Published online May 14, 2019. doi: 10.3748/wjg.v25.i18.2149
Peer-review started: February 14, 2019
First decision: March 20, 2019
Revised: March 27, 2019
Accepted: April 10, 2019
Article in press: April 11, 2019
Published online: May 14, 2019
Processing time: 91 Days and 7 Hours
The incidence of esophageal adenocarcinoma (EAC) has increased in recent decades, and its 5-year survival rate is less than 20%. As a well-established precursor, patients with Barrett's esophagus (BE) have a persistent risk of progression to EAC. Many researchers have already identified some factors that may contribute to the development of BE and EAC, and the identified risks include gastroesophageal reflux (GER), male sex, older age, central obesity, tobacco smoking, Helicobacter pylori (H. pylori) eradication, and the administration of proton pump inhibitors (PPIs) and antibiotics. The human gut harbors trillions of microorganisms, the majority of which are bacteria. These microorganisms benefit the human host in many ways, such as helping in digestion, assisting in the synthesis of certain vitamins, promoting the development of the gastrointestinal immune system, regulating metabolism and preventing invasion by specific pathogens. In contrast, microbial dysbiosis may play important roles in various diseases, such as inflammation and cancers. The composition of the microbiota located in the normal esophagus is relatively conserved without distinct microbial preferences in the upper, middle and lower esophagus. Six major phyla constitute the esophageal microbiota, including Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria and TM7, similar to the oral microbiota. Streptococcus dominates the esophageal microbiota. However, the microbiota varies in different esophageal diseases compared to that in the healthy esophagus. The type I microbiota, which is primarily composed of gram-positive bacteria, is closely associated with the normal esophagus, while type II microbiota has enriched gram-negative bacteria and is mainly associated with the abnormal esophagus. These increased gram-negative anaerobes/microaerophiles include Veillonella, Prevotella, Haemophilus, Neisseria, Granulicatella and Fusobacterium, many of which are associated with BE. The microbial diversity in the esophagus is decreased in EAC patients, and Lactobacillus fermentum is enriched compared to that in controls and BE patients. Furthermore, the microbiota may be associated with BE and EAC by interacting with their risk factors, including central obesity, GER, H. pylori, administration of PPIs and antibiotics. Therefore, a large gap in research must be bridged to elucidate the associations among these factors. Some studies have already proposed several potential mechanisms by which the microbiota participates in human carcinogenesis by complicated interactions with the human host immune system and signaling pathways. The activation of the LPS-TLR4-NF-κB pathway may contribute to inflammation and malignant transformation. This exciting field of gastrointestinal microbiota allows us to unravel the mystery of carcinogenesis from another perspective. Further studies are needed to explore whether the microbiota changes before or after disease onset, to improve our understanding of the pathogenesis, and to find novel targets for prevention, diagnosis and therapy, which could offer more cost-effective and relatively safe choices.
Core tip: Esophageal adenocarcinoma (EAC) is a malignancy with poor prognosis, and Barrett's esophagus (BE) is the only recognized precursor. As part of the human gut, the esophagus harbors distinctive microbiota, and dysbiosis may be related to BE/EAC. Many studies have attempted to characterize the esophageal microbiota in the normal esophagus and in different diseases, but more data are required. Studies on the esophageal microbiota in BE/EAC have mainly concentrated on these associations, and the underlying mechanisms remain blurred. This review focuses on the features and associations of esophageal microbiota and BE/EAC, which might provide some evidence of their relationships.
- Citation: Lv J, Guo L, Liu JJ, Zhao HP, Zhang J, Wang JH. Alteration of the esophageal microbiota in Barrett's esophagus and esophageal adenocarcinoma. World J Gastroenterol 2019; 25(18): 2149-2161
- URL: https://www.wjgnet.com/1007-9327/full/v25/i18/2149.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i18.2149
Esophageal cancer is one of the most common malignancies in the world, and the incidence of esophageal adenocarcinoma (EAC) has markedly increased in recent decades, as it accounts for almost half of all esophageal cancers[1,2]. The 5-year survival rate is less than 20%[3] because most EAC patients are first diagnosed in the advanced stages, which are not curable[4]. As a well-established risk factor for EAC, Barrett's esophagus (BE) confers a persistent risk of progression to EAC[5], increasing a patient's risk more than 30 times than the general population[6,7]. In the first global definition, namely, the Montreal definition, BE is defined as the replacement of normal squamous epithelial lining with metaplastic columnar epithelium[8]. Notably, the incidence of EAC progression from BE varies among studies. Some studies have indicated that more than 85% of newly diagnosed EAC patients have no history of BE[9,10], whereas other investigations[6,7] have reported that almost half of EAC patients have progressed from BE. Nevertheless, BE is the only well-recognized precursor of EAC, and the underlying mechanisms of pathogenesis and carcinogenesis need to be elucidated[11]. Many researchers have already shown that some factors may contribute to the development of BE and EAC, and the identified risks include, but are not limited to, gastroesophageal reflux (GER), male sex, older age, central obesity, tobacco smoking, Helicobacter pylori (H. pylori) eradication with antibiotics and acid suppression therapy[3,12-14]. However, preventive strategies are lacking.
The human gut harbors trillions of microorganisms[15-18], and the composition of the microbial communities that inhabit the mouth, esophagus, stomach and intestine are diverse and host-specific. The majority of microorganisms are bacteria and are estimated to comprise ~1014 bacterial cells, which is ten times more than the total number of human cells[19]. These microorganisms benefit the human host in many ways, such as helping in digestion, assisting in the synthesis of certain vitamins, promoting the development of the gastrointestinal immune system, regulating metabolism and preventing invasion by specific pathogens[15,16,19,20]. On the other hand, microbial dysbiosis may lead to tissue damage and play significant roles in various diseases, including inflammatory disorders and cancers[21-25]. Dysbiosis refers to an abnormal condition of the microbial ecosystem in a host[26]. Therefore, equilibrium must be achieved and maintained to support the interactions of the human host and microbiota. Before the 1990s, researchers mainly focused on the role of certain microorganisms by using protocols largely limited to culture-dependent methods, but cultivation is not suitable for defining a complicated microbial community and may induce bias as well[19]. Since the development of next-generation sequencing, which is independent of cultivation, the sensitivity of research techniques has been dramatically improved, and microbiota exploration has begun[20].
However, little is known about the relationship between the microbiota and the pathogenesis of BE and EAC. Here, we review the features of microbial communities in BE and EAC patients, which may provide some evidence of the relationships between altered esophageal microbiota and BE/EAC.
