Published online May 14, 2019. doi: 10.3748/wjg.v25.i18.2204
Peer-review started: January 7, 2019
First decision: March 13, 2019
Revised: March 25, 2019
Accepted: April 10, 2019
Article in press: April 10, 2019
Published online: May 14, 2019
Processing time: 129 Days and 1.3 Hours
The dysbiosis of the gut microbiome is evident in Crohn’s disease (CD) compared with healthy controls (HC), although the alterations from active CD to remission after treatment are unclear.
To characterize the mucosa-associated gut microbiota in patients with CD before and after the induction therapy.
The basic information was collected from the subjects and the CD activity index (CDAI) was calculated in patients. A 16S rRNA sequencing approach was applied to determine the structures of microbial communities in mucosal samples including the terminal ileal, ascending colon, descending colon and rectum. The composition and function of mucosa-associated gut microbiota were compared between samples from the same cohort of patients before and after treatment. Differential taxa were identified to calculate the microbial dysbiosis index (MDI) and the correlation between MDI and CDAI was analyzed using Pearson correlation test. Predictive functional profiling of microbial communities was obtained with PICRUSt.
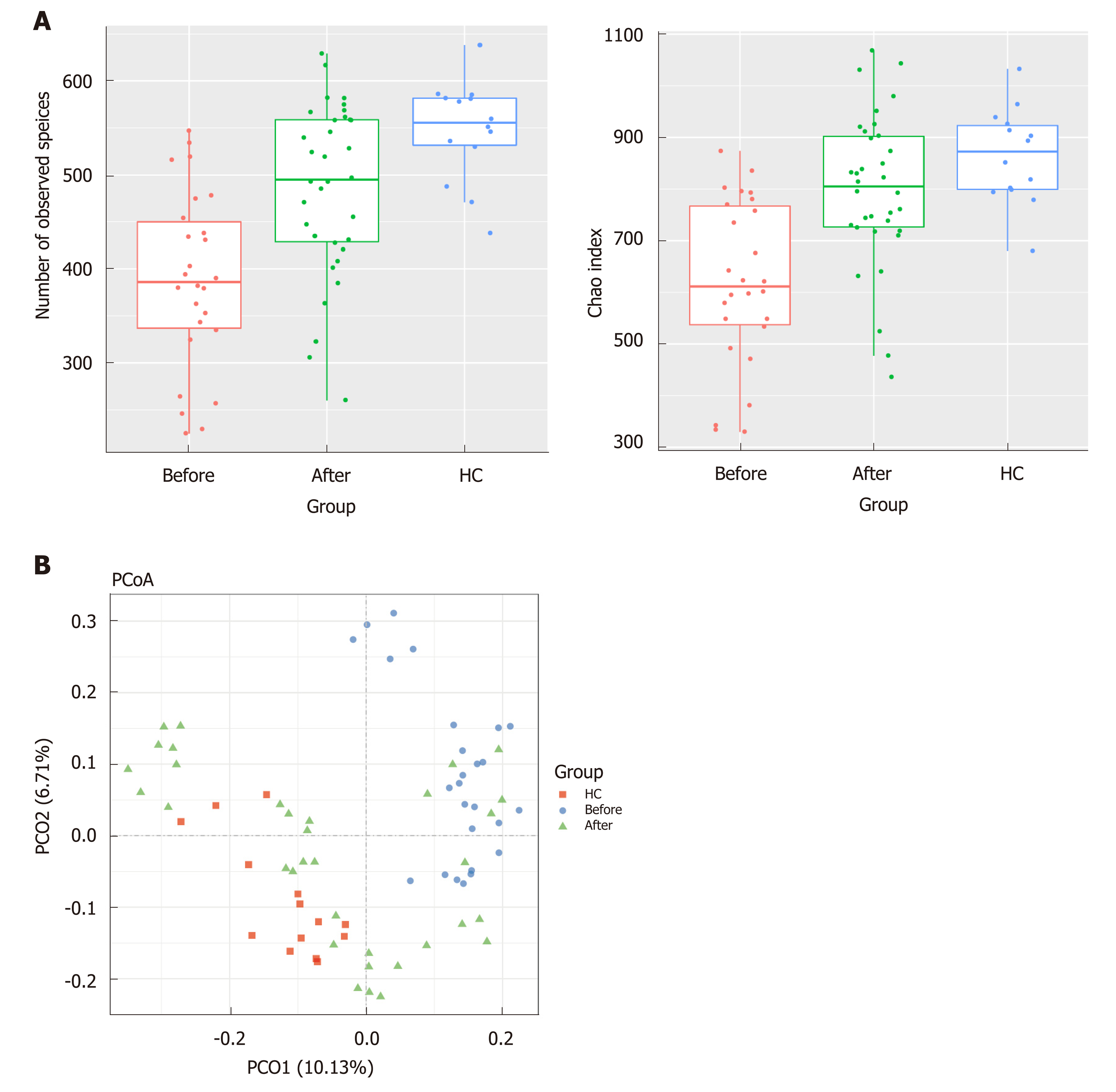
There were no significant differences in microbial richness among the four anatomical sites in individuals. Compared to active disease, the alpha diversity of CD in remission was increased towards the level of HC compared to the active stage. The principal coordinate analysis revealed that samples of active CD were clearly separated from those in remission, which clustered close to HC. Sixty-five genera were identified as differentially abundant between active and quiescent CD, with a loss of Fusobacterium and a gain of potential beneficial bacteria including Lactobacillus, Akkermansia, Roseburia, Ruminococcus and Lachnospira after the induction of remission. The combination of these taxa into a MDI showed a positive correlation with clinical disease severity and a negative correlation with species richness. The increased capacity for the inferred pathways including Lipopolysaccharide biosynthesis and Lipopolysaccharide biosynthesis proteins in patients before treatment negatively correlated with the abundance of Roseburia, Ruminococcus and Lachnospira.
