration of intestinal microorganisms. According to the traditional Chinese medicine (TCM) theory, UC can be divided into Pi-Xu-Shi-Yun syndrome (syndrome of spleen deficiency and dampness, PXSY) and Da-Chang-Shi-Re syndrome (syndrome of dampness-heat in the large intestine, DCSR). This study showed that the gut microbiota was different between patients with PXSY syndrome and those with DCSR syndrome. The genus Streptococcus was significantly more abundant in DCSR patients than in PXSY patients, while Lachnoclostridium increased in PXSY patients. Our study suggests that the gut microbiota contributes to the distinction between the two TCM syndromes of UC.
- Citation: Zhang YL, Cai LT, Qi JY, Lin YZ, Dai YC, Jiao N, Chen YL, Zheng L, Wang BB, Zhu LX, Tang ZP, Zhu RX. Gut microbiota contributes to the distinction between two traditional Chinese medicine syndromes of ulcerative colitis. World J Gastroenterol 2019; 25(25): 3242-3255
- URL: https://www.wjgnet.com/1007-9327/full/v25/i25/3242.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i25.3242
Ulcerative colitis (UC) is a form of inflammatory bowel disease (IBD). According to relevant epidemiological studies, the incidence of UC is high in Western developed countries, particularly in North America (19.2 per 100000 person-years) and Europe (24.3 per 100000 person-years)[1]. The incidence of UC has been increasing in Asia, especially in China, in recent years[2-4]. The high incidence of UC in China is particularly concentrated in coastal areas, such as Guangzhou (2.22 per 100000 person-years) and Hong Kong (1.66 per 100000 person-years)[5,6]. Notably, UC patients have a higher incidence of colorectal cancer than healthy individuals[7,8]. Moreover, UC is often difficult to treat and is accompanied by a high recurrence rate. Since di-fferent individuals have different manifestations of UC, more accurate diagnosis and treatment of UC for every patient have been emphasised, and precision medicine for UC is needed.
According to the traditional Chinese medicine (TCM) theory, the same disease can be divided into different syndromes, and different treatments would be used clinically depending on the different syndromes. For instance, UC can be divided into Pi-Xu-Shi-Yun syndrome (syndrome of spleen deficiency and dampness, PXSY) and Da-Chang-Shi-Re syndrome (syndrome of dampness-heat in the large intestine, DCSR)[9]. Shi-Re syndrome is caused by dysfunction of the Pi (“spleen”) and Wei (“stomach”), and Pi-Xu syndrome has been shown to be involved in dysfunction of the vegetative nervous system of the gastrointestinal tract and immune pathways[10]. PXSY syndrome is a deficiency syndrome, while the DCSR syndrome is a sthenia syndrome. Studies have proven that TCM works for treating UC patients[11-13] and that TCM drugs are closely associated with immunity[14,15], such as reducing proinfla-mmatory cytokines[16]. Nevertheless, the essential molecular mechanisms of TCM remain unknown.
Although the pathogenesis of UC is not yet clear, most studies discuss that the dysbiosis of commensal bacterial populations is a major factor in disease pathogenesis, which interacts with genetic susceptibility and could lead to dysregulation of the immune response to bacterial antigens in IBD[17,18]. Round et al[19] suggested that dysregulation of the gut microbiota might be the cause of UC. The microbiota in the gut affects the health of the host by interacting with genes and the environment. Some studies[20-22] found that the biodiversity of intestinal bacteria in patients with UC is significantly reduced and that the increase in destructive microbiota in the gut leads to disorders in the intestinal environment. Guo et al[23] showed that the Chinese medicine “Red Ginseng” was effective in relieving symptoms of UC and could promote the growth of probiotic bacteria in vitro. However, the relationships among gut microbiota, TCM syndromes, and UC patho-genesis are unclear.
Because the majority of studies have focused on the relationship between intestinal microbiota and the onset of UC and the contribution of gut microbiota in these two distinct TCM syndromes is still unclarified, this study compared the difference in microbial composition and function between PXSY and DCSR syndromes to determine the molecular mechanism of TCM in UC by investigation of the gut microbiota.
This study was approved by the LongHua Hospital, Shanghai University of Traditional Chinese Medicine. Three groups of adults were recruited in this study (PXSY patients, DCSR patients, and healthy controls). Volunteers under investigation in the hospital were approached if the doctors deemed that they had either UC or a normal colon at colonoscopy. Two types of TCM syndromes of UC, DCSR syndrome and PXSY syndrome, were identified according to the "dysentery" diagnosis standard of UC.
The main clinical manifestations of UC are diarrhoea, mucous pus, and blood accompanied by abdominal pain, tenesmus and back weight, and systemic symptoms of varying degrees. The course of disease is more than 4-6 wk. Mucous purulent blood is the most common symptom of UC. Currently, there is still a lack of the gold standard for UC diagnosis, which is mainly based on the combination of clinical manifestations and endoscopic, pathological, and histological findings as well as the exclusion of infectious and non-infectious colitis. We can distinguish these two syndromes according to patients' clinical manifestations, tongue coating, and pulse condition. Clinical manifestations of DCSR syndrome were abdominal pain, tenesmus, stools containing blood and white mucus, a burning sensation around the anus, and scanty, dark-yellow urine. The tongue coating was greasy and slightly yellow. Pulse was slippery and rapid. While clinical manifestations of PXSY syndrome were recurrent loose stools and increased frequency of bowel movements, especially after eating greasy food, poor appetite, gastric or abdominal distension, a sallow complexion, and lassitude. The tongue coating was pale and thin. The pulse was weak and thready[24,25].
The exclusion criteria for UC patients were: (1) Below the age of 18 or above the age of 60 years; (2) Use of microecological preparations or antibiotics within the previous two weeks; (3) Previous gastrointestinal surgery; (4) Severe hypertension, diabetes, coronary heart disease, or other internal diseases; (5) Pregnancy, lactation, or planned pregnancy; (6) Uncooperativeness; and (7) Other autoimmune diseases.
Faecal samples were collected from volunteers and stored at -80 °C before DNA isolation. After the DNA was extracted, the integrity of the sampled bacterial genome was examined using agar gel electrophoresis. The V4 hyper-variable region was PCR-amplified with forward primer 515F (5’-GTGCCAGCCMGCCGCGG-3’) and reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’). Unique barcodes were attached to the primers to allow multiplex deep sequencing. The 25-μL reaction system contained 2 µL DNA extract, 0.4 µL forward primer, 0.4 µL reverse primer, 0.2 µL Taq DNA, and 2.5 µL 10 × buffer. PCR cycling parameters were initial denaturation at 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 1 min; and a final extension at 72 °C for 10 min. The PCR product was electrophoresed on a 2.0% agarose gel for 30 min at 120 V, followed by staining with ethidium bromide (0.5 µg/mL) for 20 min, and the brightness of the image strip was observed.
Trimmomatic[26] (version 0.36) and QIIME[27] software (version 1.8.1) were applied for sequence processing and analysis. In this study, reads with a length less than 200 bp were excluded from further processing and analysis. All the joined sequences were identified as chimaeras with USEARCH[28], and chimaeric sequences were also filtered. Operational taxonomic units (OTUs) were grouped by a de novo strategy against the Greengenes database with an identity > 97%. Microbiota taxonomy was assigned by RDP classifier. In each group, OTUs with reads of more than half the sample size of that group were kept. Good’s coverage index was used to detect the sequencing depth. The Chao1 index was calculated to evaluate the microbial diversity. Principal coordinate analysis (PCoA) was performed to determine the degree of dissimilarity among groups using the unweighted UniFrac method.
The microbial 16S rRNA sequencing data were used to make OTU tables via the closed reference OTU picking method. The microbial community metagenome prediction was done with Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt)[29]. OTU tables were normalized by 16S rRNA copy number, followed by metagenome predictions against Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology and compilation into KEGG pathways. Finally, linear discriminant analysis effect size (LEfSe)[30] on galaxy online (http://huttenhower.sph.harvard.edu/ galaxy)[31] was carried out to detect inner function differentials among groups. Linear discriminant analysis scores larger than 2 and P-values from the Kruskal-Wallis test less than 0.05 were considered significant.