The colon has the largest microbiota in the body[20], whereas the esophagus has far fewer microbes. Although bacterial communities have been observed with high inter- and intra-individual variations, an overlapping community has been identified between sites[27,28]. Previous studies[20,29-31] have suggested that Firmicutes (Clostridium, Ruminococcus, Eubacterium, Peptostreptococcus, Peptococcus, Lactobacillus-L), Bacteroidetes, Proteobacteria (Enterobacteriaceae) and Actinobacteria (Bifidobacterium-BF) phyla constitute the majority of the human gut microbiota. The composition of the microbiota located in the normal esophagus is relatively conserved, and the estimated resident microbes are mainly composed of more than 100 species, most of which have already been identified[32,33]. The dominant microbes that colonize the normal esophagus are Streptococcus[32,33], and Dong L et al.[28] reported that no distinct microbial preference exists in the upper, middle and lower esophagus. However, the microbiota varies in different esophageal diseases compared to the healthy esophagus[34].
In 2004, Pei Z et al.[32] found that six major phyla constituted the esophageal microbiota, namely, Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria and TM7, which are comparable to the oral microbiota. The genus Streptococcus dominates the esophageal microbiota[32]. Since then, more studies have emerged, and a classification for the esophageal microbiota was proposed[33]. In 2009, Yang L et al.[33] reported that type I microbiota, which is mainly composed of gram-positive bacteria, is closely associated with the normal esophagus and is dominated by the Firmicutes phylum. Consistent with previous studies, Streptococcus was the most dominant genus, and its relative abundance was higher. The type II microbiota is enriched in gram-negative bacteria (more than 50%) and is mainly associated with the abnormal esophagus. The relative abundances of 24 other genera are increased in the type II microbiota, many of which are relevant to BE. These increased gram-negative anaerobes/microaerophiles include Veillonella, Prevotella, Haemophilus, Neisseria, Granulicatella and Fusobacterium. Moreover, one study[19] conducted in Japan showed that the numbers of bacteria were similar in control, esophagitis and BE groups, despite the changes in the relative abundance of taxa. In patients with esophagitis or BE, the microbial diversity changed, and the abundance of Streptococcus species was reduced[33]. Gram-negative anaerobes/microaerophiles occupied greater proportions[33], such as Veillonella, Prevotella, Fusobacterium and Neisseria[19,33]. This shift from a gram-positive aerobic microbiota to a gram-negative anaerobic microbiota may be influenced by microenvironmental changes and related to abnormal disease states[33]. These consistent observations suggested that the altered microbiota is reliable in BE and could be further studied. Of note, Macfarlane S et al.[35] found that Campylobacter colonized the esophagus of the majority of BE patients and could not be identified in the control group. Moreover, Amir I et al.[36] strongly suggested that the family Enterobacteriaceae (mainly the genus Escherichia) is associated with esophageal abnormalities, such as esophagitis and BE, and may have a possible role in the pathogenesis of inflammation and metaplasia. Therefore, a large-scale joint multi-center, multi-region, multi-race study about the alteration of the esophageal microbiota in BE/EAC is needed to provide more evidence.
Damage of the esophageal epithelium could affect the normal barrier and induce the translocation of other bacteria, thus influencing the microenvironment and immune homeostasis[11]. Although studies of specific bacteria in the development of EAC in addition to H. pylori[3] are rare, more attention has been paid to the local microbiota changes. The microbial diversity in the esophagus is decreased in EAC patients[12], regardless of the exact sampling locations. The decreased genera included some gram-negative and gram-positive taxa, such as Veillonella and Granulicatella. In contrast, Lactobacillus fermentum was found to be enriched in EAC patients compared to controls and BE patients. Notably, lactic acid bacteria could be dominant and affect the microenvironment[12]. A low microenvironmental pH may facilitate the growth of Lactobacillus spp. and Streptococcus spp. in the tumor niche[12]. Fermentation could produce more factors to inhibit the proliferation of other competitor microbes as well. Then, Lactobacillus may dominate the environment of the lower esophagus. Moreover, some specific species were demonstrated to have higher abundance. At the phylum level, the proportional abundance of Tenericutes was higher. At the genus level, the proportional abundances of Fusobacterium, Megasphaera, Campylobacter, Capnocytophaga, and Dialister were greater[12]. However, Blackett KL et al.[37] did not identify any specific taxa with significant differences, and Zaidi AH et al.[38] reported that Streptococcus pneumonia was present at a relatively higher abundance in control and BE groups compared to EAC in rat BE and EAC models. Interestingly, Peters BA et al.[39] found that the oral microbiota composition could reflect the prospective risk for EAC, and the genus Neisseria and the species Streptococcus pneumoniae were associated with EAC risk, which is consistent with the findings in the studies above. It was reasonable to conclude that the esophageal microbiota is largely influenced by the oral microbiota and that the oral microbiota composition could provide some evidence of EAC progression[17].
Furthermore, the microbiota may be associated with BE and EAC by interacting with their risk factors. One notable example is the case of obesity. As a chronic systemic disease and a proposed risk factor, obesity, particularly central obesity, is closely related to BE and EAC[20,40-42]. The linear pattern between increasing body mass index (BMI) and increasing risks of BE and EAC has been verified in several studies[43-46], which partially accounts for the increasing prevalence of EAC. Central obesity is closely related to EAC, even after adjustment for BMI[47,48], whereas the association between BMI and EAC risk disappeared after adjustment for central obesity. Moreover, the relationship between central obesity and BE has a similar pattern. Therefore, adiposity distribution may play an important role in BE and EAC pathogenesis. However, it is unclear whether weight loss could contribute to a reduced risk of BE and EAC. The possible mechanisms by which central obesity contributes to BE and EAC have been explored and discussed in several aspects. First, the increased abdominal adipose tissue might increase intra-abdominal pressure and gastric compression, disrupting the normal function of the gastroesophageal junction and promoting GER, which is also a well-recognized risk factor for BE and EAC[3]. Second, excess adipose tissue could secrete pro-inflammatory cytokines and adipokines[20], and these active factors could provoke inflammatory and metabolic changes in the body[40], such as stimulation of cell proliferation, apoptosis inhibition and neoplastic transformation. Third, the gut microbiota is altered in obese patients and has been associated with the activation of inflammation, which may play an important role in the development of BE and EAC[49]. Streptococcus and Prevotella species are the dominant bacteria in the upper gastrointestinal tract, and their ratio may be associated with central obesity and hiatal hernia length[27], which are two known risks of BE and EAC. In addition, the gut microbiota may be adjusted concomitantly along with dietary changes that humans experience and that are the main cause of central obesity, but some 'lost' taxa may be difficult to regain over generations[50]. Therefore, a large gap in research must be bridged to elucidate the associations among central obesity, microbiota and EAC[51].