The dysbiosis of mucosa-associated microbiota was associated with the disease phenotype and may become a potential diagnostic tool for the recurrence of disease.
Core tip: The dysbiosis of gut microbiome is associated with the development of Crohn’s disease (CD), although the alteration from active CD to remission after treatment is unclear. This study illustrated that the composition of mucosa-associated gut microbiota in active CD significantly changed after the induction of remission regardless of drugs used, which got close to healthy subjects. We speculate that the maintenance of gut microbiota balance may be potential therapeutic target for reducing the risk of disease relapse.
- Citation: He C, Wang H, Liao WD, Peng C, Shu X, Zhu X, Zhu ZH. Characteristics of mucosa-associated gut microbiota during treatment in Crohn’s disease. World J Gastroenterol 2019; 25(18): 2204-2216
- URL: https://www.wjgnet.com/1007-9327/full/v25/i18/2204.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i18.2204
Crohn’s disease (CD), a subtype of inflammatory bowel disease (IBD), has become a global disease with accelerating incidence over the last few decades[1]. CD is characterized by multiple episodes of exacerbation and remission, with clinical manifestations of diarrhea, abdominal pain, fistulas and perianal lesions, which may affect the whole digestive tract and cause systemic symptoms[2]. Current therapies for IBD include anti-inflammatory and immunomodulatory treatments such as 5-aminosalicylates, corticosteroids, thiopurines, thalidomide and anti-tumor necrosis factor alpha, all of which aim to achieve clinical remission and mucosal healing[3,4]. The pathogenesis of CD is multifactorial and involves the interplay of host genetics, immune dysregulation and environmental factors resulting in an aberrant immune response and subsequent intestinal inflammation[5].
Recent progress in understanding the composition and function of human microbiota has revealed the important role of microbiota in immune homeostasis[6]. Accumulating studies using culture-independent techniques have shown the dysbiosis of gut microbiota in patients with CD, including decreased bacterial diversity, with an expansion of putative aggressive groups (such as Enterobacteriaceae, Fusobacterium) combined with decreases in protective groups (such as Faecalibacterium, Roseburia)[7,8]. In addition to observing the characteristics of gut microbiota in CD, a study by Wang et al[9] evaluated their dynamic changes after infliximab (IFX) therapy and found that the dysbiosis could be corrected in patients with a sustained therapeutic response. Furthermore, a prospective study assessed the stool metagenomes of IBD patients starting biologic therapy and demonstrated a higher abundance of butyrate producers at baseline in therapy-responsive CD patients, indicating the predicative effect of the gut microbiome in treatment response[10]. Due to the dysregulated microbiota in the pathogenesis of IBD, several studies have reported the potential effect of restoring dysbiotic gut microbiota, including the use of probiotics and unprocessed donor feces, in the management of IBD[11]. It is necessary to clarify the key bacteria that play a role in disease remission and relapse, and then, precise manipulation of these bacteria may become a therapeutic target in the future.
To date, most studies investigating the gut microbiota of CD have typically used fecal samples since they are readily obtained[7-9]. However, the composition of fecal microbiota has been shown to be significantly different from mucosal microbiota; this difference is believed to directly affect epithelial and mucosal function and to be more deeply involved in the pathophysiology of CD[12,13]. To collect sufficient mucosal samples, we processed endoscopically uninflamed mucosa, which was thicker than the inflamed mucosa and probably more appropriate for microbial analysis[14]. In addition, only limited differences in microbiota composition were observed between inflamed and uninflamed mucosa in patients[15]. The previous cross-sectional study of the alterations between CD patients and healthy controls (HC) could be misread based on the interindividual differences, which make it difficult to characterize the critical bacteria in CD. Thus, we investigated the mucosal-associated microbiome in paired samples from CD patients before and after clinical treatment by 16S rRNA gene sequencing to determine the association between gut microbiota and disease activity.
A prospective study was performed in nine CD patients who were enrolled in flare at baseline and then induced remission after clinical therapy. Inclusion criteria were a diagnosis of CD confirmed by endoscopy and histology and the activity of the disease was measured by the CD activity index (CDAI). Six HC without previous history of chronic disease were also recruited in the study from the First Affiliated Hospital of Nanchang University, China. Exclusion criteria for the two groups included severe concomitant disease involving the liver, heart, lung or kidney, pregnancy or breast-feeding, and treatment with antibiotics and prebiotics during the previous 4 wk. The mucosal samples were collected during the colonoscopy and both the patients and healthy subjects underwent intestinal washing before the examination. We did not collect both inflamed and noninflamed tissues since a previous study showed that the mucosal microbiota of inflamed and noninflamed regions of the gastrointestinal tract in CD or ulcerative colitis (UC) were indistinguishable, with virtually no taxa demonstrating disproportional abundances at a significant threshold nor any significant diversity differences observed[15]. Written informed consent was obtained from all the subjects and this study was approved by the Medical Ethics Committee of Nanfang Hospital.
A total of 74 mucosal biopsies were collected from 15 participants, including 9 patients with CD and 6 healthy individuals. Specimens of terminal ileum, ascending colon, descending colon and rectum in noninflamed mucosa were taken during colonoscopic examination. Sampling included both active and remission stages for each patient who underwent clinical treatment. All the samples were immediately put in liquid nitrogen and stored at - 80 °C before processing.