Since the distribution of each clinical characteristic followed a normal distribution except of age, the P-value of each clinical characteristic was calculated by the chi-square test, while age was estimated by the rank sum test. A Kruskal-Wallis test with post hoc Dunn’s test was performed to compare relative abundances of taxa and microbial diversity among the three groups. To examine significant differences between the UC and control groups, a Wilcoxon rank-sum test with two-tailed distribution was performed. Differential taxa were identified with adjusted P-values < 0.05 (adjusted by the Bonferroni correction) for multiple comparisons, and a false discovery rate < 0.05 was considered significantly different.
Clinical characteristics of the 93 human subjects are shown in Table 1. Of these subjects, 63 were diagnosed with UC based on colonoscopy at LongHua Hospital, Shanghai University of Traditional Chinese Medicine. Among these volunteers, 31 UC patients were diagnosed with DCSR syndrome, 32 UC patients were diagnosed with PXSY syndrome, and 30 individuals had no specific digestive system diseases and were recruited as healthy controls. All volunteers provided informed consent before participating in the experiment. Among all the individuals, there was no significant difference in any clinical factor, such as gender, age, or inflammation location (P > 0.05) (Table 1). PCoA plots in Appendix Figure suggests that gender and age had no obvious influence on sample clustering.
| HC | DCSR | PXSY | |
| Gender | |||
| Male | 15 | 15 | 17 |
| Female | 15 | 16 | 15 |
| Age (mean ± SD), yr | 43.13 ± 10.98 | 44.16 ± 11.49 | 41.94 ± 13.42 |
| Duration of disease | |||
| < 1 yr | 4 | 4 | |
| 1-3 yr | 11 | 13 | |
| 3-5 yr | 12 | 11 | |
| > 5 yr | 4 | 4 | |
| Inflammation location | |||
| Rectum | 5 | 5 | |
| Left colon | 21 | 22 | |
| Whole colon | 5 | 5 | |
| Disease activity | |||
| Clinical remission | 7 | 9 | |
| Mild activity | 9 | 9 | |
| Moderate activity | 7 | 7 | |
| Severe activity | 8 | 7 | |
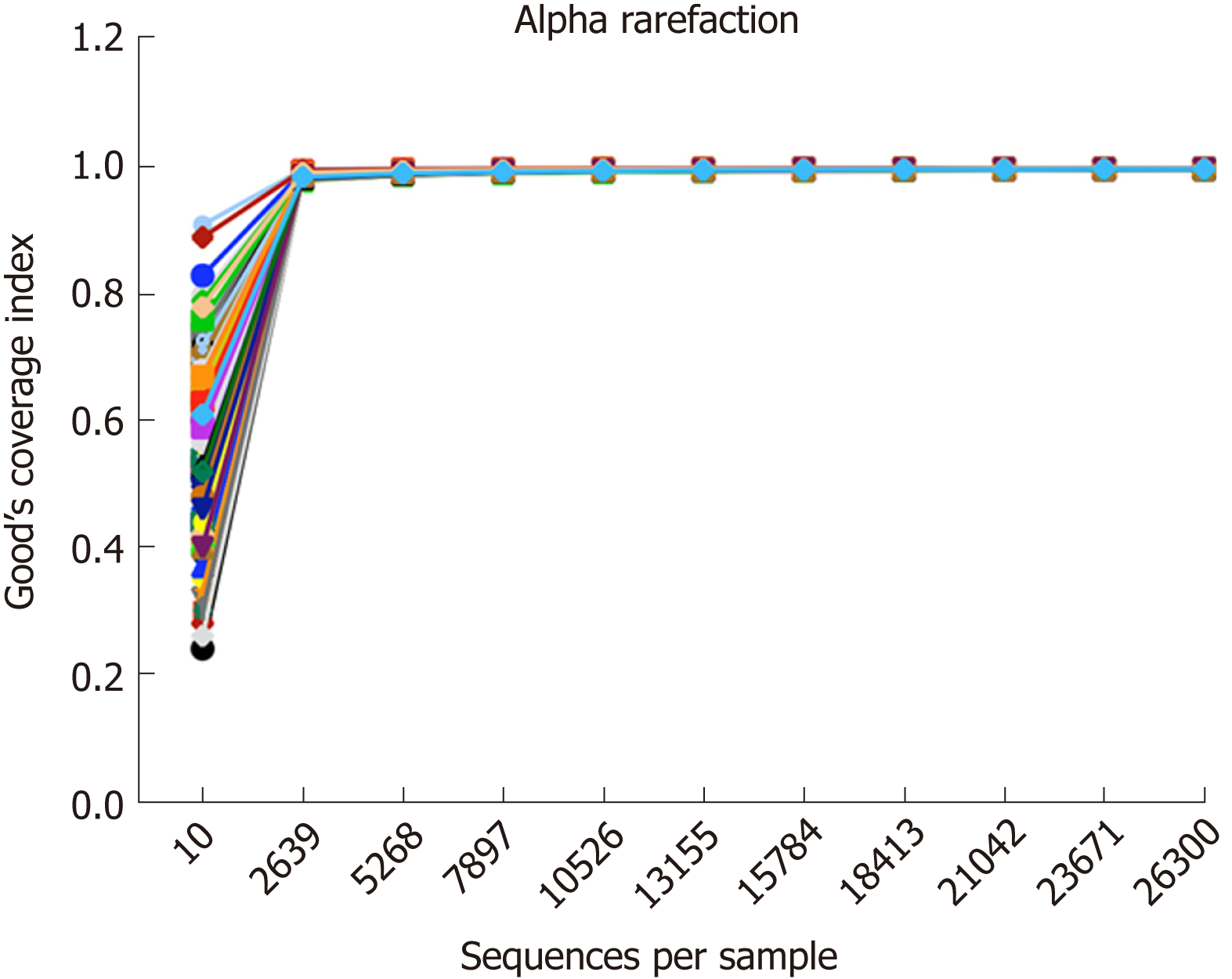
Gut microbiomes of 93 samples (Table 1) were analysed by 16S rRNA pyrosequencing, and a total of 3494198 sequences were obtained after quality control. Good’s coverage estimator was determined to measure the sequencing depth of each sample. The alpha rarefaction curve (Figure 1) of each sample reached at the platform extracted approximately 2700 sequences per sample, and Good’s coverage estimator of each group approached 100% (>98.9%), indicating that the sequencing depth was sufficient to reflect the majority of bacterial sequences in all samples.