Ultimately, the optimal method for esophageal microbiota sampling is fundamental and needs to be explored. Many studies have used invasive endoscopy to obtain focal tissues that are similar to those obtained by biopsy or endoscopic brushing to examine the microbiota in a larger area. Gall A et al.[27] showed that mucosal brush samples could enhance the detection of bacterial diversity in the esophagus and stomach and the microbiota compositions were similar after replicate sampling. Fillon SA et al.[52] proposed another minimally invasive sampling method, namely the overnight esophageal string test, and suggested that the compositions of the esophageal microbiota were similar between the traditional biopsy and the string test. Due to the lack of standard sampling methods, Elliott DRF et al.[12] studied the values of the Cytosponge prototype as a non-endoscopic sampling device to seek minimally invasive methods for sampling the esophageal microbiota. For comparison, endoscopic biopsies, brushes and throat swabs were also included. Nevertheless, most of the microbial species overlapped among different sampling methods. However, the sampling area was larger with the Cytosponge, and the microbial DNA yield and total microbial abundance were higher. Moreover, the Cytosponge also provided clues for histological data, thus making it a valuable sampling method.
The important findings discussed above are summarized in Table 1. Nevertheless, some of these data were drawn from a specific population, and the sample sizes were not large. Consequently, these findings still need to be verified before any application to the general population, and more effort should be made to reveal the exact alteration of the microbiota in different diseases.
| Publication year | Sample population | Sequencing approach | Related notable findings | Ref |
| 2004 | Four patients with normal esophagus | 16S rDNA | 1 Members of six phyla, Firmicutes, Bacteroides, Actinobacteria, Proteobacteria, Fusobacteria, and TM7, were represented. | [32] |
| 2 Streptococcus (39%), Prevotella (17%), and Veilonella (14%) were most prevalent. | ||||
| 2007 | Seven subjects without BE and seven patients with BE | 16S rRNA | Campylobacter colonized the esophagus in the majority of BE patients and could not be identified in the control group. | [35] |
| 2009 | Thirty-four patients with normal, esophagitis, or Barrett's esophagus | 16S rRNA | 1 Esophageal microbiomes can be classified into two types. | [33] |
| 2 The type I microbiome was mainly composed of gram-positive bacteria, dominated by the genus Streptococcus and concentrated in the phenotypically normal esophagus. | ||||
| 3 The type II microbiome contained a greater proportion of gram-negative anaerobes/microaerophiles and primarily correlated with esophagitis (Odds Ratio: 15.4) and BE (Odds Ratio: 16.5). | ||||
| 2012 | Fifteen subjects | 16S rRNA | The compositions of the esophageal microbiota were similar between the traditional biopsy and the overnight esophageal string test. | [52] |
| 2014 | Thirteen patients with esophagitis, six patients with BE, fifteen normal controls | 16S rRNA | The Enterobacteriaceae family (mainly the genus Escherichia) is associated with esophageal abnormalities, such as esophagitis and BE. | [36] |
| 2015 | Twelve participants enrolled in the Seattle Barrett's Esophagus Research Program | 16S rRNA | 1 Streptococcus and Prevotella species dominate the upper GI and the ratio of these two species is associated with waist-to-hip ratio and hiatal hernia length, two known EAC risk factors in Barrett’s esophagus. | [27] |
| 2 Mucosal brush samples enhanced the detection of bacterial diversity in the esophagus and stomach, and the microbiota compositions were similar after replicate sampling. | ||||
| 2017 | Twenty normal controls, twenty-four non- dysplastic BE twenty-three dysplastic BE | 16S rRNA | 1 The microbial diversity in the esophagus is decreased in EAC patients, regardless of the exact sampling locations. | [12] |
| 2 Lactobacillus fermentum was enriched in EAC patients, and lactic acid bacteria dominated and affected the microenvironment. | ||||
| 2018 | Twenty-seven dental and esophageal disease-free individuals | 16S rRNA | 1 The phyla Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, and TM7 were most abundant in both the oral cavity and the esophagus. | [28] |
| 2 The genera Streptococcus, Neisseria, Prevotella, Actinobacillus, and Veillonella were most abundant in both oral cavity and esophagus, and Streptococcus in the esophagus. | ||||
| 3 No site-specific bacteria were found for three different segments (upper, middle, and lower) of the esophagus. |
It has been reported that more than 15% of human malignant cancers are related to infection or infection-associated inflammation[53], and many relevant studies are mainly focused on single microorganisms. There are two main theories to explain bacterial diseases[33]. Koch proposed the classic pathogen theory, which requires the presence of specific pathogens[54]. The other theory of microecological diseases is a new concept in which the whole microbiota contributes to pathogenicity[26]. As the only bacteria considered a class I human carcinogen, H. pylori is closely related to the progression of gastritis, gastric ulcer, gastric atrophy and gastric adenocarcinoma[55,56]. The bacterium has infected more than 50% of the world's population[57] and continues to spread. Although H. pylori is primarily localized to the gastric mucosa, its colonization could affect the gastric and esophageal microbiota[11,58], and the microbial composition in the esophagus and stomach overlaps to a certain extent[27].
The increasing incidence of BE and EAC may be inversely associated with H. pylori infection[59,60], and the inverse correlation between H. pylori and EAC risk has been well documented[10,27]. Meta-analyses[61-63] based on epidemiological and observational studies showed that EAC coincides with H. pylori eradication[49]. That is, H. pylori might affect carcinogenesis in the lower esophagus[27]. However, the inner mechanisms have only been partially revealed[10,27]. H. pylori harbors some factors that lead to chronic inflammation and cancer, such as cytotoxin-associated gene A (CagA), vacuolating cytotoxin (VAC) and adhesins. The bacterium could promote inflammatory responses by activating nuclear factor kappa B (commonly known as NF-κB)[56] and may induce the production of certain cytokines such as IL-1β, IL-2, IL-8 and tumor necrosis factor-α (TNF-α), which trigger inflammatory responses in the gastric epithelium. H. pylori may also directly damage host DNA, dysregulate DNA transcription factors such as caudal type homeobox 2 (Cdx2), and induce epithelial injury and acid secretory functions[55,56,58]. Another possible mechanism is that H. pylori-induced gastric atrophy causes a reduction in gastric acid, which is the main source of GER substances[20]. Some studies suggest that H. pylori eradication could increase the serum level of ghrelin, which may lead to obesity and affect gastric emptying[61], subsequently initiating the risks of BE and EAC. Moreover, H. pylori may stimulate apoptosis of EAC cells via the Fas-caspase cascade, which may account for another protective mechanism[61].