Microbial DNA was extracted from the mucosal biopsies using the E.Z.N.A. stool DNA kit (Omega Biotek, Norcross, GA, United States) according to the manufacturer’s protocols. The 16S rDNA V3-V4 region of the Eukaryotic ribosomal RNA gene was amplified by PCR (95 °C for 2 min, followed by 27 cycles at 98 °C for 10 s, 62 °C for 30 s, and 68 °C for 30 s and a final extension at 68 °C for 10 min) using primers 341F: CCTACGGGNGGCWGCAG; 806R: GGACTACHVGGGTATCTAAT, where the barcode is an eight-base sequence unique to each sample. PCRs were performed in triplicate 50 μL mixture containing 5 μL of 10 × KOD Buffer, 5 μL of 2.5 mM dNTPs, 1.5 μL of each primer (5 μM), 1 μL of KOD Polymerase, and 100 ng of template DNA.
Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions and qualified using QuantiFluor-ST (Promega, United States). Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) using Illumina Hiseq 2500 following standard protocols. The raw reads were deposited into the NCBI Sequence Read Archive database (Accession Number: SRP157001).
The raw data were filtered to obtain clean reads by eliminating the adapter pollution and low-quality sequences. Paired end clean reads were merged as raw tags using FLASH (Fast Length Adjustment of Short reads, v 1.2.11) with a minimum overlap of 10 bp and mismatch error rates of 2%[16]. Noisy sequences of raw tags were filtered by QIIME (v1.9.1) pipeline under specific filtering conditions to obtain high-quality clean tags[17]. The tags were then clustered as Operational Taxonomic Unit (OTU) by scripts of USEARCH (v 7.0.1090) software with a 97% similarity threshold[18]. The representative OTU sequences were taxonomically classified using Ribosomal Database Project classifier v.2.2 trained on the Greengenes database[19,20]. Finally, an OTU table and a phylogenetic tree were generated for diversity analysis. To estimate the diversity of the microbial community of the sample, we calculated the within-sample (alpha) diversity by Wilcoxon rank test for two groups and multiple group comparisons were made using Kruskal-Wallis test. Beta diversity was estimated by computing weighted Unifrac distance and was visualized with principal coordinate analysis (PCoA). Statistical differences (P < 0.05) between the two groups in the relative abundance of bacterial phyla and genera were evaluated using Metastats (Kruskal-Wallis test for more than two groups).
According to the genera average abundance in patients before and after treatment, genera were divided into before-enriched and after-enriched. The correlation network of genera differentially enriched in before and after group was constructed by Pearson correlation test based on the abundance. The correlation network was visualized using Cytoscape (version 3.3.0). Pearson correlation test was also performed for investigating microbial dysbiosis index (MDI) and CDAI, as well as differential genera and predicted pathways.
The metagenomes of the gut microbiome were imputed from 16S rRNA sequences with PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States). This method predicts the gene family abundance from the phylogenetic information with an estimated accuracy of 0.8. The closed OTU table was used as the input for metagenome imputation and was first rarefied to an even sequencing depth prior to the PICRUSt analysis. Next, the resulting OTU table was normalized by 16S rRNA gene copy number. The gene content was predicted for each individual. Then, the predicted functional composition profiles were collapsed into level 3 of the KEGG database pathways. The output file was further analyzed using Statistical Analysis of Metagenomic Profiles (STAMP) software package[21].
To determine the effect of disease activity on microbial composition, we collected four parts of mucosal samples including ileum, ascending colon, descending colon and rectum from nine patients in active and remission stages. Patients characteristics are described in Supplement Table 1. The CDAI was significantly decreased in the After group compared to the Before group, indicating that the remission had been induced after clinical treatments (Figure S1). The Before group included samples from three anatomical sites (ileum, ascending colon, descending colon) and the After group supplemented with rectum mucosa, except for three samples that were unqualified for sequencing. Given the ethical issue, we did not collect mucosa from the four sites in healthy subjects, but the samples in each site were relatively uniform (3 from ileum, 4 from ascending colon, 3 from descending colon and 4 from rectum). After filtering and bioinformatic processing, a median yield of 275041 high-quality reads were obtained per sample.
The gut microbiota richness, measured by observed species and Shannon index, was not significantly different among ileum, ascending colon, descending colon and rectum in both CD and HC (Figure S2, numbers of observed species, P = 0.12 for CD and P = 0.49 for HC; Shannon index, P = 0.78 for CD and P = 0.91 for HC, Kruskal-Wallis test). The analysis of beta diversity based on the unweighted UniFrac distances showed that there was no significant difference among the four regions of intestinal tract in both CD and HC (Adonis analysis, P = 0.85 for CD, P = 0.94 for HC). Interestingly, analysis of alpha diversity as calculated by number of observed species and Chao1 index revealed that the microbial biodiversity of the patients in remission was significantly increased towards the HC compared to their active stage before treatment (Figure 1A, numbers of observed species, P < 0.0001; Chao1 index, P < 0.0001; Kruskal-Wallis test). Beta diversity represented by PCoA analysis clearly showed that the samples from patients after treatment, which clustered separately from those before treatment, tended to approach that of HC (Figure 1B, Adonis analysis, P = 0.001).