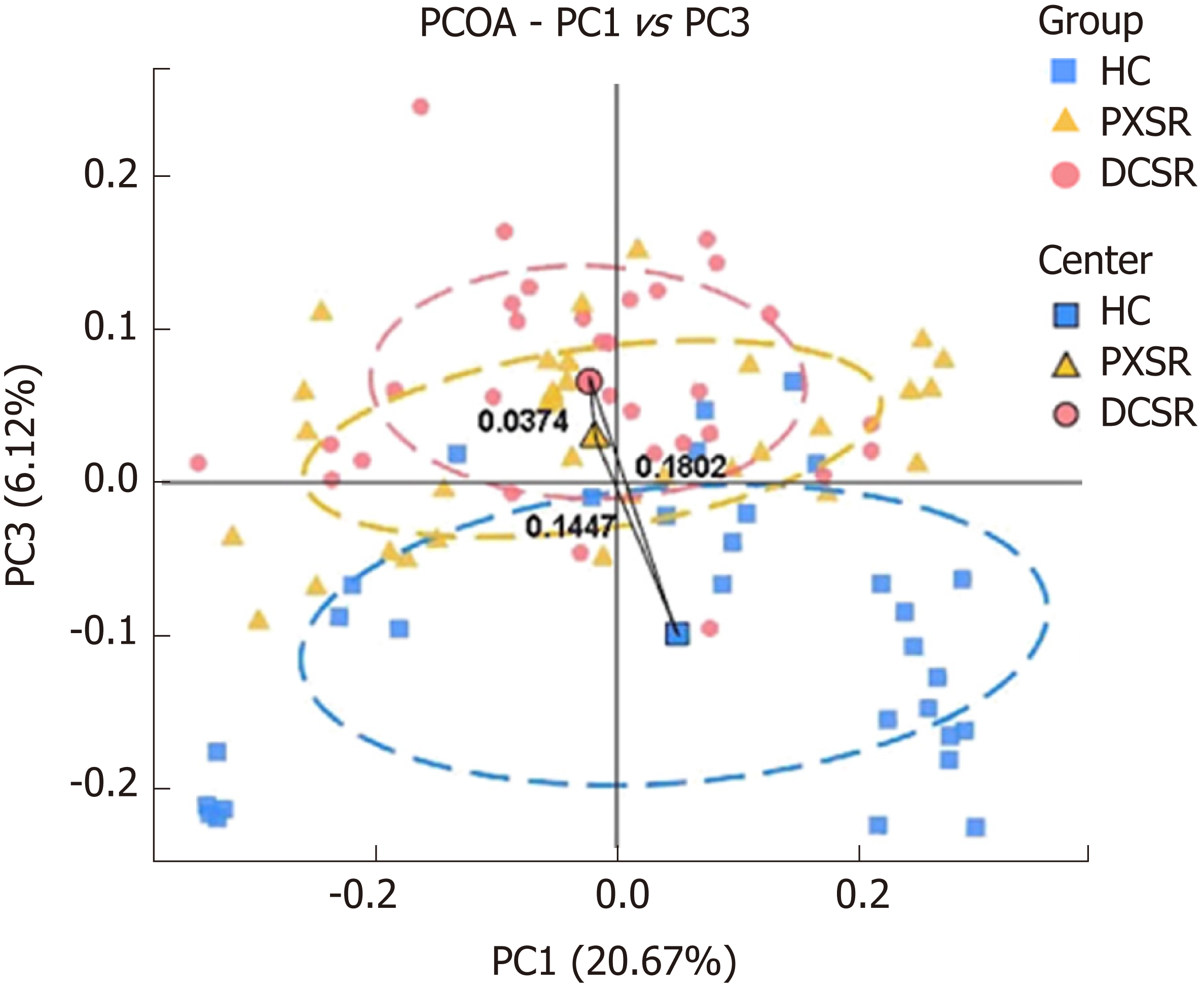
The beta diversities of the healthy control, PXSY syndrome, and DCSR syndrome cohorts were assessed by UniFrac analysis. The PCoA plot (Figure 2) showed that the majority of samples were clustered by health status but not by age or gender (Appendix Figure), reflecting that an association between health status and gut microbiomes existed. Moreover, the UC patients were clustered into two groups according to the two TMC syndromes, PXSY and DCSR, which indicated that the gut microbiota may contribute to distinguishing these two syndromes.
Non-parametric statistical testing methods were used to analyse the differential microbes between UC patients and healthy subjects. The microbes with an average abundance greater than 1% in any of the groups (Table 2) are as follows:
| HC | UC | PXSY | DCSR | UC-H FDR | P-H P1 | D-H P1 | D-P P1 | |
| Bacteroidetes | 47.02 | 41.3 | 49.09 | 33.26 | 0.25 | 1.00 | 0.03 | 0.01 |
| Paraprevotellaceae | 1.04 | 0.18 | 0.35 | 0.00 | 0.02 | 0.07 | 0.02 | 1.00 |
| Porphyromonadaceae | 4.8 | 1.86 | 2.55 | 1.15 | 0.01 | 0.04 | 0.01 | 1.00 |
| Parabacteroides | 4.79 | 1.83 | 2.55 | 1.09 | 0.01 | 0.05 | 0.00 | 1.00 |
| Rikenellaceae | 1.66 | 0.71 | 1.15 | 0.25 | 0.04 | 0.11 | 0.04 | 1.00 |
| Alistipes | 1.53 | 0.59 | 0.94 | 0.24 | 0.03 | 0.06 | 0.03 | 1.00 |
| Firmicutes | 43.74 | 48.23 | 41.1 | 55.59 | 0.38 | 1.00 | 0.06 | 0.01 |
| Enterococcaceae | 0.03 | 0.68 | 0.31 | 1.05 | 0.01 | 0.02 | 0.03 | 1.00 |
| Enterococcus | 0.03 | 0.68 | 0.31 | 1.05 | 0.02 | 0.02 | 0.03 | 1.00 |
| Streptococcaceae | 0.7 | 2.57 | 1.25 | 3.95 | 0.01 | 0.79 | 0.00 | 0.01 |
| Streptococcus | 0.67 | 2.54 | 1.23 | 3.89 | 0.01 | 0.58 | 0.00 | 0.01 |
| Lachnospiraceae | 24.18 | 15.78 | 13.91 | 17.71 | 0.01 | 0.01 | 0.03 | 1.00 |
| Uncultured | 2.99 | 0.94 | 0.5 | 1.39 | 0.00 | 0.00 | 0.02 | 0.38 |
| Dorea | 8.87 | 2.83 | 2.51 | 3.17 | 0.01 | 0.00 | 0.02 | 1.00 |
| Lachnoclostridium* | 1.16 | 1.04 | 1.65 | 0.41 | 0.2 | 0.01 | 1.00 | 0.01 |
| Unknown | 2.28 | 1.23 | 1.2 | 1.26 | 0.01 | 0.02 | 0.02 | 1.00 |
| Ruminococcaceae | 8.46 | 14.56 | 10.72 | 18.52 | 0.08 | 1.00 | 0.01 | 0.17 |
| Faecalibacterium | 4.24 | 10.75 | 6.48 | 15.15 | 0.00 | 0.11 | 0.00 | 0.09 |
| Oscillospira | 0.61 | 1.4 | 1.65 | 1.13 | 0.08 | 0.01 | 1.00 | 0.17 |
| Ruminococcus | 1.63 | 0.42 | 0.45 | 0.39 | 0.00 | 0.01 | 0.00 | 0.84 |
| Veillonellaceae | 6.4 | 8.44 | 10.24 | 6.58 | 0.37 | 0.6 | 1.00 | 1.00 |
| Dialister | 0.81 | 3.43 | 5.3 | 1.51 | 0.2 | 0.03 | 1.00 | 0.05 |
| Phascolarctobacterium | 1.13 | 0.68 | 0.5 | 0.86 | 0.04 | 0.01 | 0.26 | 0.61 |
| Proteobacteria | 7.1 | 8.6 | 7.61 | 9.63 | 0.63 | 1.00 | 1.00 | 1.00 |
| Alcaligenaceae | 1.77 | 1.29 | 0.95 | 1.64 | 0.04 | 0.02 | 0.19 | 1.00 |
| Sutterella | 1.77 | 1.29 | 0.95 | 1.64 | 0.05 | 0.02 | 0.19 | 1.00 |
| Enterobacteriaceae | 4.47 | 5.97 | 5.61 | 6.34 | 0.76 | 1.00 | 1.00 | 1.00 |
| Unknown | 0.14 | 0.62 | 0.22 | 1.03 | 0.06 | 0.38 | 0.02 | 0.65 |
At the family level, the relative abundance of Paraprevotellaceae, Porphy-romonadaceae, Rikenellaceae, and Lachnospiraceae decreased in UC patients compared to healthy individuals, while Enterococcaceae and Streptococcaceae were significantly abundant in UC patients. At the genus level, the abundance of Para-bacteroides, Alistipes, Ruminococcus, Phascolarctobacterium, Dorea, Sutterella, and an uncultured genus belonging to the family Lachnospiraceae decreased in the UC group compared to healthy controls. On the other hand, the genera Enterococcus, Streptococcus, and Faecalibacterium were more significantly abundant in UC patients than in the healthy cohort. Notably, the families Porphyromonadaceae and Lachno-spiraceae exhibited a similar depleted trend in UC patients, as some studies have previously reported[32-34]. It has also been reported that the genera Enterococcus and Streptococcus were differentially abundant in UC, while Ruminococcus was also significant in the healthy cohort[24,35].