Acid suppression therapies have been highly effective in acid-induced diseases, such as gastritis, esophagitis, BE and EAC. Proton pump inhibitors (PPIs) are considered benign and are commonly used in clinical practice[64]. The suppression of acid secretion could affect gastric acidity, volume and GER and even the bacterial composition in the stomach and esophagus, with potential consequences for human health and diseases[36,64]. The administration of PPIs could change the microbial composition in the esophagus and stomach in BE patients[34,36], which may contribute to the pathogenesis of BE, though this has not been well established. Studies have suggested that PPIs may directly target certain bacteria that contain P-type ATPase enzymes as part of their proton pumps, such as Streptococcus pneumoniae and H. pylori[65]. Moreover, the microenvironment could also be affected by the increased pH in the stomach and esophagus after PPI therapy. PPIs could reduce the number of gram-negative bacteria and decrease the risk for neoplasia in the esophagus[64]. To detail the changes, Amir I et al.[36] collected esophageal samples before and after 8 wk of PPI treatment in BE patients and found that PPI usage changed the esophageal microbiota. At the family level, Comamonadaceae was decreased, whereas other families, such as Clostridiaceae and Lachnospiraceae, were increased. However, long-term PPI therapy induced hypergastrinemia, which may upregulate cyclooxygenase-2 (COX-2) expression, cell proliferation and esophageal carcinogenesis[66]. PPI administration plays an important role in H. pylori eradication therapy. A study[67] conducted by Fischbach LA et al. showed that PPIs augment anti-H. pylori activity, and H. pylori appears to exert a protective role in esophageal neoplasia. The possible reason might be that PPIs have some direct protective effects in BE, which extend far beyond other effects[64]. At present, there is no direct evidence of a relationship between PPIs and esophageal carcinogenesis, and long-term preclinical and clinical studies with larger samples are needed to reveal the precise associations among PPIs, H. pylori and BE/EAC.
The introduction and worldwide application of antibiotics might also have contributed to the increasing incidence of BE and EAC[20]. The discovery and wide usage of antibiotics have cured many infectious diseases, but several unexpected effects have appeared, some of which may influence the progression of BE and EAC. Antibiotics may definitively change the gastrointestinal microbiota[3], and alteration of the microbial abundance and/or diversity might contribute to disease pathology[68]. Cho I et al.[69] found that the sub-therapeutic administration of antibiotics increased the abundance of Firmicutes and decreased the abundance of Bacteroidetes, which are the two main phyla in the colonic microbiota. Moreover, the overweight population harbored a higher ratio of Firmicutes to Bacteroidetes than the controls[26], and weight loss increased the abundance of Bacteroidetes. These data[26,69] indicated that long-term exposure to antibiotics changes the colonic microbiota, which may induce obesity and GER. In the same way, the use of antibiotics can change the microbiota in the esophagus. Tian ZY et al.[70] have suggested that H. pylori infection and antibiotic treatment changes the microbiota composition in the esophagus in a mouse model. In addition, an altered esophageal microbiota might play a more direct role than H. pylori or obesity in inflammation and in BE and EAC carcinogenesis[20]. On the other hand, antibiotics may help restore the normal esophageal microbiota to type I from type II by increasing the relative abundance of Streptococcus. Interestingly, antibiotics are another important part of H. pylori eradication therapy. The complicated associations among these factors need further investigation and validation[20].
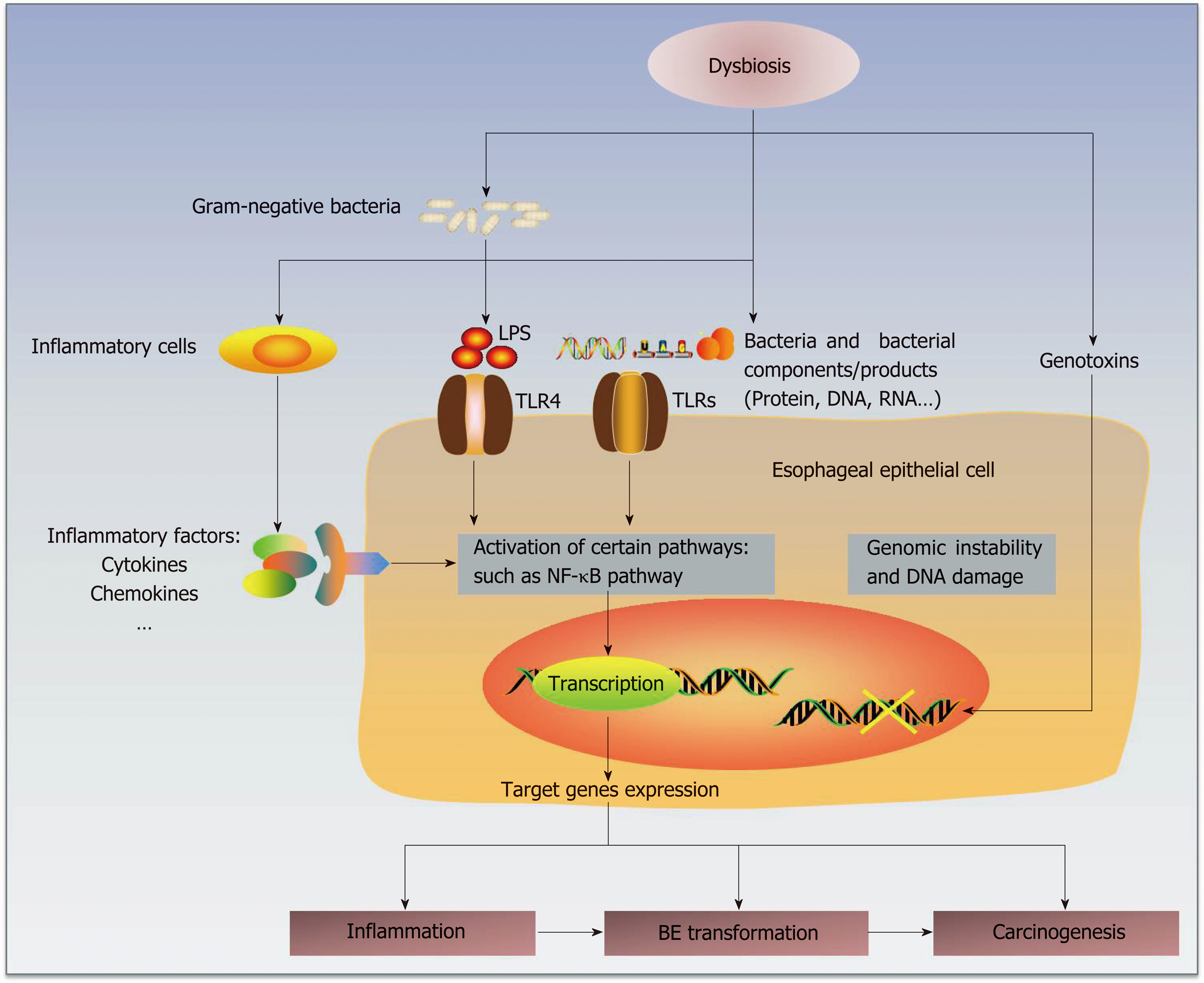
Some studies have already proposed several potential mechanisms by which the microbiota participates in human carcinogenesis[20] by complicated interactions with the human host immune system and signaling pathways[56] (Figure 1). First, alteration of the microbiota may result in inflammation, and persistent chronic inflammation may promote carcinogenesis[71]. The disequilibrium between human immunity and microbiota may change the compositions of essential bacterial molecules at certain organs or sites and then form microorganism-associated molecular patterns (MAMPs)[71], such as toll-like receptors (TLRs) and nucleotide-binding-oligomerization-domain (NOD)-like receptors[72,73]. Then, the subsequent activation of related pathways may lead to the production and release of some target genes involved in inflammation[73], such as cytokines, chemokines and other inflammatory factors. For example, specific bacterial invasion could promote the production of IL-17 and IL-23 and then induce an inflammatory response[74].