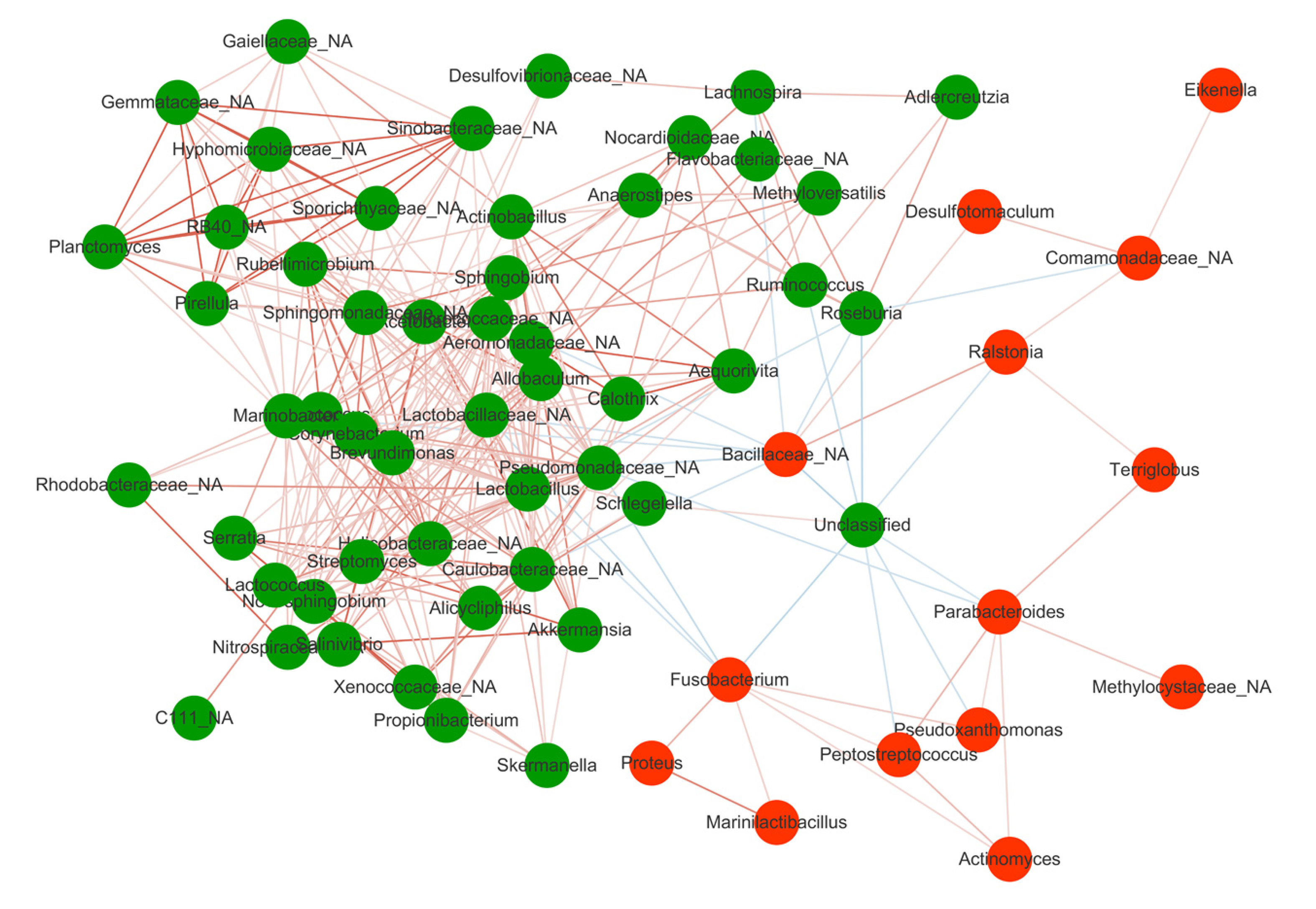
To investigate the specific changes of microbiota in patients with active and remission CD, we assessed the relative abundance of taxa before and after treatment. At the phylum level, the Fusobacteria was lower in the After group than the Before group (Figure 2A). In addition, the Firmicutes was significantly decreased in CD whereas the Proteobacteria was overrepresented in CD relative to HC. At the genus level, we observed 65 bacterial taxa that displayed different abundance between Before and After group. Compared with the After group, 14 bacterial taxa were enriched in the Before group, while 51 bacterial taxa were depleted in the Before group. The Before-enriched bacterial taxa included Fusobacterium, Streptococcus, Bacillaceae, etc. Bacterial taxa that were depleted in the Before group included Anaerostipes, Roseburia, Ruminococcus, Lactobacillus, Akkermansia, Lachnospira, etc, which were significantly more abundant in HC than CD (Figure 2B). Then, we used these taxa to calculate the MDI, which is the log of (total abundance in organisms increased in Before group) over (total abundance of organisms decreased in Before group) for the samples. The MDI showed a positive correlation with clinical disease severity (CDAI), and a negative correlation with species richness (Figure 2C), suggesting the close association between gut microbiota disorder and disease activity.
We further compared the effect of different medications including thalidomide, azathioprine (AZA), AZA plus prednisolone and IFX on gut microbiota. First, the observed species and Chao1 index, which represents alpha diversity, tended to increase after treatment in four groups, although without significance in IFX and AZA (Figure S3, numbers of observed species, P < 0.05 for thalidomide, P < 0.01 for AZA plus prednisolone, P > 0.05 for AZA and IFX; Chao1 index, P < 0.05 for thalidomide, P < 0.01 for AZA plus prednisolone, P > 0.05 for AZA and IFX, t test). The analysis of beta diversity based on the unweighted UniFrac distances showed that the overall microbial composition of the After group was significantly different from the Before group regardless of treatment drugs (Adonis analysis, P = 0.005 for thalidomide, P = 0.019 for AZA, P = 0.004 for AZA plus prednisolone, P = 0.018 for IFX). Furthermore, the differential taxa between After and Before groups were identified according to each treatment. The relative abundance of Roseburia, Ruminococcus and Anaerostipes was significantly increased after IFX treatment while the number of Fusobacterium and Streptococcus was lower. The relative abundance of Roseburia, Ruminococcaceae and Lachnospira was significantly increased while Fusobacteriaceae was decreased after patients were treated with AZA. In the AZA plus prednisolone group, the relative abundance of Lactobacillus, Ruminococcus and Lachnospiraceae was increased while that of Fusobacterium and Bacillaceae was decreased after treatment. In the thalidomide group, the relative abundance of Lactobacillus and Roseburia was also increased after treatment (Supplement Table 2). Collectively, these alterations in microbiota composition between Before and After group were similar among the four medications.