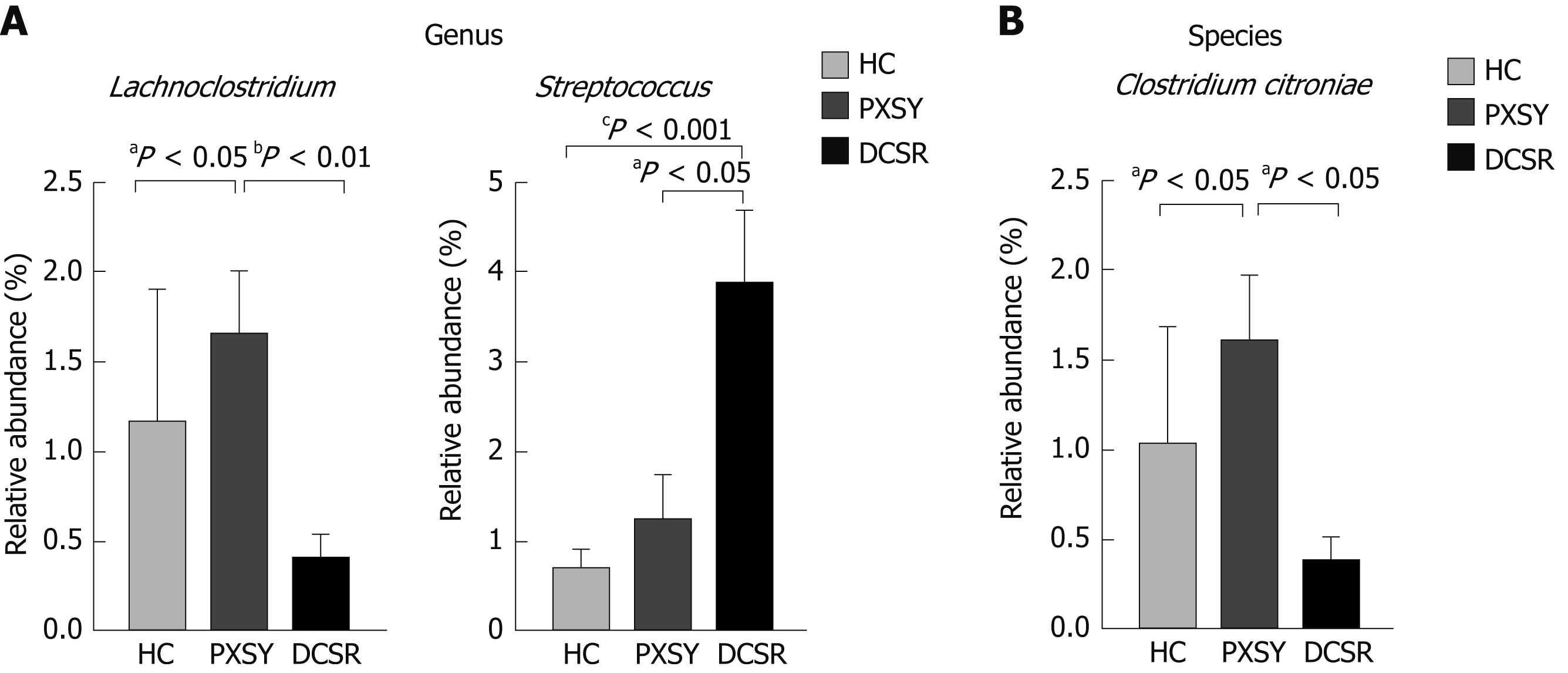
Intestinal microbiomes that were significantly different between PXSY and DCSR as determined by the Kruskal-Wallis test are highlighted in Table 2. Bacteroides and Firmicutes were the dominant phyla in these samples, and differed significantly between the PXSY and DCSR groups. Firmicutes was significantly abundant in the DCSR cohort, while Bacteroides was differentially abundant in the PXSY cohort. The differential taxa at the family and genus levels are shown in Figure 3. Within the phylum Firmicutes, the family Streptococcaceae was significantly different between DCSR syndrome and PXSY syndrome patients. At the genus level, Streptococcus and Lachnoclostridium were significantly different between DCSR and PXSY patients. Known as a lactate producer, Streptococcus was more abundant in DCSR patients than in PXSY patients and healthy controls, indicating that DCSR patients might have more lactate in the gut than PXSY patients and controls. Lachnoclostridium is more abundant in PXSY patients than in DCSR patients and healthy controls. The species Clostridium citroniae, which belongs to the genus Lachnoclostridium, also exhibited the same tendency.
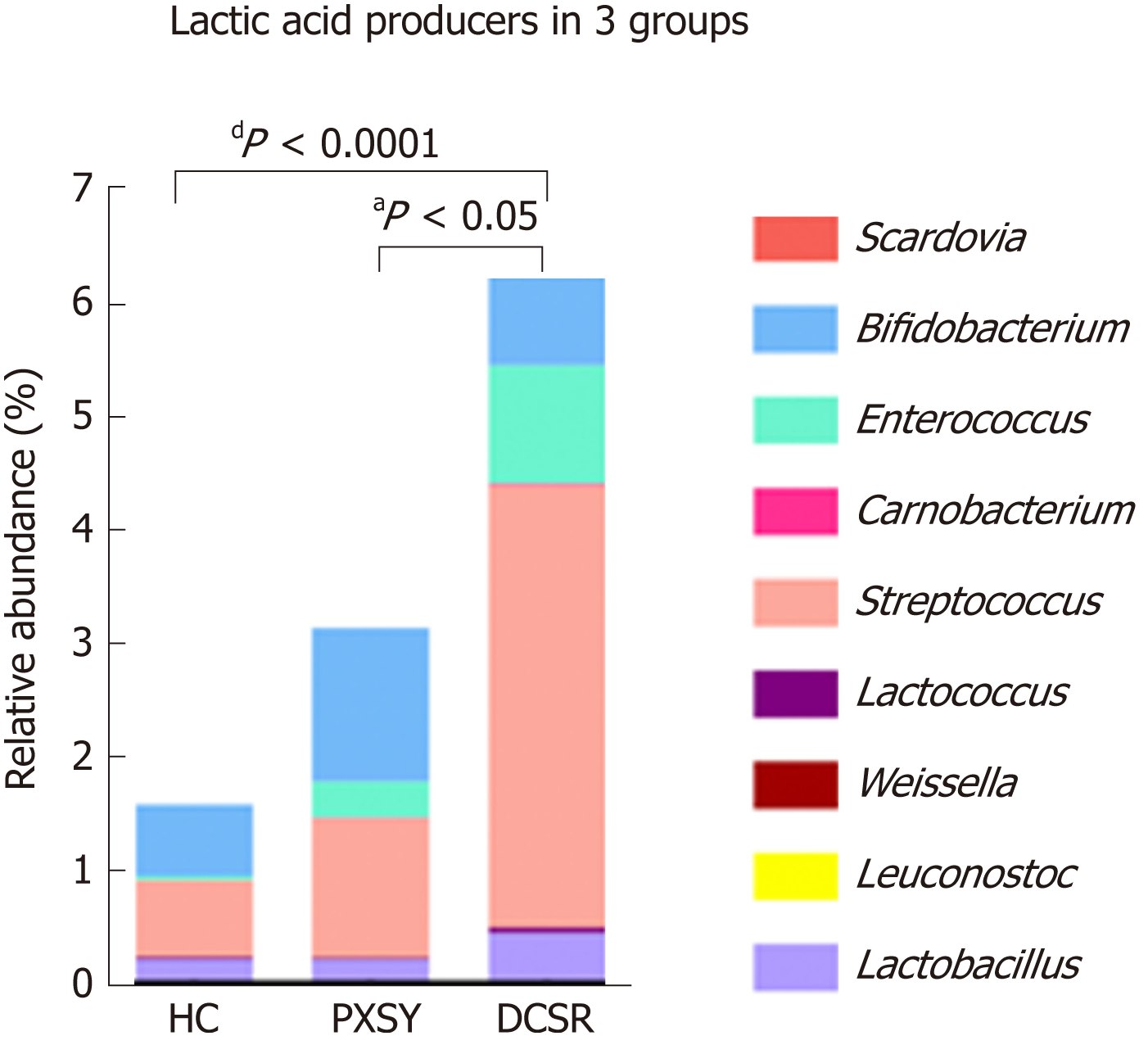
The lactate producer Streptococcus was increased in the DCSR group, which led us to explore the lactate-producing taxa in PXSY and DCSR syndromes. The relative abundance of the main lactate producers in the gut is shown in Figure 4. All of the main lactate producers showed a trend of increased relative abundance. Compared to healthy controls and PXSY patients, DCSR syndrome patients had significantly abundant lactic acid bacteria, suggesting that more lactic acid might accumulate in the guts of DCSR patients than in the guts of PXSY patients.