Bacterial products, or even the microbes themselves, could be sensed by some receptors on the epithelial membranes. The esophageal type II microbiota could produce larger amounts of gram-negative bacterial components, involving lipopolysaccharides (LPS)[19,75]. LPS could delay gastric emptying via COX1/2[76] and contribute to the development of GER by increasing the intra-gastric pressure. Notably, LPS could also affect the function of the lower esophageal sphincter, which may promote GER and carcinogenesis[75].
TLRs recognize known molecules from microbes[77,78] and have a well-recognized role in carcinogenesis[38]. As part of the pathogen-associated molecular patterns (PAMPs) or danger-associated molecular patterns (DAMPs), TLRs serve as receptors of various ligands, such as bacterial cell wall components and DNA and viral double-stranded RNA[77,78]. Therefore, TLRs mediate the interaction of the immune system with the microbiota[79]. Due to the connective roles between innate and adaptive immune responses, TLRs represent an important linking factor between inflammation and cancers[38,77,78,80]. As the natural ligand of LPS, TLR4 is expressed in the human esophageal epithelium, and its expression increases in BE and EAC[75]. TLR4 activation triggers the NF-κB pathway, which is related to inflammation-associated carcinogenesis[81] and mediates the initial metaplastic BE changes[82]. COX-2 is also upregulated as one of the downstream genes, which is related to gastric emptying[75] and occurs along the progression of BE and EAC[83]. Therefore, the activation of the LPS-TLR4-NF-κB pathway may contribute to inflammation and malignant transformation[20,75]. Similarly, activation of the Wnt signaling pathway could induce defects in cellular tight junction proteins and decrease the production of some mucins, which may have positive roles in the protection from carcinogenesis[84].
Second, bacteria may generate genotoxins that could cause genomic damage. For instance, the cytolethal distending toxin, which may be produced by gram-negative bacteria, can induce DNA damage and genomic instability[23,85]. Some bacterial products have tumor-promoting effects[86], such as CagA and VacA, and some bacteria may also activate procarcinogens to provoke inflammation and cancer develop-ment[87].
The number of studies on how microbial communities contribute to the pathogenesis of BE and EAC is increasing, and greater attention has been paid to the etiology and molecular mechanisms[49]. Although some observations are promising, the sample sizes of the related studies are not large enough, and the existing data are too limited to draw any convincing conclusion. The most appropriate methods and controls need to be proven as well. The mechanisms by which the microbiota affects the pathogenesis of BE and EAC are still not clear, and further studies are required. Whether the direct interactions between microbes and epithelia or the released products from microbes regulate local inflammation and immunity are under investigation[88]. Furthermore, it is important to identify when changes occur in the microbial composition during disease progression.
Without a doubt, exploring BE pathogenesis is a good way to study the carcinogenesis of EAC[11]. However, the alterations of microbial diversity in BE and EAC are modest[36], and the particular species of bacteria that can discriminate BE and EAC from controls have not yet been identified. Moreover, some low-abundance genera might be difficult to detect. Amir I et al.[36] reported that they could not identify any biomarker taxa for distinguishing BE from controls. Yang L et al.[33] showed that some gram-negative bacteria are enriched in BE and might thus be related to BE. Moreover, some genera might contribute to the etiology and pathogenesis of BE and EAC, or result from BE and EAC[11]. We still need to search for distinct microbial species that could be biomarkers for BE and EAC in individuals at higher risk, and the subtypes of EAC should be taken into consideration.
Nevertheless, finding the true causal mechanisms of dysbiosis is complicated, and the identification of a single species or a collection of species responsible for a particular disorder is sophisticated. The introduction of new techniques, such as next-generation sequencing, will definitely assist in revealing the mechanisms of the gastrointestinal microbiome in BE and EAC development, which might provide some evidence of their relations and the pathogenesis of BE and EAC. The microbiota in BE and EAC patients remains to be explored, particularly with the adjustment of other risks, such as sex and central obesity. It is crucial to improve our understanding of the process by which the microbial composition may promote disorder progression[20].
The prognosis of EAC is poor, and the most pivotal factor is the tumor stage at diagnosis[3]. Given this situation in EAC patients, screening certain individuals with higher risks could be useful for early diagnosis. The cost-effectiveness and feasibility of endoscopic usage urge us to seek a less invasive and effective way of screening and early detection for BE and EAC[89]. Early detection of EAC will improve the survival rate and quality of life. Identification of BE patients who have higher risks of developing EAC could provide diagnostic clues and avoid unnecessary procedures, and the medical resources could be re-distributed to those who truly need attention. Understanding the pathogenesis and exploring biomarkers could also lead to early detection and prevention and improve the survival of some EAC patients. The therapeutic manipulation of the microbiota, such as prebiotics, probiotics or microbiota transplants, could be a potent approach in the management of inflammation and cancers[56].
The convincing associations between the microbiota and BE/EAC have indicated the importance of these studies. This exciting field of gastrointestinal microbiota allows us to unravel the mystery of carcinogenesis from another perspective, and the integration of different biomarkers may lead us to rapid advances. The introduction of animal models could be the proverbial icing on the cake. Future perspective studies with sophisticated techniques are needed to explore whether the microbiota changes before or after disease onset, to improve our understanding of the pathogenesis, and to find novel targets for prevention, diagnosis and therapy, which could offer more cost-effective and relatively safe choices[19,56,90].