Spearman correlation test was performed to evaluate the relationships among the genera identified in MDI. Significant positive correlations were found in the genera depleted in the Before group including Lachnospira, Roseburia, Anaerostipes, Ruminococcus, which were abundant in HC, suggesting their synergy as commensal bacteria in maintaining gut microbiota homeostasis and promoting the mucosal healing process (Figure 3). On the other hand, the genera enriched in the Before group such as Fusobacterium and Bacillaceae showed negative correlations with those depleted in the Before group, indicating an antagonistic relationship between harmful bacteria in the active stage and beneficial bacteria in the remission stage.
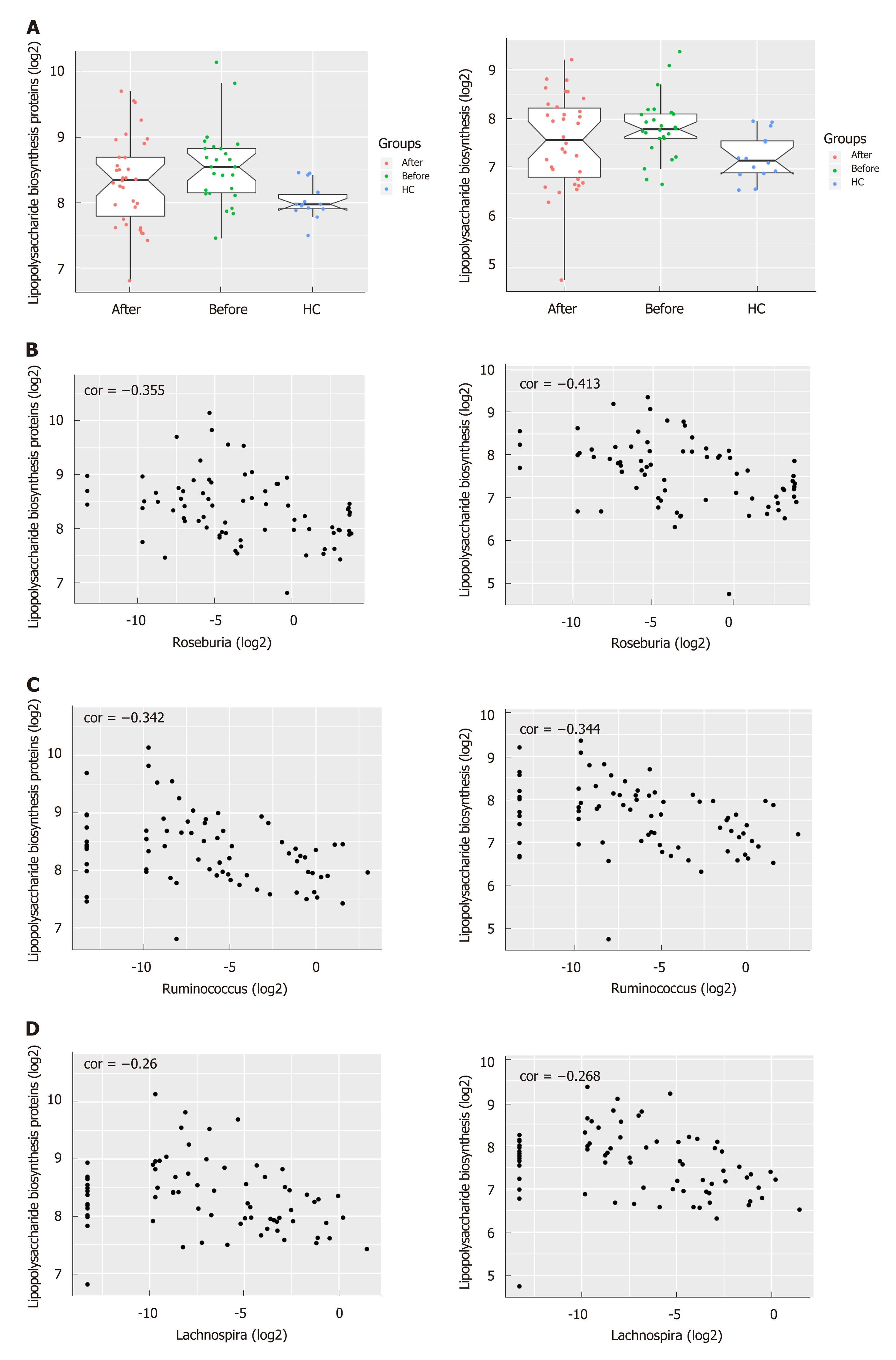
To infer the metagenome functional content based on the microbial community profiles obtained from the 16S rRNA gene sequences, we used PICRUSt[22]. The pathways including Lipopolysaccharide (LPS) biosynthesis proteins and LPS biosynthesis, which were enriched in patients before treatment compared to HC, tended to decrease after the induction of remission (Figures 4A and S4). On the other hand, the pathways including Folate biosynthesis, Starch and sucrose metabolism as well as Glycolysis/Gluconeogenesis which were deficient in active CD patients tended to increase after treatment and approached that of HC. Intriguingly, the abundance of LPS biosynthesis proteins and LPS biosynthesis were negatively correlated with three genera including Roseburia, Ruminococcus and Lachnospira, which were more abundant in HC and patients in remission, suggesting their potential role in anti-inflammation in CD (Figure 4B-D).
In the current study, we analyzed, using a 16S rRNA sequencing approach, the alterations of the gut mucosal microbiota in CD patients during their treatment. The comparison from the same cohort showed that the composition of mucosa-associated microbiota changed significantly after the induction of remission, with increased diversity as well as a restoration of potential beneficial bacteria. The MDI that was identified using differential microbiota between patients before and after treatment showed the correlation of microbial dysbiosis with disease activity.
Accumulating evidence has demonstrated the disorder of gut microbiota in CD, and this disorder is considered as an essential factor in driving inflammation[7,23]. The gut microbial community of CD patients is characterized by reduced diversity as well as compositional changes in phylum level, including a decreased abundance of Firmicutes and an increased abundance of Proteobacteria when compared to HC[24]. However, it remains unclear whether these alterations of gut microbiota are associated with disease activity. Consistent with previous studies, we found the dysbiosis of mucosa-associated microbiota in patients with CD and then analyzed its composition before and after the induction of remission. Due to the distinct microbiome signatures in different sub-phenotypes of CD, we enrolled the patients with the same behavioral phenotype to exclude bias[25]. Both the alpha and beta diversity showed that the structure of gut microbiota in patients before treatment was significantly different from those after treatment, approaching that of HC and indicating the partial restoration of microbial balance after treatment. A previous study by Wang et al[9] reported the dynamic changes of fecal microbiota during IFX therapy in pediatric CD, and IFX has been demonstrated to diminish CD-associated gut microbial dysbiosis. Another earlier study using a polymerase chain reaction-denaturing gradient gel electrophoresis also found that treatment with Adalimumab induced short-term changes in the microbiota composition, and these changes seem to parallel the partial recovery of the gut bacterial ecology[26]. AZA is the most commonly employed 6-mercaptopurines drug, and these drugs have been found to exert anti-inflammatory effects by targeting not only human macrophages but also the gut microbiota linked to CD[27]. Consistent with these previous findings, the present study showed that the mucosa-associated gut microbiota changed after treatment, regardless of the drug used, and thus we speculate that the alterations of gut microbiota may be associated with the change in disease status from active to remission. The causal relationship between disease activity and microbiota, however, needs further investigation using a germ-free animal model.
To further clarify the critical taxa that may be associated with the activity of CD, we identified 65 genera showing significant difference before and after treatment, and these genera were used to calculate the MDI. Interestingly, the positive correlation between MDI and CDAI indicates that the activity of CD may be associated with the dysbiosis of gut microbiota. A previous study in treatment-naïve children with CD also described the MDI using the differential taxa between CD and healthy subjects, and then demonstrated that the MDI characterized CD severity[28]. Some MDI-associated taxa were common to both studies including Fusobacterium, Lachnospira, Ruminococcus and Parabacteroides. The relative abundance of Lachnospira, Ruminococcus and Roseburia, which are known to produce short chain fatty acids (SCFAs) was significantly increased in patients with clinical remission compared to those with active CD. SCFAs, which are mainly composed of acetate, propionate and butyrate, are the end products of the fermentation of dietary fiber by the gut microbiota and have been shown to exert multiple beneficial effects on mammalian energy metabolism[29]. Recently, numerous studies have shown the loss of SCFA-producing taxa in CD and that IFX treatment was able to restore their levels in responsive patients, indicating their association with disease severity[9,24,30]. The beneficial effect of SCFAs on CD is probably due to their anti-inflammation capacity which is documented both in vitro and in vivo[31]. Chronic inflammation is a hallmark of CD and results from the recruitment and activation of immune cells from the circulation. Butyrate elicits anti-inflammatory effects via the inhibition of IL-12 and the upregulation of IL-10 production in human monocytes, repressing production of pro-inflammatory molecules TNF-α, IL-1β, nitric oxide, and reducing NF-κB activation[32,33]. A clinical study explored the therapeutic effect of butyrate on patients with CD and found that the administration of butyrate induced clinical improvement and remission in 53% of patients, in whom butyrate successfully downregulated mucosal levels of NF-κB and IL-1β[34]. Thus, it is conceivable to modulate the dysbiotic gut microbiota using probiotics, prebiotics and fecal microbiota transplantation (FMT) for the management of CD. Several studies have reported the potential of FMT for CD treatment, and we speculate that it may helpful to select donors with a high abundance of the beneficial bacteria, which are deficient in patients to improve the therapeutic efficacy[35].
Analysis of the inferred metagenome in our study showed that the abundance of LPS biosynthesis proteins and the LPS biosynthesis pathway in patients with CD were significantly decreased after treatment while the abundance of Folate biosynthesis, glycolysis/gluconeogenesis, starch and sucrose metabolism were increased, suggesting that the induction of remission could partially rectify the dysbiosis of gut microbiota and restore the homeostasis of metabolic function. LPS, one important component in the outer membrane of gram-negative bacteria, plays a critical role in triggering inflammatory responses that could further result in various diseases such as CD[36]. The serum levels of LPS were demonstrated to increased markedly in active CD patients compared with patients in remission and HC and were positively correlated with the severity of the disease[37]. Moreover, the blockade of intestinal mucosal inflammation with IFX could reduce the levels of LPS and IFX has been reported to diminish the CD-associated gut microbial dysbiosis[9,37]. Analysis of microbiota genes in this study showed a prominent upregulation of LPS-related pathways in patients before treatment, and this upregulation may be due to the overgrowth of LPS-producing bacteria such as Fusobacterium and Eikenella. Folate deficiency is common in patients with IBD, mostly in CD patients. Up to 80% of patients with CD present with low levels of serum folate, and this is more common in active disease than at times of remission[38,39]. Folate is produced in large quantities by the colonic microbiota, mainly probiotics such as Lactobacillus and Bifidobacterium[40]. The enrichment of Lactobacillus in CD patients after treatment as shown in our study may promote the biosynthesis of folate whose deficiency is associated with disease severity.
This study has some limitations. First, the number of patients was somewhat small. Therefore, the characteristics of mucosa-associated gut microbiota during CD treatment and the predictive value of MDI in disease activity warrant verification in large populations. Second, the patients in this study received different drugs for inducing remission, which may cause some bias. Our results showed that the composition of gut microbiota in patients before treatment was significantly different from those after treatment regardless of the drug used. Due to the small sample size in each treatment group, the effect of drugs on gut microbiota needs further investigation.
In conclusion, the results in this study presented that both the structure and function of mucosa-associated bacterial microbiota in patients with CD changed significantly during treatment, approaching healthy status. The dysbiosis of gut microbiota, which is associated with disease activity, is partially restored after the induction of remission with characteristics including increased biodiversity and an increase in SCFA-producing bacteria. Therefore, the disturbance of gut microbiota, especially the overgrowth of pathogenic bacteria and the depletion of beneficial bacteria may act as a potential therapeutic target for CD treatment.
Accumulating evidence demonstrated the alterations of gut microbiota in patients with Crohn’s disease (CD) compared to healthy subjects. However, comparative analysis of mucosal microbiota in the same cohort of patients before and after treatment remains limited. The different characteristics of mucosa-associated gut microbiota between active and quiescent CD may provide as a predictive tool for disease relapse as well as a potential therapeutic target for treatment.
Most studies investigating the gut microbiota of CD have used fecal samples while only few studies have investigated the mucosal microbiota, which is believed to directly affect epithelial function and may be more deeply involved in the pathogenesis of CD. Although the dysbiosis of gut microbiota have been reported in patients with CD as compared with healthy controls, the microbial changes during treatment and their association with disease activity are largely unknown.
To illustrate the global alterations of mucosa-associated microbiota in patients with active CD before and after treatment.
A total of 74 mucosal biopsies were collected from 15 participants including 9 patients with CD and 6 healthy individuals. Sampling included both active and remission stages for each patient who underwent clinical treatment. The gut microbiota was sequenced by 16S rRNA analysis.
Our results showed that the structure of gut microbiota in patients with active CD changed significantly after the induction of remission, including the decreased abundance of pathogenic bacteria and increased abundance of beneficial bacteria.
The dysbiosis of mucosa-associated gut microbiota in active CD was partially restored after treatment, indicating the association of microbiota and disease activity.
The variations of gut microbiota may act as a tool to supervise and predict the recurrence of CD, and the maintenance of microbial homeostasis could become a potential therapeutic target for the disease.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kolios G S-Editor: Yan JP L-Editor: A E-Editor: Zhang YL
| 1. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet. 2018;390:2769-2778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2677] [Cited by in RCA: 4103] [Article Influence: 512.9] [Reference Citation Analysis (110)] |
| 2. | Nikolaus S, Schreiber S. Diagnostics of inflammatory bowel disease. Gastroenterology. 2007;133:1670-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 292] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 3. | Ruemmele FM, Veres G, Kolho KL, Griffiths A, Levine A, Escher JC, Amil Dias J, Barabino A, Braegger CP, Bronsky J, Buderus S, Martín-de-Carpi J, De Ridder L, Fagerberg UL, Hugot JP, Kierkus J, Kolacek S, Koletzko S, Lionetti P, Miele E, Navas López VM, Paerregaard A, Russell RK, Serban DE, Shaoul R, Van Rheenen P, Veereman G, Weiss B, Wilson D, Dignass A, Eliakim A, Winter H, Turner D; European Crohn's and Colitis Organisation; European Society of Pediatric Gastroenterology, Hepatology and Nutrition. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn's disease. J Crohns Colitis. 2014;8:1179-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 843] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 4. | Fishman SJ, Feins NR, D'Amoto RJ, Folkman J. Thalidomide for Crohn's disease. Gastroenterology. 2000;119:596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 5. | de Souza HS, Fiocchi C. Immunopathogenesis of IBD: Current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1119] [Article Influence: 124.3] [Reference Citation Analysis (1)] |
| 6. | Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1428] [Cited by in RCA: 2248] [Article Influence: 281.0] [Reference Citation Analysis (1)] |
| 7. | Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, Martinez X, Varela E, Sarrabayrouse G, Machiels K, Vermeire S, Sokol H, Guarner F, Manichanh C. A microbial signature for Crohn's disease. Gut. 2017;66:813-822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 454] [Cited by in RCA: 597] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 8. | He Q, Gao Y, Jie Z, Yu X, Laursen JM, Xiao L, Li Y, Li L, Zhang F, Feng Q, Li X, Yu J, Liu C, Lan P, Yan T, Liu X, Xu X, Yang H, Wang J, Madsen L, Brix S, Wang J, Kristiansen K, Jia H. Two distinct metacommunities characterize the gut microbiota in Crohn's disease patients. Gigascience. 2017;6:1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 9. | Wang Y, Gao X, Ghozlane A, Hu H, Li X, Xiao Y, Li D, Yu G, Zhang T. Characteristics of Faecal Microbiota in Paediatric Crohn's Disease and Their Dynamic Changes During Infliximab Therapy. J Crohns Colitis. 2018;12:337-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 129] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 10. | Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe. 2017;21:603-610.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 327] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 11. | McIlroy J, Ianiro G, Mukhopadhya I, Hansen R, Hold GL. Review article: The gut microbiome in inflammatory bowel disease-avenues for microbial management. Aliment Pharmacol Ther. 2018;47:26-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 139] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 12. | Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes. 2015;6:173-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 157] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 13. | Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, Ishikawa E, Shima T, Hara T, Kado S, Jinnohara T, Ohno H, Kondo T, Toyooka K, Watanabe E, Yokoyama S, Tokoro S, Mori H, Noguchi Y, Morita H, Ivanov II, Sugiyama T, Nuñez G, Camp JG, Hattori M, Umesaki Y, Honda K. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 665] [Cited by in RCA: 815] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 14. | Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017;152:327-339.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 611] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 15. | Forbes JD, Van Domselaar G, Bernstein CN. Microbiome Survey of the Inflamed and Noninflamed Gut at Different Compartments Within the Gastrointestinal Tract of Inflammatory Bowel Disease Patients. Inflamm Bowel Dis. 2016;22:817-825. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 16. | Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957-2963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7679] [Cited by in RCA: 9233] [Article Influence: 659.5] [Reference Citation Analysis (0)] |
| 17. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23739] [Article Influence: 1582.6] [Reference Citation Analysis (0)] |
| 18. | Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8951] [Cited by in RCA: 9974] [Article Influence: 831.2] [Reference Citation Analysis (0)] |
| 19. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12842] [Cited by in RCA: 13186] [Article Influence: 732.6] [Reference Citation Analysis (0)] |
| 20. | DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069-5072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8383] [Cited by in RCA: 7557] [Article Influence: 397.7] [Reference Citation Analysis (0)] |
| 21. | Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123-3124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2193] [Cited by in RCA: 2719] [Article Influence: 247.2] [Reference Citation Analysis (0)] |
| 22. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5694] [Cited by in RCA: 6298] [Article Influence: 524.8] [Reference Citation Analysis (0)] |
| 23. | Khanna S, Raffals LE. The Microbiome in Crohn's Disease: Role in Pathogenesis and Role of Microbiome Replacement Therapies. Gastroenterol Clin North Am. 2017;46:481-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (1)] |
| 24. | Nishino K, Nishida A, Inoue R, Kawada Y, Ohno M, Sakai S, Inatomi O, Bamba S, Sugimoto M, Kawahara M, Naito Y, Andoh A. Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J Gastroenterol. 2018;53:95-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 312] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 25. | Dovrolis N, Drygiannakis I, Filidou E, Kandilogiannakis L, Arvanitidis K, Tentes I, Kolios G, Valatas V. Gut Microbial Signatures Underline Complicated Crohn's Disease but Vary Between Cohorts; An In Silico Approach. Inflamm Bowel Dis. 2019;25:217-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Busquets D, Mas-de-Xaxars T, López-Siles M, Martínez-Medina M, Bahí A, Sàbat M, Louvriex R, Miquel-Cusachs JO, Garcia-Gil JL, Aldeguer X. Anti-tumour Necrosis Factor Treatment with Adalimumab Induces Changes in the Microbiota of Crohn's Disease. J Crohns Colitis. 2015;9:899-906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Migliore F, Macchi R, Landini P, Paroni M. Phagocytosis and Epithelial Cell Invasion by Crohn's Disease-Associated Adherent-Invasive Escherichia coli Are Inhibited by the Anti-inflammatory Drug 6-Mercaptopurine. Front Microbiol. 2018;9:964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2354] [Article Influence: 214.0] [Reference Citation Analysis (0)] |
| 29. | den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325-2340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2408] [Cited by in RCA: 3283] [Article Influence: 273.6] [Reference Citation Analysis (3)] |
| 30. | Assa A, Butcher J, Li J, Elkadri A, Sherman PM, Muise AM, Stintzi A, Mack D. Mucosa-Associated Ileal Microbiota in New-Onset Pediatric Crohn's Disease. Inflamm Bowel Dis. 2016;22:1533-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 31. | Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: A study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13:2826-2832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 543] [Cited by in RCA: 622] [Article Influence: 34.6] [Reference Citation Analysis (5)] |
| 32. | Segain JP, Raingeard de la Blétière D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottière HM, Galmiche JP. Butyrate inhibits inflammatory responses through NFkappaB inhibition: Implications for Crohn's disease. Gut. 2000;47:397-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 886] [Cited by in RCA: 974] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 33. | Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1076] [Cited by in RCA: 1615] [Article Influence: 146.8] [Reference Citation Analysis (0)] |
| 34. | Di Sabatino A, Morera R, Ciccocioppo R, Cazzola P, Gotti S, Tinozzi FP, Tinozzi S, Corazza GR. Oral butyrate for mildly to moderately active Crohn's disease. Aliment Pharmacol Ther. 2005;22:789-794. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 176] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 35. | Li P, Zhang T, Xiao Y, Tian L, Cui B, Ji G, Liu YY, Zhang F. Timing for the second fecal microbiota transplantation to maintain the long-term benefit from the first treatment for Crohn's disease. Appl Microbiol Biotechnol. 2019;103:349-360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 36. | Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 168] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 37. | Guo Y, Zhou G, He C, Yang W, He Z, Liu Z. Serum Levels of Lipopolysaccharide and 1,3-β-D-Glucan Refer to the Severity in Patients with Crohn's Disease. Mediators Inflamm. 2015;2015:843089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Weisshof R, Chermesh I. Micronutrient deficiencies in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care. 2015;18:576-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 181] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 39. | Bermejo F, Algaba A, Guerra I, Chaparro M, De-La-Poza G, Valer P, Piqueras B, Bermejo A, García-Alonso J, Pérez MJ, Gisbert JP. Should we monitor vitamin B12 and folate levels in Crohn's disease patients? Scand J Gastroenterol. 2013;48:1272-1277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 40. | Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3:118-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 410] [Article Influence: 29.3] [Reference Citation Analysis (0)] |