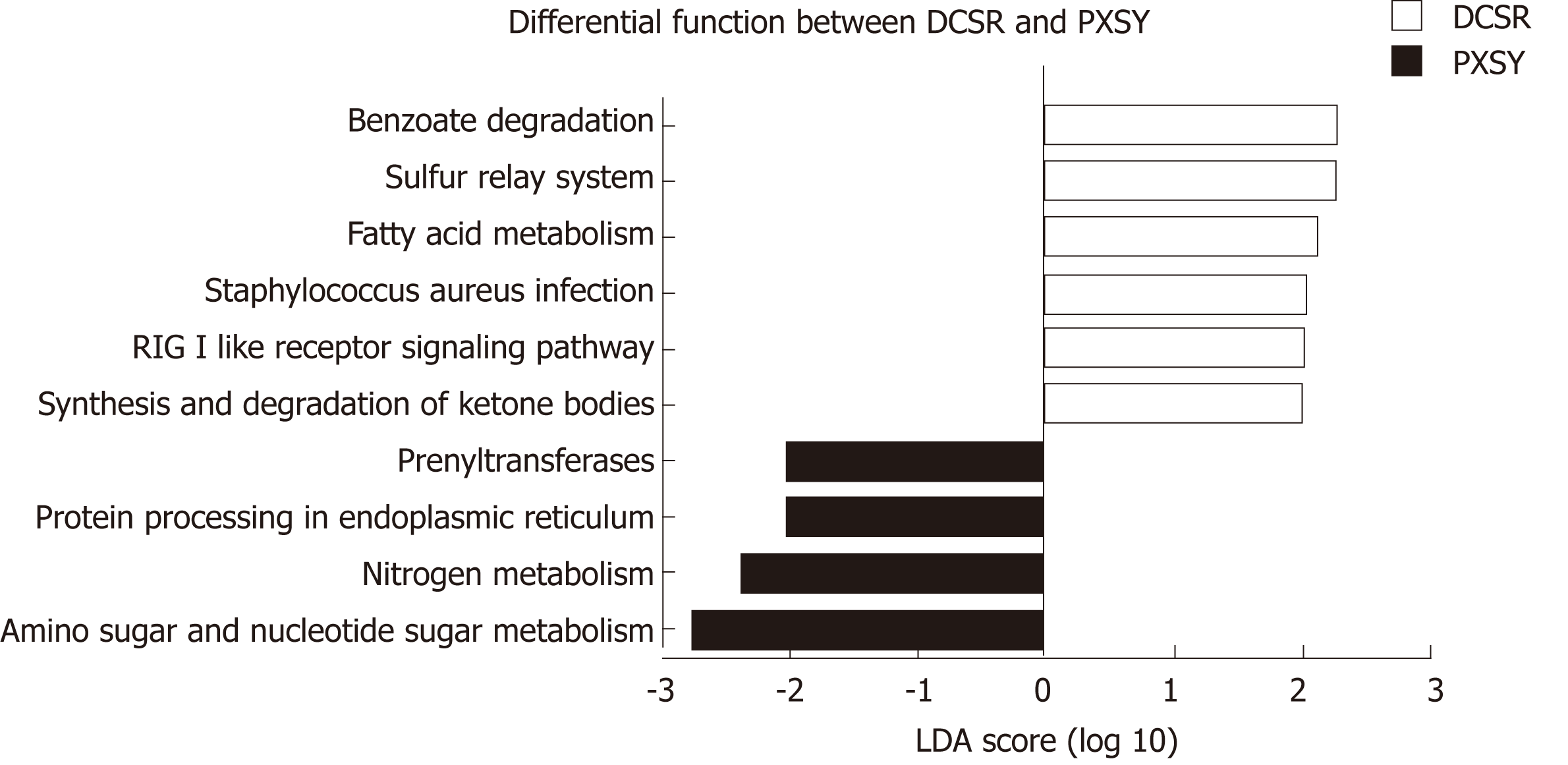
The gut microbiome composition of all groups was determined based on 16S rRNA gene sequencing data. KEGG pathways were categorized using PICRUSt, and ten differential functions were identified with LEfSe. The functional pathways of gut microbiomes between the PXSY and DCSR groups are shown in Figure 5. After differential function analysis, the functions related to liquid metabolism, such as synthesis and degradation of ketone bodies and fatty acid metabolism, were more significant in DCSR patients. Moreover, the RIG-I-like receptor (RLR) signalling pathway and benzoate degradation were also more abundant in DCSR patients. In the PXSY cohort, the genetic information processing functions of protein processing in the endoplasmic reticulum and the carbohydrate metabolism (amino sugar and nucleotide sugar metabolism) were abundant. In addition, nitrogen metabolism and prenyltransferase functions were abundant in the PXSY cohort.
The gut microbiota is closely associated with the host, and its dysregulation could lead to diseases and inflammation[36-39], such as IBD. Scaldaferri et al[40] found that the most severe inflammation was concentrated in the gut of IBD patients at the site with the largest number of microbes, suggesting that changes in the gut microbiota are closely related to intestinal inflammation and that the microbiota affects the host through the immune system[41]. This study is pilot research about the contribution of the gut microbiota to the distinction between the two TCM syndromes of UC and the corresponding molecular mechanism.
According to the TCM theory, PXSY syndrome is a deficiency syndrome, while DCSR syndrome is a sthenia syndrome. Clinical manifestations of the DCSR syndrome focus on the empirical symptoms of intestinal inflammation and injury, while the clinical manifestations of PXSY syndrome focus on the deficiency symptoms of weakened digestion and absorption function. The main clinical symptoms of DCSR syndrome are abdominal pain, diarrhoea, mucous purulent stool, yellowish fur, and thready and slippery pulse. Some of these patients can also show symptoms such as burning pain in the anus, tenesmus, body heat, short red urine, dry or bitter mouth, and ozostomia. The main clinical manifestations in patients with PXSY syndrome include thin sloppy stool, stool mucus that is white rather than red, white frozen, pink tongue, tooth mark around the tongue, and white and greasy tongue coating. Some of these patients may have the symptoms such as dull abdominal pain, abdominal fullness and distention, poor appetite, tiredness, thready and weak pulse, and thready and slippery pulse.
The microbiota in the guts of the healthy cohort and UC patients was investigated in this study, which showed that the richness and diversity of the gut microbiota were different between patients and healthy controls. We found that the intestinal microflora was different between the PXSY and DCSR groups, which is essential for distinguishing the two TCM syndromes of UC and potentially contributes to personalized medicine.
Using comparisons among PXSY patients, DCSR patients, and healthy subjects, our study illustrated that the genus Streptococcus significantly increased in UC patients, especially in patients with DCSR syndrome. The excessive growth of Streptococcus in DCSR syndrome patients might indicate the pathological mechanisms of DCSR syndrome.
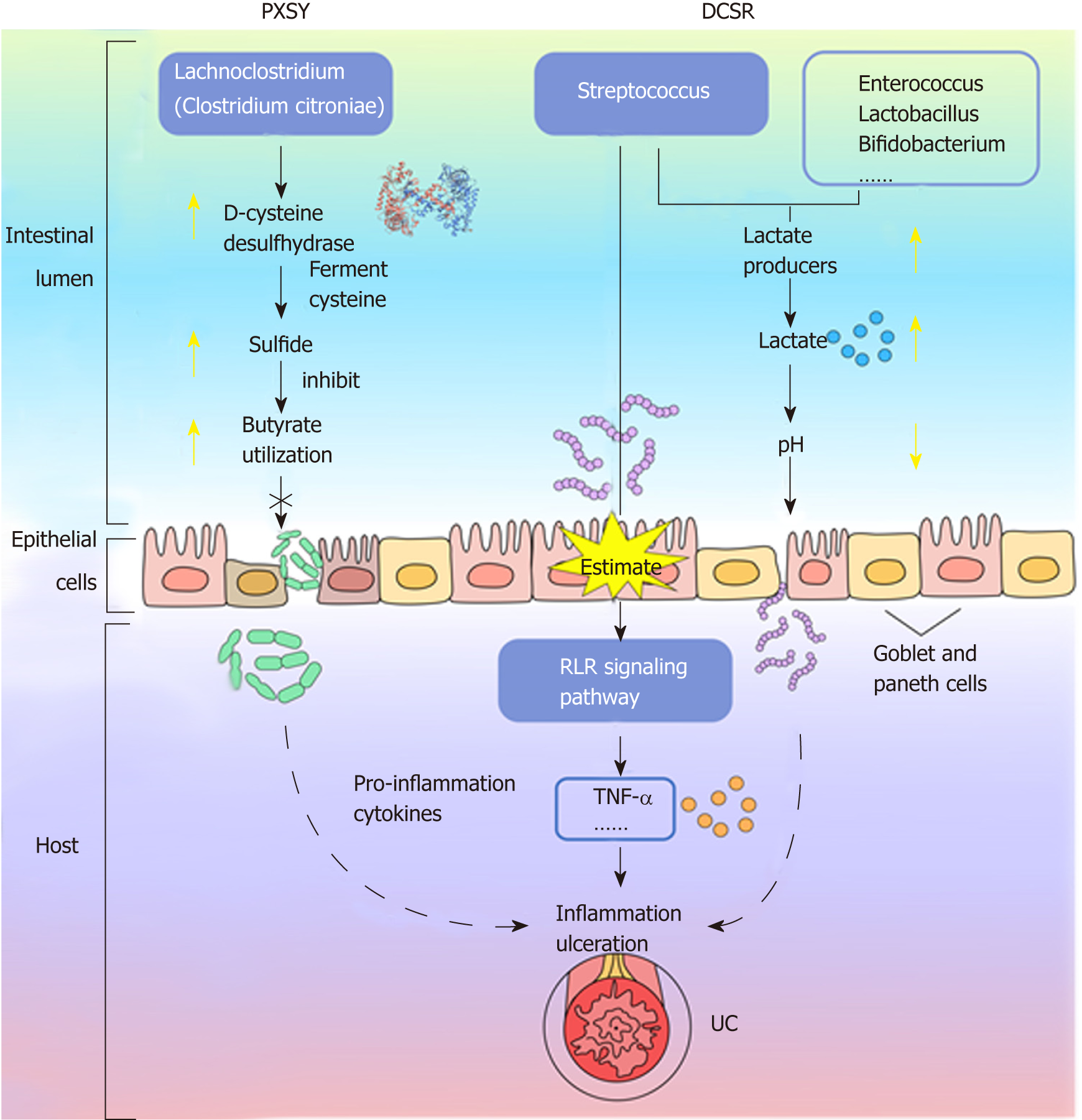
Dysbiosis of gut microbiota stimulates the inflammatory response of the host and even induces serious complications[42-44]. The RLR signalling pathway is associated with inflammation[45-47]. Here, Streptococcus, together with other pathogens in the gut, could stimulate RLRs and produce pro-inflammatory cytokines such as TNF-α via the RLR signalling pathway to promote inflammation (Figure 6). However, the continuous stimulation of the RLR signalling pathway results in an excessive inflammatory response. This excessive inflammatory response leaves the intestinal barrier function impaired, and then more microorganisms in the intestinal lumen will pass through an already incomplete epithelial barrier. Finally, these microbiotas will cause inflammation and damage to the intestinal epithelium layer.
The majority of gut microbiota can ferment carbohydrates to produce short-chain fatty acids (SCFAs), lactate, and some inorganic compounds. Some of these products are beneficial to the health of the host, but some harm the intestinal tract, such as by causing damage to the intestinal epithelial barrier or promoting bowel inflammation.
Lactate can be produced by some microbes such as Streptococcus in the intestine. The lactate producers can ferment carbohydrates to produce lactic acid, and the accumulation of lactic acid can reduce the pH of the intestinal tract. It has been reported that the over-accumulation of lactate in the intestine of patients leads to a decrease in the pH[48-49]. In disease status, disorder of the gut microbiota appears, and blooms of lactate producers are significant in DCSR syndrome. Streptococcus in the presence of other lactate producers (Enterococcus, Lactobacillus, Bifidobacterium and so on)[50-54] can produce lactate. Nevertheless, the utilization rate of lactate by other microbes is decreased in disease status, which causes the over-accumulation of lactic acid in the gut so that the intestinal pH is reduced and the distribution and abundance of intestinal microbiota are changed. Additionally, the accumulation of lactate can stimulate the intestinal tract and aggravate inflammation.
This study showed that the abundance of Lachnoclostridium in the PXSY group was higher than that in the DCSR group. Members of Lachnoclostridium ferment some monosaccharides and disaccharides to acetate as the major SCFA products[55]. Within the genus Lachnoclostridium, the differential microbes between DCSR and PXSY patients, the main differential species is Clostridium citroniae, which also increased in DCSR patients. Clostridium citroniae contains D-cysteine desulfatase[56] and can ferment cysteine to sulfide, pyruvate, and ammonia as end products[57], and more Clostridium citroniae in the PXSY group suggested that more D-cysteine desulfatase might be present in the intestine of spleen-deficient PXSY patients than in DCSR patients.
The carbohydrate fermentation conducted by bacteria in the colon gives rise to the luminal SCFAs, including acetate, propionate, and butyrate. Among these SCFAs, but-yrate is considered to be the preferred source of nutrition and energy for colonic epithelial cells and is, to a major extent, metabolized by these cells[58]. In addition, butyrate is considered to have anti-inflammatory capacity, and the increase in sulfide may inhibit the oxidation of butyrate, which could damage the epithelial barrier and even result in more severe inflammation.
The gut microbiota could also activate a variety of pathogen-associated molecular patterns on the surface of intestinal epithelial cells and downstream signalling pathways, inducing epithelial cells to secrete anti-inflammatory substances[59,60]. However, the molecular and metabolic functions of most bacteria are still unknown[60].
Clostridium citroniae contains D-cysteine desulfhydrase, which can ferment cysteine to produce sulfides. Here, Clostridium citroniae was more abundant in PXSY patients than in DCSR patients and healthy subjects. While in patients with PXSY status, increased Clostridium citroniae results in increased D-cysteine desulfhydrase, which results in an increase in sulfides. Finally, sulfides inhibit the oxidization of butyric acid so that the epithelial cells cannot utilize the butyrate in the gut and cause epithelial cell apoptosis. Damage to colonic epithelial cells is reflected in the incomplete intestinal barrier, and gut microbes pass through the intestinal barrier to induce severe inflammation and ulceration. In brief, Clostridium citroniae might be detrimental to the health of the intestinal tract by inhibiting the utilization of butyrate.
PXSY syndrome reflects a kind of deficiency in immune activity, while DCSR syndrome is a kind of dysfunction in gut immunity. These differential trends in the microbiota between the two TCM syndromes of UC indicated that different effects were exerted on the mucosal immune activity and the epithelial barrier function by themselves. The genus Streptococcus, which could lead to inflammation, was significantly more abundant in DCSR patients. It is suggested that Streptococcus and other microbes in the gut might have a certain relationship with DCSR syndrome symptoms such as diarrhoea mixed with blood. These differential microbes are expected to be the markers and therapeutic targets for the clinical precision detection of UC.
There were several weaknesses in the study, like small sample size, non-blinding, and cross-sectional nature. In the following studies, the sample size needs to be further expanded to further strengthen and enrich the accuracy and stability of the research results. This study is a cross-sectional study, and it is difficult to determine the causal relationship between the results. In future studies, the intervention factors can be considered and the dynamic observation before and after the intervention can be carried out.
In conclusion, the gut microbiota is different between patients with PXSY syndrome and those with DCSR syndrome. The genus Streptococcus is significantly more abundant in DCSR patients than in PXSY patients, while Lachnoclostridium increases in PXSY patients. Dysbiosis of the gut microbiota influences the immune responses of the host, which results in inflammation, and the microbial analysis of the two TCM syndromes essentially reflects different immune activities in the human body, but they all point to promotion of inflammation in the gut.
Ulcerative colitis (UC) is considered to be closely associated with alteration of intestinal microorganisms. According to the traditional Chinese medicine (TCM) theory, UC can be divided into Pi-Xu-Shi-Yun syndrome (syndrome of spleen deficiency and dampness, PXSY) and Da-Chang-Shi-Re syndrome (syndrome of dampness-heat in the large intestine, DCSR). PXSY syndrome is a deficiency syndrome, while the DCSR syndrome is a sthenia syndrome. However, the relationships among gut microbiota, TCM syndromes, and UC pathogenesis are unclear.
The majority of studies have focused on the relationship between intestinal microbiota and the onset of UC, and the contribution of gut microbiota in these two distinct TCM syndromes is still unclarified. This study aimed to compare the difference in microbial composition and function between PXSY and DCSR syndromes to determine the molecular mechanism of TCM in UC by investigation of the gut microbiota.
The objective of this study was to investigate the role of gut microbiota in UC and the distinction of microbiota dysbiosis between PXSY and DCSR syndromes.
We analysed gut microbiome composition of stool samples by 16S rRNA pyrosequencing. We assessed the beta diversity by UniFrac analysis. We also processed the high-throughput se-quencing reads with QIIME, and further predicted biological functions using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States.
We determined the composition of gut bacterial communities in 93 stool samples (30 healthy controls, 32 patients with PXSY syndrome, and 31 patients with DCSR syndrome) by 16S rRNA pyrosequencing. Beta diversity showed that the composition of the microbiota was different among the three groups. We found that Porphyromonadaceae, Rikeneliaceae, and Lach-nospiraceae significantly decreased while Enterococcus, Streptococcus, and other potential pathogens significantly increased in UC patients compared to healthy subjects at the family level. We further found that Parabacteroides, Dorea, and Ruminococcus decreased while Faecalibacterium showed increased abundance in UC compared to healthy controls at the genus level. Five differential taxa were identified between PXSY and DCSR syndromes. We observed a significantly increased abundance of Streptococcus in DCSR patients at the genus level, while Lachnoclostridium increased in PXSY patients. Additionally, we found that the differential fun-ctional pathways of the gut microbiome between the PXSY and DCSR groups mainly included lipid metabolism, immunity, and the metabolism of polypeptides.
The present study identified that the gut microbiota is different between patients with PXSY syndrome and those with DCSR syndrome. The genus Streptococcus is significantly more abundant in DCSR patients than in PXSY patients, while Lachnoclostridium increases in PXSY patients. The microbial analysis of the two TCM syndromes essentially reflects different immune activities in the human body, but they all point to promotion of inflammation in the gut.
The relationship between TCM syndromes and intestinal flora is an interesting and important research topic. Our study preliminarily explored the characteristics and differences of intestinal flora of patients with two different TCM syndrome. Further studies are required to confirm our findings and to clarify the precise mechanism of intestinal flora characteristics related to TCM deficiency and positive syndrome.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Day AS, Madnani MA, Serban ED, Tsujikawa T, Vasudevan A S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3789] [Cited by in RCA: 3526] [Article Influence: 271.2] [Reference Citation Analysis (5)] |
| 2. | Ng SC. Epidemiology of inflammatory bowel disease: Focus on Asia. Best Pract Res Clin Gastroenterol. 2014;28:363-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 3. | Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 771] [Cited by in RCA: 895] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 4. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet. 2018;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4097] [Article Influence: 512.1] [Reference Citation Analysis (110)] |
| 5. | Kaplan GG. The global burden of IBD: From 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1157] [Cited by in RCA: 1868] [Article Influence: 186.8] [Reference Citation Analysis (1)] |
| 6. | Ng SC, Tang W, Ching JY, Wong M, Chow CM, Hui AJ, Wong TC, Leung VK, Tsang SW, Yu HH, Li MF, Ng KK, Kamm MA, Studd C, Bell S, Leong R, de Silva HJ, Kasturiratne A, Mufeena MNF, Ling KL, Ooi CJ, Tan PS, Ong D, Goh KL, Hilmi I, Pisespongsa P, Manatsathit S, Rerknimitr R, Aniwan S, Wang YF, Ouyang Q, Zeng Z, Zhu Z, Chen MH, Hu PJ, Wu K, Wang X, Simadibrata M, Abdullah M, Wu JC, Sung JJY, Chan FKL; Asia–Pacific Crohn's and Colitis Epidemiologic Study (ACCESS) Study Group. Incidence and phenotype of inflammatory bowel disease based on results from the Asia-pacific Crohn's and colitis epidemiology study. Gastroenterology. 2013;145:158-165.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 597] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 7. | Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: A meta-analysis. Gut. 2001;48:526-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1985] [Cited by in RCA: 2077] [Article Influence: 86.5] [Reference Citation Analysis (1)] |
| 8. | Desai D, Shah S, Deshmukh A, Abraham P, Joshi A, Gupta T, Deshpande R, Khandagale V, George S. Colorectal cancers in ulcerative colitis from a low-prevalence area for colon cancer. World J Gastroenterol. 2015;21:3644-3649. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 9. | Lu YH, Cong LL. [Study on the Chinese medical syndrome distribution of ulcerative colitis]. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2012;32:450-454. [PubMed] |
| 10. | Chen Z, Wang P. Clinical Distribution and Molecular Basis of Traditional Chinese Medicine ZHENG in Cancer. Evid Based Complement Alternat Med. 2012;2012:783923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 11. | Dai YC, Zhang YL, Wang LJ, Guo Q, Yang K, Ye RH, Tang ZP. Clinical presentation and treatment strategies for ulcerative colitis: A retrospective study of 247 inpatients. Chin J Integr Med. 2016;22:811-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Dai YC, Zheng L, Zhang YL, Chen X, Chen DL, Tang ZP. Effects of Jianpi Qingchang decoction on the quality of life of patients with ulcerative colitis: A randomized controlled trial. Medicine (Baltimore). 2017;96:e6651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Zheng L, Zhang YL, Dai YC, Chen X, Chen DL, Dai YT, Tang ZP. Jianpi Qingchang decoction alleviates ulcerative colitis by inhibiting nuclear factor-κB activation. World J Gastroenterol. 2017;23:1180-1188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 42] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 14. | Lin X, Yi Z, Diao J, Shao M, Zhao L, Cai H, Fan Q, Yao X, Sun X. ShaoYao decoction ameliorates colitis-associated colorectal cancer by downregulating proinflammatory cytokines and promoting epithelial-mesenchymal transition. J Transl Med. 2014;12:105. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 15. | Liu DY, Guan YM, Zhao HM, Yan DM, Tong WT, Wan PT, Zhu WF, Liu HN, Liang XL. The protective and healing effects of Si Shen Wan in trinitrobenzene sulphonic acid-induced colitis. J Ethnopharmacol. 2012;143:435-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Zou Y, Lin J, Li W, Wu Z, He Z, Huang G, Wang J, Ye C, Cheng X, Ding C, Zheng X, Chi H. Huangqin-tang ameliorates dextran sodium sulphate-induced colitis by regulating intestinal epithelial cell homeostasis, inflammation and immune response. Sci Rep. 2016;6:39299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 17. | Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, Ott S, Kupcinskas L, Doré J, Raedler A, Schreiber S. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology. 2011;141:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 452] [Article Influence: 32.3] [Reference Citation Analysis (1)] |
| 18. | Becker C, Neurath MF, Wirtz S. The Intestinal Microbiota in Inflammatory Bowel Disease. ILAR J. 2015;56:192-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 137] [Article Influence: 15.2] [Reference Citation Analysis (1)] |
| 19. | Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3922] [Cited by in RCA: 3462] [Article Influence: 216.4] [Reference Citation Analysis (0)] |
| 20. | Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17:577-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 225] [Article Influence: 22.5] [Reference Citation Analysis (1)] |
| 21. | Stephani J, Radulovic K, Niess JH. Gut microbiota, probiotics and inflammatory bowel disease. Arch Immunol Ther Exp (Warsz). 2011;59:161-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Cucchiara S, Stronati L, Aloi M. Interactions between intestinal microbiota and innate immune system in pediatric inflammatory bowel disease. J Clin Gastroenterol. 2012;46 Suppl:S64-S66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Guo M, Ding S, Zhao C, Gu X, He X, Huang K, Luo Y, Liang Z, Tian H, Xu W. Red Ginseng and Semen Coicis can improve the structure of gut microbiota and relieve the symptoms of ulcerative colitis. J Ethnopharmacol. 2015;162:7-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Nishikawa J, Kudo T, Sakata S, Benno Y, Sugiyama T. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009;44:180-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 25. | Branch of Gastrointestinal Diseases. China Association of Chinese Medicine. [Consensus on Chinese medical diagnosis and treatment of ulcerative colitis (2009)]. Zhongguo Zhong Xi Yi Jie He Za Zhi. 2010;30:527-532. [PubMed] |
| 26. | Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-2120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30322] [Cited by in RCA: 41063] [Article Influence: 3733.0] [Reference Citation Analysis (1)] |
| 27. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23733] [Article Influence: 1582.2] [Reference Citation Analysis (0)] |
| 28. | Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194-2200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9953] [Cited by in RCA: 9743] [Article Influence: 695.9] [Reference Citation Analysis (0)] |
| 29. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5694] [Cited by in RCA: 6297] [Article Influence: 524.8] [Reference Citation Analysis (0)] |
| 30. | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11842] [Cited by in RCA: 10307] [Article Influence: 736.2] [Reference Citation Analysis (0)] |
| 31. | Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3-W10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1416] [Cited by in RCA: 1311] [Article Influence: 145.7] [Reference Citation Analysis (0)] |
| 32. | Lavelle A, Lennon G, O'Sullivan O, Docherty N, Balfe A, Maguire A, Mulcahy HE, Doherty G, O'Donoghue D, Hyland J, Ross RP, Coffey JC, Sheahan K, Cotter PD, Shanahan F, Winter DC, O'Connell PR. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut. 2015;64:1553-1561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 210] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 33. | Mar JS, LaMere BJ, Lin DL, Levan S, Nazareth M, Mahadevan U, Lynch SV. Disease Severity and Immune Activity Relate to Distinct Interkingdom Gut Microbiome States in Ethnically Distinct Ulcerative Colitis Patients. MBio. 2016;7:pii: e01072-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 79] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 34. | Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780-13785. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3731] [Cited by in RCA: 3430] [Article Influence: 190.6] [Reference Citation Analysis (1)] |
| 35. | Ohkusa T, Koido S. Intestinal microbiota and ulcerative colitis. J Infect Chemother. 2015;21:761-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 36. | Jiao N, Baker SS, Nugent CA, Tsompana M, Cai L, Wang Y, Buck MJ, Genco RJ, Baker RD, Zhu R, Zhu L. Gut microbiome may contribute to insulin resistance and systemic inflammation in obese rodents: A meta-analysis. Physiol Genomics. 2018;50:244-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 206] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 37. | Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ, Zhu R, Zhu L. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67:1881-1891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 514] [Article Influence: 73.4] [Reference Citation Analysis (1)] |
| 38. | Zhu L, Liu W, Alkhouri R, Baker RD, Bard JE, Quigley EM, Baker SS. Structural changes in the gut microbiome of constipated patients. Physiol Genomics. 2014;46:679-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 266] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 39. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1279] [Article Influence: 106.6] [Reference Citation Analysis (1)] |
| 40. | Scaldaferri F, Gerardi V, Lopetuso LR, Del Zompo F, Mangiola F, Boškoski I, Bruno G, Petito V, Laterza L, Cammarota G, Gaetani E, Sgambato A, Gasbarrini A. Gut microbial flora, prebiotics, and probiotics in IBD: Their current usage and utility. Biomed Res Int. 2013;2013:435268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 41. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3003] [Cited by in RCA: 2986] [Article Influence: 229.7] [Reference Citation Analysis (0)] |
| 42. | Tahara T, Arisawa T. Pathogenesis of CpG island methylator phenotype-positive colorectal cancers: Role of genetic alteration and colonic flora. Epigenomics. 2014;6:249-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 43. | Cui DJ, Yang XL, Hu M, Yang LC. Effects of Preoperative Methotrexate on Complications After Surgery for Inflammatory Bowel Disease. Inflamm Bowel Dis. 2017;23:E2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 44. | Van Der Sloot KW, Joshi AD, Bellavance DR, Gilpin KK, Stewart KO, Lochhead P, Garber JJ, Giallourakis C, Yajnik V, Ananthakrishnan AN, Alizadeh BZ, Xavier RJ, Khalili H. Visceral Adiposity, Genetic Susceptibility, and Risk of Complications Among Individuals with Crohn's Disease. Inflamm Bowel Dis. 2017;23:82-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 57] [Article Influence: 7.1] [Reference Citation Analysis (1)] |
| 45. | Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 415] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 46. | Errett JS, Gale M. Emerging complexity and new roles for the RIG-I-like receptors in innate antiviral immunity. Virol Sin. 2015;30:163-173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 47. | Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: Sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 48. | Vernia P, Caprilli R, Latella G, Barbetti F, Magliocca FM, Cittadini M. Fecal lactate and ulcerative colitis. Gastroenterology. 1988;95:1564-1568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 184] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 49. | Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: A metabolic key player in cancer. Cancer Res. 2011;71:6921-6925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 819] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 50. | Le Blay G, Michel C, Blottière HM, Cherbut C. Prolonged intake of fructo-oligosaccharides induces a short-term elevation of lactic acid-producing bacteria and a persistent increase in cecal butyrate in rats. J Nutr. 1999;129:2231-2235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 143] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 51. | Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol. 2007;73:6526-6533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 163] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 52. | de Matos BM, Brighenti FL, Do T, Beighton D, Koga-Ito CY. Acidogenicity of dual-species biofilms of bifidobacteria and Streptococcus mutans. Clin Oral Investig. 2017;21:1769-1776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 53. | Belenguer A, Holtrop G, Duncan SH, Anderson SE, Calder AG, Flint HJ, Lobley GE. Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol Ecol. 2011;77:107-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 54. | Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15:2631-2641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 429] [Cited by in RCA: 323] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 55. | Feng Y, Stams AJM, de Vos WM, Sánchez-Andrea I. Enrichment of sulfidogenic bacteria from the human intestinal tract. FEMS Microbiol Lett. 2017;364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 56. | Nagasawa T, Ishii T, Kumagai H, Yamada H. D-Cysteine desulfhydrase of Escherichia coli. Purification and characterization. Eur J Biochem. 1985;153:541-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 57. | Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: A study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13:2826-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 58. | Cawich SO, Harnarayan P, Budhooram S, Naraynsingh V. Axillary Artery Injury Accompanying Humeral Neck Fracture. Int J Angiol. 2015;24:296-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 59. | Gupta AK, Harris JD, Erickson BJ, Abrams GD, Bruce B, McCormick F, Nicholson GP, Romeo AA. Surgical management of complex proximal humerus fractures-a systematic review of 92 studies including 4500 patients. J Orthop Trauma. 2015;29:54-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 152] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 60. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J; MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7836] [Article Influence: 522.4] [Reference Citation Analysis (4)] |