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Conflict of interest statement: There is no conflict of interest associated with any of the authors that contributed to this manuscript.
P-Reviewer: Ehrenpreis EDD, Eleftheriadis N S-Editor: Yan JP L-Editor: Filipodia E-Editor: Zhang YL
| 1. | Bird-Lieberman EL, Fitzgerald RC. Early diagnosis of oesophageal cancer. British journal of cancer. 2009;101:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (1)] |
| 2. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11831] [Article Influence: 845.1] [Reference Citation Analysis (4)] |
| 3. | Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:390-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 380] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 4. | Hur C, Miller M, Kong CY, Dowling EC, Nattinger KJ, Dunn M, Feuer EJ. Trends in esophageal adenocarcinoma incidence and mortality. Cancer. 2013;119:1149-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 395] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 5. | Cook MB, Coburn SB, Lam JR, Taylor PR, Schneider JL, Corley DA. Cancer incidence and mortality risks in a large US Barrett's oesophagus cohort. Gut. 2018;67:418-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 6. | Desai TK, Krishnan K, Samala N, Singh J, Cluley J, Perla S, Howden CW. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett's oesophagus: a meta-analysis. Gut. 2012;61:970-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 413] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 7. | Sharma P. Clinical practice. Barrett's esophagus. The New England journal of medicine. 2009;361:2548-2556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 182] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 8. | Vakil N, van Zanten SV, Kahrilas P, Dent J, Jones R, Global Consensus G. The Montreal definition and classification of gastroesophageal reflux disease: a global evidence-based consensus. The American journal of gastroenterology. 2006;101:1900-1920; quiz 1943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2368] [Cited by in RCA: 2448] [Article Influence: 128.8] [Reference Citation Analysis (2)] |
| 9. | Cooper SC, El-agib A, Dar S, Mohammed I, Nightingale P, Murray IA, Cooper BT, Trudgill NJ. Endoscopic surveillance for Barrett's oesophagus: the patients' perspective. European journal of gastroenterology & hepatology. 2009;21:850-854. [PubMed] |
| 10. | Rubenstein JH, Shaheen NJ. Epidemiology, Diagnosis, and Management of Esophageal Adenocarcinoma. Gastroenterology. 2015;149:302-317 e301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 260] [Article Influence: 26.0] [Reference Citation Analysis (1)] |
| 11. | Quante M, Graham TA, Jansen M. Insights Into the Pathophysiology of Esophageal Adenocarcinoma. Gastroenterology. 2018;154:406-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 12. | Elliott DRF, Walker AW, O'Donovan M, Parkhill J, Fitzgerald RC. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. The lancet Gastroenterology & hepatology. 2017;2:32-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 13. | Cook MB, Wild CP, Forman D. A systematic review and meta-analysis of the sex ratio for Barrett's esophagus, erosive reflux disease, and nonerosive reflux disease. American journal of epidemiology. 2005;162:1050-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 165] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 14. | Quante M, Abrams JA, Wang TC. The rapid rise in gastroesophageal junction tumors: is inflammation of the gastric cardia the underwater iceberg? Gastroenterology. 2013;145:708-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3003] [Cited by in RCA: 2985] [Article Influence: 229.6] [Reference Citation Analysis (0)] |
| 16. | Karlsson F, Tremaroli V, Nielsen J, Backhed F. Assessing the human gut microbiota in metabolic diseases. Diabetes. 2013;62:3341-3349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 311] [Article Influence: 25.9] [Reference Citation Analysis (1)] |
| 17. | Gorkiewicz G, Moschen A. Gut microbiome: a new player in gastrointestinal disease. Virchows Archiv : an international journal of pathology. 2018;472:159-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 18. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9101] [Cited by in RCA: 7820] [Article Influence: 521.3] [Reference Citation Analysis (4)] |
| 19. | Yang L, Chaudhary N, Baghdadi J, Pei Z. Microbiome in reflux disorders and esophageal adenocarcinoma. Cancer journal. 2014;20:207-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 20. | Neto AG, Whitaker A, Pei Z. Microbiome and potential targets for chemoprevention of esophageal adenocarcinoma. Seminars in oncology. 2016;43:86-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Goodman AL, Gordon JI. Our unindicted coconspirators: human metabolism from a microbial perspective. Cell metabolism. 2010;12:111-116. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, Housseau F, Pardoll DM, Sears CL. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature medicine. 2009;15:1016-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1113] [Cited by in RCA: 1301] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 23. | Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1374] [Cited by in RCA: 1660] [Article Influence: 127.7] [Reference Citation Analysis (1)] |
| 24. | Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, Jin C, Wunderlich C, Wunderlich T, Eisenbarth SC, Flavell RA. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9862-9867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 255] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 25. | Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1339] [Cited by in RCA: 1375] [Article Influence: 80.9] [Reference Citation Analysis (1)] |
| 26. | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7222] [Cited by in RCA: 6397] [Article Influence: 336.7] [Reference Citation Analysis (0)] |
| 27. | Gall A, Fero J, McCoy C, Claywell BC, Sanchez CA, Blount PL, Li X, Vaughan TL, Matsen FA, Reid BJ, Salama NR. Bacterial Composition of the Human Upper Gastrointestinal Tract Microbiome Is Dynamic and Associated with Genomic Instability in a Barrett's Esophagus Cohort. PloS one. 2015;10:e0129055. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 28. | Dong L, Yin J, Zhao J, Ma SR, Wang HR, Wang M, Chen W, Wei WQ. Microbial Similarity and Preference for Specific Sites in Healthy Oral Cavity and Esophagus. Frontiers in microbiology. 2018;9:1603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Rogers CJ, Prabhu KS, Vijay-Kumar M. The microbiome and obesity-an established risk for certain types of cancer. Cancer journal. 2014;20:176-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 30. | Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65-80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1071] [Cited by in RCA: 920] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 31. | Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M'Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174-180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5822] [Cited by in RCA: 5017] [Article Influence: 358.4] [Reference Citation Analysis (2)] |
| 32. | Pei Z, Bini EJ, Yang L, Zhou M, Francois F, Blaser MJ. Bacterial biota in the human distal esophagus. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4250-4255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 337] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 33. | Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588-597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 398] [Cited by in RCA: 359] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 34. | Pei Z, Yang L, Peek RM, Jr Levine SM, Pride DT, Blaser MJ. Bacterial biota in reflux esophagitis and Barrett's esophagus. World journal of gastroenterology. 2005;11:7277-7283. [PubMed] |
| 35. | Macfarlane S, Furrie E, Macfarlane GT, Dillon JF. Microbial colonization of the upper gastrointestinal tract in patients with Barrett's esophagus. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2007;45:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 36. | Amir I, Konikoff FM, Oppenheim M, Gophna U, Half EE. Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environmental microbiology. 2014;16:2905-2914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 143] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 37. | Blackett KL, Siddhi SS, Cleary S, Steed H, Miller MH, Macfarlane S, Macfarlane GT, Dillon JF. Oesophageal bacterial biofilm changes in gastro-oesophageal reflux disease, Barrett's and oesophageal carcinoma: association or causality? Alimentary pharmacology & therapeutics. 2013;37:1084-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 38. | Zaidi AH, Kelly LA, Kreft RE, Barlek M, Omstead AN, Matsui D, Boyd NH, Gazarik KE, Heit MI, Nistico L, Kasi PM, Spirk TL, Byers B, Lloyd EJ, Landreneau RJ, Jobe BA. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC cancer. 2016;16:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 39. | Peters BA, Wu J, Pei Z, Yang L, Purdue MP, Freedman ND, Jacobs EJ, Gapstur SM, Hayes RB, Ahn J. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer research. 2017;77:6777-6787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 287] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 40. | Lagergren J. Influence of obesity on the risk of esophageal disorders. Nature reviews Gastroenterology & hepatology. 2011;8:340-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 41. | Chak A, Falk G, Grady WM, Kinnard M, Elston R, Mittal S, King JF, Willis JE, Kondru A, Brock W, Barnholtz-Sloan J. Assessment of familiality, obesity, and other risk factors for early age of cancer diagnosis in adenocarcinomas of the esophagus and gastroesophageal junction. The American journal of gastroenterology. 2009;104:1913-1921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 42. | Ali AS, Ali S, Ahmad A, Bao B, Philip PA, Sarkar FH. Expression of microRNAs: potential molecular link between obesity, diabetes and cancer. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2011;12:1050-1062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 43. | Whiteman DC, Sadeghi S, Pandeya N, Smithers BM, Gotley DC, Bain CJ, Webb PM, Green AC, Australian Cancer S. Combined effects of obesity, acid reflux and smoking on the risk of adenocarcinomas of the oesophagus. Gut. 2008;57:173-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 215] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 44. | Hoyo C, Cook MB, Kamangar F, Freedman ND, Whiteman DC, Bernstein L, Brown LM, Risch HA, Ye W, Sharp L, Wu AH, Ward MH, Casson AG, Murray LJ, Corley DA, Nyren O, Pandeya N, Vaughan TL, Chow WH, Gammon MD. Body mass index in relation to oesophageal and oesophagogastric junction adenocarcinomas: a pooled analysis from the International BEACON Consortium. International journal of epidemiology. 2012;41:1706-1718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 45. | Turati F, Tramacere I, La Vecchia C, Negri E. A meta-analysis of body mass index and esophageal and gastric cardia adenocarcinoma. Annals of oncology : official journal of the European Society for Medical Oncology. 2013;24:609-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (1)] |
| 46. | Petrick JL, Kelly SP, Liao LM, Freedman ND, Graubard BI, Cook MB. Body weight trajectories and risk of oesophageal and gastric cardia adenocarcinomas: a pooled analysis of NIH-AARP and PLCO Studies. British journal of cancer. 2017;116:951-959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 47. | Steffen A, Huerta JM, Weiderpass E, Bueno-de-Mesquita HB, May AM, Siersema PD, Kaaks R, Neamat-Allah J, Pala V, Panico S, Saieva C, Tumino R, Naccarati A, Dorronsoro M, Sanchez-Cantalejo E, Ardanaz E, Quiros JR, Ohlsson B, Johansson M, Wallner B, Overvad K, Halkjaer J, Tjonneland A, Fagherazzi G, Racine A, Clavel-Chapelon F, Key TJ, Khaw KT, Wareham N, Lagiou P, Bamia C, Trichopoulou A, Ferrari P, Freisling H, Lu Y, Riboli E, Cross AJ, Gonzalez CA, Boeing H. General and abdominal obesity and risk of esophageal and gastric adenocarcinoma in the European Prospective Investigation into Cancer and Nutrition. International journal of cancer. 2015;137:646-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 48. | Singh S, Sharma AN, Murad MH, Buttar NS, El-Serag HB, Katzka DA, Iyer PG. Central adiposity is associated with increased risk of esophageal inflammation, metaplasia, and adenocarcinoma: a systematic review and meta-analysis. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2013;11:1399-1412 e1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 249] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 49. | Aleman JO, Eusebi LH, Ricciardiello L, Patidar K, Sanyal AJ, Holt PR. Mechanisms of obesity-induced gastrointestinal neoplasia. Gastroenterology. 2014;146:357-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 150] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 50. | Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 965] [Cited by in RCA: 1165] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 51. | Kaakoush NO, Morris MJ. The oesophageal microbiome: an unexplored link in obesity-associated oesophageal adenocarcinoma. FEMS microbiology ecology. 2016;92(10). [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Fillon SA, Harris JK, Wagner BD, Kelly CJ, Stevens MJ, Moore W, Fang R, Schroeder S, Masterson JC, Robertson CE, Pace NR, Ackerman SJ, Furuta GT. Novel device to sample the esophageal microbiome--the esophageal string test. PloS one. 2012;7:e42938. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 53. | de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. The Lancet Oncology. 2012;13:607-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1583] [Cited by in RCA: 1714] [Article Influence: 131.8] [Reference Citation Analysis (1)] |
| 54. | Kaufmann SH, Schaible UE. 100th anniversary of Robert Koch's Nobel Prize for the discovery of the tubercle bacillus. Trends in microbiology. 2005;13:469-475. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 55. | Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. European journal of immunology. 2015;45:17-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 202] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 56. | Tozun N, Vardareli E. Gut Microbiome and Gastrointestinal Cancer: Les liaisons Dangereuses. Journal of clinical gastroenterology. 2016;50:S191-S196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 57. | Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1361] [Cited by in RCA: 2040] [Article Influence: 255.0] [Reference Citation Analysis (0)] |
| 58. | Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiological reviews. 2010;90:859-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2501] [Cited by in RCA: 2741] [Article Influence: 182.7] [Reference Citation Analysis (1)] |
| 59. | Kamangar F, Dawsey SM, Blaser MJ, Perez-Perez GI, Pietinen P, Newschaffer CJ, Abnet CC, Albanes D, Virtamo J, Taylor PR. Opposing risks of gastric cardia and noncardia gastric adenocarcinomas associated with Helicobacter pylori seropositivity. Journal of the National Cancer Institute. 2006;98:1445-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 232] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 60. | Anderson LA, Murphy SJ, Johnston BT, Watson RG, Ferguson HR, Bamford KB, Ghazy A, McCarron P, McGuigan J, Reynolds JV, Comber H, Murray LJ. Relationship between Helicobacter pylori infection and gastric atrophy and the stages of the oesophageal inflammation, metaplasia, adenocarcinoma sequence: results from the FINBAR case-control study. Gut. 2008;57:734-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 61. | Nie S, Chen T, Yang X, Huai P, Lu M. Association of Helicobacter pylori infection with esophageal adenocarcinoma and squamous cell carcinoma: a meta-analysis. Diseases of the esophagus : official journal of the International Society for Diseases of the Esophagus. 2014;27:645-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 62. | Xie FJ, Zhang YP, Zheng QQ, Jin HC, Wang FL, Chen M, Shao L, Zou DH, Yu XM, Mao WM. Helicobacter pylori infection and esophageal cancer risk: an updated meta-analysis. World journal of gastroenterology. 2013;19:6098-6107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 108] [Cited by in RCA: 123] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 63. | Rokkas T, Pistiolas D, Sechopoulos P, Robotis I, Margantinis G. Relationship between Helicobacter pylori infection and esophageal neoplasia: a meta-analysis. Clinical gastroenterology and hepatology:. the official clinical practice journal of the American Gastroenterological Association. 2007;5:1413-1417, 1417 e1411-1412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 150] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 64. | Freedberg DE, Lebwohl B, Abrams JA. The impact of proton pump inhibitors on the human gastrointestinal microbiome. Clinics in laboratory medicine. 2014;34:771-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 65. | Vesper BJ, Jawdi A, Altman KW, Haines GK, 3rd, Tao L, Radosevich JA. The effect of proton pump inhibitors on the human microbiota. Current drug metabolism. 2009;10:84-89. [PubMed] |
| 66. | Singh S, Garg SK, Singh PP, Iyer PG, El-Serag HB. Acid-suppressive medications and risk of oesophageal adenocarcinoma in patients with Barrett's oesophagus: a systematic review and meta-analysis. Gut. 2014;63:1229-1237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 198] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 67. | Fischbach LA, Graham DY, Kramer JR, Rugge M, Verstovsek G, Parente P, Alsarraj A, Fitzgerald S, Shaib Y, Abraham NS, Kolpachi A, Gupta S, Vela MF, Velez M, Cole R, Anand B, El Serag HB. Association between Helicobacter pylori and Barrett's esophagus: a case-control study. The American journal of gastroenterology. 2014;109:357-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 68. | Snider EJ, Freedberg DE, Abrams JA. Potential Role of the Microbiome in Barrett's Esophagus and Esophageal Adenocarcinoma. Digestive diseases and sciences. 2016;61:2217-2225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 69. | Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, Li H, Alekseyenko AV, Blaser MJ. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1092] [Cited by in RCA: 1208] [Article Influence: 92.9] [Reference Citation Analysis (0)] |
| 70. | Tian Z, Yang Z, Gao J, Zhu L, Jiang R, Jiang Y. Lower esophageal microbiota species are affected by the eradication of Helicobacter pylori infection using antibiotics. Experimental and therapeutic medicine. 2015;9:685-692. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 71. | Schwabe RF, Jobin C. The microbiome and cancer. Nature reviews Cancer. 2013;13:800-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1054] [Cited by in RCA: 1228] [Article Influence: 102.3] [Reference Citation Analysis (2)] |
| 72. | Verbeek RE, Siersema PD, Vleggaar FP, Ten Kate FJ, Posthuma G, Souza RF, de Haan J, van Baal JW. Toll-like Receptor 2 Signalling and the Lysosomal Machinery in Barrett's Esophagus. Journal of gastrointestinal and liver diseases : JGLD. 2016;25:273-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 73. | Kinnebrew MA, Pamer EG. Innate immune signaling in defense against intestinal microbes. Immunological reviews. 2012;245:113-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 74. | Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, Datz C, Feng Y, Fearon ER, Oukka M, Tessarollo L, Coppola V, Yarovinsky F, Cheroutre H, Eckmann L, Trinchieri G, Karin M. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1035] [Cited by in RCA: 1035] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 75. | Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2138-2144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 76. | Calatayud S, Garcia-Zaragoza E, Hernandez C, Quintana E, Felipo V, Esplugues JV, Barrachina MD. Downregulation of nNOS and synthesis of PGs associated with endotoxin-induced delay in gastric emptying. American journal of physiology Gastrointestinal and liver physiology. 2002;283:G1360-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 77. | Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 822] [Cited by in RCA: 948] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 78. | Wagner H. Innate immunity's path to the Nobel Prize 2011 and beyond. European journal of immunology. 2012;42:1089-1092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 79. | Baghdadi J, Chaudhary N, Pei Z, Yang L. Microbiome, innate immunity, and esophageal adenocarcinoma. Clinics in laboratory medicine. 2014;34:721-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 80. | Thakur KK, Bolshette NB, Trandafir C, Jamdade VS, Istrate A, Gogoi R, Cucuianu A. Role of toll-like receptors in multiple myeloma and recent advances. Experimental hematology. 2015;43:158-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 81. | Maeda S, Omata M. Inflammation and cancer: role of nuclear factor-kappaB activation. Cancer science. 2008;99:836-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 174] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 82. | Souza RF, Krishnan K, Spechler SJ. Acid, bile, and CDX: the ABCs of making Barrett's metaplasia. American journal of physiology Gastrointestinal and liver physiology. 2008;295:G211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 135] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 83. | Buskens CJ, Van Rees BP, Sivula A, Reitsma JB, Haglund C, Bosma PJ, Offerhaus GJ, Van Lanschot JJ, Ristimaki A. Prognostic significance of elevated cyclooxygenase 2 expression in patients with adenocarcinoma of the esophagus. Gastroenterology. 2002;122:1800-1807. [PubMed] |
| 84. | Jovov B, Shaheen NJ, Orlando GS, Djukic Z, Orlando RC. Defective barrier function in neosquamous epithelium. The American journal of gastroenterology. 2013;108:386-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 85. | Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11537-11542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 611] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 86. | Plottel CS, Blaser MJ. Microbiome and malignancy. Cell host & microbe. 2011;10:324-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 482] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 87. | Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355-1359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3709] [Cited by in RCA: 3187] [Article Influence: 167.7] [Reference Citation Analysis (0)] |
| 88. | Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nature immunology. 2013;14:668-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 414] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 89. | di Pietro M, Canto MI, Fitzgerald RC. Endoscopic Management of Early Adenocarcinoma and Squamous Cell Carcinoma of the Esophagus: Screening, Diagnosis, and Therapy. Gastroenterology. 2018;154:421-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 90. | Suerbaum S. Microbiome analysis in the esophagus. Gastroenterology. 2009;137:419-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |