Published online Apr 14, 2019. doi: 10.3748/wjg.v25.i14.1697
Peer-review started: January 14, 2019
First decision: February 21, 2019
Revised: March 6, 2019
Accepted: March 15, 2019
Article in press: March 16, 2019
Published online: April 14, 2019
Processing time: 91 Days and 1 Hours
Long non-coding RNAs (lncRNAs) are widely involved in tumor regulation. Nevertheless, the role of the lncRNA cancer susceptibility 19 (CASC19) in colorectal cancer (CRC) has yet to be fully clarified.
To explore the effect of CASC19 on proliferation and metastasizing ability of CRC cells.
CASC19 expression in human CRC tissues, pair-matched adjacent normal colon tissues, and CRC cells was detected using quantitative real-time PCR (qRT-PCR). CASC19 expression, as well as its relation to overall survival, was extrapolated by Kaplan-Meier survival analysis together with multivariable Cox regression assay. In vitro experiments were performed to confirm whether CASC19 regulates CRC cell invasion, migration, proliferation, and apoptosis.
CASC19 expression was markedly upregulated in CRC tissues and CRC cell lines (P < 0.05). qRT-PCR revealed that CASC19 expression was higher in 25 tissue samples from patients with aggressive CRC compared with the 27 tissue samples from patients with nonaggressive CRC (P < 0.05). Higher CASC19 expression was associated with poorer patient prognoses. Furthermore, in vitro experiments demonstrated that CASC19 overexpression enhanced CRC cell invasion, migration, and proliferation. CASC19 overexpression enhanced the expression of cell migration inducing hyaluronidase 1 (CEMIP) and epithelial-mesenchymal transition markers. MiR-140-5p was found to be able to bind directly to CASC19 and CEMIP. Overexpression of miR-140-5p reversed the effect of CASC19 on cell proliferation and tumor migration, as well as suppressed CASC19-induced CEMIP expression.
CASC19 positively regulates CEMIP expression through targeting miR-140-5p. CASC19 may possess an oncogenic function in CRC progression, highlighting its potential as an essential biomarker in CRC diagnosis and therapy.
Core tip: The long non-coding RNA cancer susceptibility 19 is highly expressed in colorectal cancer tissues. In vitro studies have shown that the long non-coding RNA cancer susceptibility 19 may regulate the proliferation, epithelial-mesenchymal transition, and metastasizing ability of colorectal cancer cells by regulating microRNA-140-5p, as well as cell migration by inducing hyaluronidase 1.
- Citation: Wang XD, Lu J, Lin YS, Gao C, Qi F. Functional role of long non-coding RNA CASC19/miR-140-5p/CEMIP axis in colorectal cancer progression in vitro. World J Gastroenterol 2019; 25(14): 1697-1714
- URL: https://www.wjgnet.com/1007-9327/full/v25/i14/1697.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i14.1697
Colorectal cancer (CRC) is a tumor that is increasingly common in the modern world[1]. Tumor metastasis is one of the most important causes of poor prognosis for patients with CRC. At the time of diagnosis, approximately 20%-25% of patients with CRC are found to have liver metastasis. At the same time, liver metastasis occurs in up to 40%-50% of patients after resection of primary CRC[2]. Although current methods for the diagnosis and therapy of CRC have achieved remarkable progress, tumor metastasis remains an important factor affecting the survival of patients[3]. In recent years, gene therapy has become an intense focus of research. Transporting tumor suppressor genes or non-coding RNAs via nanocarriers may be a new option for cancer therapeutics[4]. Therefore, a thorough understanding of the molecular pathophysiological pathways underlying CRC is crucial to developing an effective therapeutic strategy.
Non-coding RNAs include microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). MiRNAs bind to the 3’-untranslated regions (3’-UTR) of the message RNA (mRNA) of the target genes, resulting in mRNA degradation and inhibition of the translation process. LncRNAs are RNAs that are longer than 200 nucleotides. The existing literature primarily investigates the regulatory roles of lncRNAs in several biological processes[5,6]. Dysregulation of lncRNAs is observed in various types of cancers, including breast cancer[7], oesophageal cancer[8], hepatocellular carcinoma[9-11], lung cancer[12], gastric cancer[13], and CRC[14-18]. LncRNA dysregulation has been discovered to be closely related to cancer progression. For example, overexpression of the lncRNA n335586 contributes to cell migration and invasion in hepatocellular carcinoma[19], while the lncRNA CASP5 facilitates the migration and invasion of human glioblastoma cells[20]. The regulatory mechanism of lncRNAs is still not clearly understood, and its possible role in cancer has been hypothesized to be as a competing endogenous RNA (ceRNA) for sponge miRNAs. For instance, the lncRNA UCA1 may adsorb microRNA (miRNA/miR)-182, thereby affecting the expression of its downstream target gene PFKFB2 and promoting glioma metastasis[21]. The lncRNA PVT1 enhances colon cancer metastasis by altering the miR-30d-5p/RUNX2 axis[22]. CRC progression has recently been discovered to be associated with endogenous lncRNA sponges. The cancer susceptibility 19 (CASC19) is a 324 bp lncRNA that is located on chromosome 8q24.21. Several lines of evidence suggest that the expression of CASC19 is overregulated in CRC, and this may play an oncogenic role in CRC progression[23-25]. However, the mechanism by which CASC19 regulates CRC progression is not fully understood.
The cell migration inducing hyaluronidase 1 (CEMIP) gene is located on chromosome 15q25 and encodes a 150 kDa protein. CEMIP is originally described as an inner ear protein and its mutation leads to hearing loss[26]. CEMIP has traditionally been linked to hyaluronic acid depolymerization[27]. Recent findings indicate that CEMIP may be involved in tumor development and may promote tumor cell proliferation and metastasis. For instance, the high expression of CEMIP is associated with a poor prognosis of prostate cancer[28], gastric cancer[29], and CRC[28,30-34]. These reports suggest that CEMIP contributes to cancer heterogeneity and may be a potential therapeutic target.
Our present study demonstrated that CRC possesses a characteristic alteration in CASC19 expression profile that is related to CRC progression. Overexpression of CASC19 promotes CRC progression. In addition, mechanism analysis showed that CASC19 positively regulates CEMIP expression via sponge miR-140-5p, thereby exerting a carcinogenic effect in CRC progression. Future therapies targeting the CASC19/miR-140-5p/CEMIP axis may be beneficial in CRC.
This study included 52 patients who were pathologically diagnosed as having CRC and received surgical treatment between January 2015 and December 2016 at Tianjin Medical University General Hospital. Dissected tumor and adjacent normal colonic mucosal tissues (as samples taken from areas well close to the section margin) were immediately frozen in liquid nitrogen and stored at -80 °C. All the patients did not receive neoadjuvant therapy. All patients provided written informed consent. This study was approved by the Medical Ethics Committee of Tianjin Medical University General Hospital (Ethical No. IRB2015-YX-018) and was conducted in strict compliance with the Declaration of Helsinki.
Healthy colon epithelial cell line (FHC) and CRC cell lines (HCT-116, SW480, Lovo, and SW620) were purchased from the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in DMEM medium supplemented with 10% FBS in a controlled environment of 5% CO2 at 37 °C. Cell medium was replaced every 2 d. Before transfection, a total of 2.5 × 104 cells per well were seeded onto 6-well plates and incubated for 24 h, then the culture medium was discarded and 100 nmol/L of CASC19 overexpressing plasmid (CASC19-p; Genechem, Shanghai, China), 200 nmol/L of siRNA mixture, 100 nmol/L of miR-140-5p mimic (Ribobio, Guangzhou, China), or 200 nmol/L of miR-140-5p inhibitor (Ribobio, Guangzhou, China) was used for cell transfection. The pcDNA 3.1 negative control (CASC19-p NC), pcNDA3.1-CASC19 (CASC19-p), CASC19 siRNAs (siCASC19), CEMIP siRNAs (siCEMIP), and pcNDA3.1-CEMIP (CEMIP-p) were purchased from Genechem (Shanghai, China). MiR-140-5p mimic, miR-140-5p inhibitor, mimic NC, and inhibitor NC were obtained from Ribobio (Guangzhou, China).
The luciferase reporter system was constructed by Genechem (Shanghai, China) as previously described[35]. Briefly, site-directed mutations were introduced into the CASC19 or CEMIP binding site of miR-140-5p (QuikChange Lightning Site-Directed Mutagenesis Kit, Stratagene, United States). The 3’-UTR fragment of wild-type (Wt) and mutant (Mut) CASC19 or CEMIP was then sub-cloned into the pGL3 luciferase vector (Promega, United States) by PCR, respectively. The miR-140-5p mimic was co-transfected with the vector into 293T cell lines for 12 h in 96-well plates using an enhanced infection solution (containing 5 μg/mL polybrene) (Genechem, Shanghai, China). The 293T cells were then cultured for 24 h and lysed to analyze the luciferase activity. Renilla (Promega, United States) activity was used as the internal control.
A flow cytometric technique was used to perform cell cycle analysis as detailed in a prior study[36]. In brief, HCT-116 and SW480 cells were harvested after transfection, and cell fixation was done with 70% pre-cooled ethanol at -20 °C overnight. Subsequently, cell staining was carried out using propidium iodide (PI, 50 μg/mL; Solarbio, Tianjin, China) and RNase A (0.1 mg/mL, Sigma, United States) at room temperature for 30 min. Then, the cells were analyzed with a flow cytometer (BD FACSCato II Flow Cytometer, BD Biosciences, Franklin Lakes, United States). All data pertaining to cell cycle studies are described in terms of the percentage of cell distributions in different phases (G0/G1, S, and G2/M).
The cell counting kit-8 (CCK-8) assay was used to assess the ability of SW480 and HCT-116 cells to proliferate. Cells (2 × 103) were added to each well of a 96-well plate with 100 μL of culture medium. CCK-8 solution (10 μL; Dojindo Laboratories, Japan) was applied into each well from day 0 to day 5. The cells were left to incubate for 3 h at 37 °C. A microplate luminometer (Bio-Rad Laboratories, United States) was then used to quantify absorbance values at 450 nm.
The abilities of cells to migrate and invade were assessed by wound-healing assay and Transwell assay with Matrigel (BD Bioscience, United States). Briefly, for the wound-healing assay, transfected cells were allowed to proliferate in 6-well plates until a confluent monolayer was achieved. The cell layer was gently scratched with a 200 μL pipette tip to inflict a wound. PBS was used to rinse the cells to remove debris and wound healing was allowed to take place for the next 24 h. The area of open wound covered by cells was described in terms of “percent wound closure (%)” [(Scratch distance0h - Scratch distance24h)/Scratch distance0h × 100%, the distances were measured with Image J]. For the transwell assay, 200 μL of medium without serum containing 1 × 105 cells was applied onto the upper chamber, while 600 μL of medium with 10% fetal bovine serum (FBS) was added to the bottom chamber. The cells were left to incubate for 24 h at 37 °C. The membranes were then removed and cells were fixated with 4% paraformaldehyde before undergoing a 10-min staining period with 0.1% crystal violet. An inverted microscope was used to count the total cell number.
For the apoptosis assay, 200 μL of PBS was used to suspend cells. PI and fluorescein isothiocyanate (FITC) conjugated Annexin V (BD Biosciences, Franklin Lakes, NJ, United States) were then added into the suspension and left to incubate for 20 minutes at room temperature. A flow cytometer (FACSVerse, BD Biosciences, San Diego, CA, United States) was used to analyze cells.
Total RNA was extracted with an RNA extraction kit (QIAGEN, Shanghai, China) and 1 μg of total RNA was added to a 20 μL system. The GoScript Reverse Transcription system (Promega, United States) was used for reverse transcription of CASC19 and CEMIP, and miR-140-5p was reverse transcribed by the stem-loop method (QIAGEN, Shanghai, China). GAPDH and U6 served as internal controls. The primer sequences are as follows: forward, 5'-GAGGAAGGCAGCACAATGATG-3' and reverse, 5'-CTTGCCAGTGTCTTCTCCTGA-3' for CASC19; forward, 5'-GCTCTGGGATTTAAGGCAGC-3' and reverse, 5'-ATTGGAGCCATGGACTGTGA-3' for CEMIP; forward, 5'-CAGTGGTTTTACCCTATGGTAG-3' and universal primer, 5’-TGGTGTCGTGGAGTCG-3’ for miR-140-5p; forward, 5'-TGCACCACCAACTGCTTAGC-3' and reverse 5'-GGCATGGACTGTGGTCATGAG-3' for GAPDH; forward, 5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3' for U6. All PCR reactions lasted 40 cycles, and the relative fold change of gene expression was derived using the 2-ΔΔCt method.
Cellular localization of CASC19 was determined by subcellular fractionation using the PARIS Kit (Thermo Fisher Scientific, United States). Briefly, cytoplasmic and nuclear RNA were first extracted. The expression of CASC19 in the cytoplasmic and nuclear fractions was determined by qRT-PCR. GAPDH and U6 served as the cytoplasmic and nuclear controls, respectively.
RIP assay (Magna RIPTM RNA Binding Protein Immunoprecipitation Kit, Millipore, United States) was utilized to verify the binding association between miR-140-5p and endogenous CASC19 in CRC cells. Briefly, harvested HCT-116 and SW480 cells were resuspended in cell RIP lysis buffer and kept on ice for 20 min. The RIP lysate was then centrifuged and 100 μL of the supernatant was removed and added to each bead-antibody [anti-Argonaute 2 (Ago2) or negative control IgG] complex in RIP immunoprecipitation buffer. All tubes were left to incubate overnight in a rotator at 4 °C. The next day, immunoprecipitation tubes were centrifuged briefly and placed on a magnetic separator to collect the bead-antibody-RNA complexes. Protein K was used to purify the RNA. Purified RNA was then analyzed by RT-PCR.
Specimens and CRC cells were lysed in RIPA to obtain total protein, and 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate 40 μg of each protein sample with the proteins subsequently reapplied to polyvinylidene fluoride membranes (Millipore, United States). These membranes containing blotted proteins were exposed to 0.5% bovine serum albumin to block endogenous reactions, followed by overnight probing at 4 °C with anti-CEMIP (1:1500, Novus, United States), anti-Vimentin (1:1500, CST, United States), anti-N-cadherin (1:1500, CST, United States), anti-E-cadherin [1:1500, Cell Signaling Technology (CST), United States], anti-poly AD-ribose polymerase (PARP) (1:2000, CST, United States), anti-caspase 3 (1:2000, CST, United States), anti-caspase 7 (1:2000, CST, United States), and anti-β-actin (1:3000, CST, United States) antibodies. The next day, membranes were rinsed and subjected to 2-h room temperature incubation with diluted secondary horseradish peroxidase (HRP)-marked antibodies (1:2500, CST, United States). An enhanced chemiluminescence detection kit (Millipore, United States) was used to detect immunoreactive protein bands. Images were acquired with a G-Box system (Syngene, United States) and analyzed with Image J (National Institute of Mental Health, United States).
Immunohistochemistry assay was used to determine CEMIP expression in CRC tissues and adjacent normal colonic mucosal tissues as described in our previous study[37]. Briefly, tissue samples were fixed in 4% paraformaldehyde, embedded in paraffin, and sliced into 4 μm sections. Dehydration was carried out with xylene and a gradient of ethanol solution before endogenous peroxidase blocking was carried out with a 3% H2O2 solution. Antigen retrieval was performed with heated sodium citrate solution (92 °C-95 °C, 10 nmol/L, pH 6.0) for 5 min and 1% goat serum was added to the sections at room temperature for 2 min to block the sections. Rabbit anti-human CEMIP antibody (1:150; CST, United States) was applied onto the sections and left overnight to incubate at 4 °C. After incubation with the secondary antibody for 1 h at RT, freshly prepared diaminobenzidine was then added to the sections that were then stained with hematoxylin. A light microscope (Leica Microsystems, Wetzlar, Germany) was used to visualize slides and brown particles in the cytoplasm or cytomembranes were taken to indicate positive staining.
All data are expressed in terms of the mean ± SD. Continuous data were analyzed using the t-test or the one-way analysis of variance (ANOVA). The relationship between CASC19 expression and clinicopathological features was determined by one-way analysis of variance, the Chi-squared test, or the Fisher’s exact test. The prognostic value of CASC19 in CRC patients was explored by Cox regression analysis. Kaplan-Meier methods and log-rank tests were used to carry out survival analyses. The correlations between CASC19, miR-140-5p, and CEMIP were analyzed using Spearman or Pearson correlation analysis. All statistical analyses were carried out with the use of GraphPad Prism (GraphPad Software, Version 5.0, United States) and SigmaPlot software (SPSS 22.0, United States). P-values < 0.05 were considered statistically significant. All results are representative of three repeated experiments.
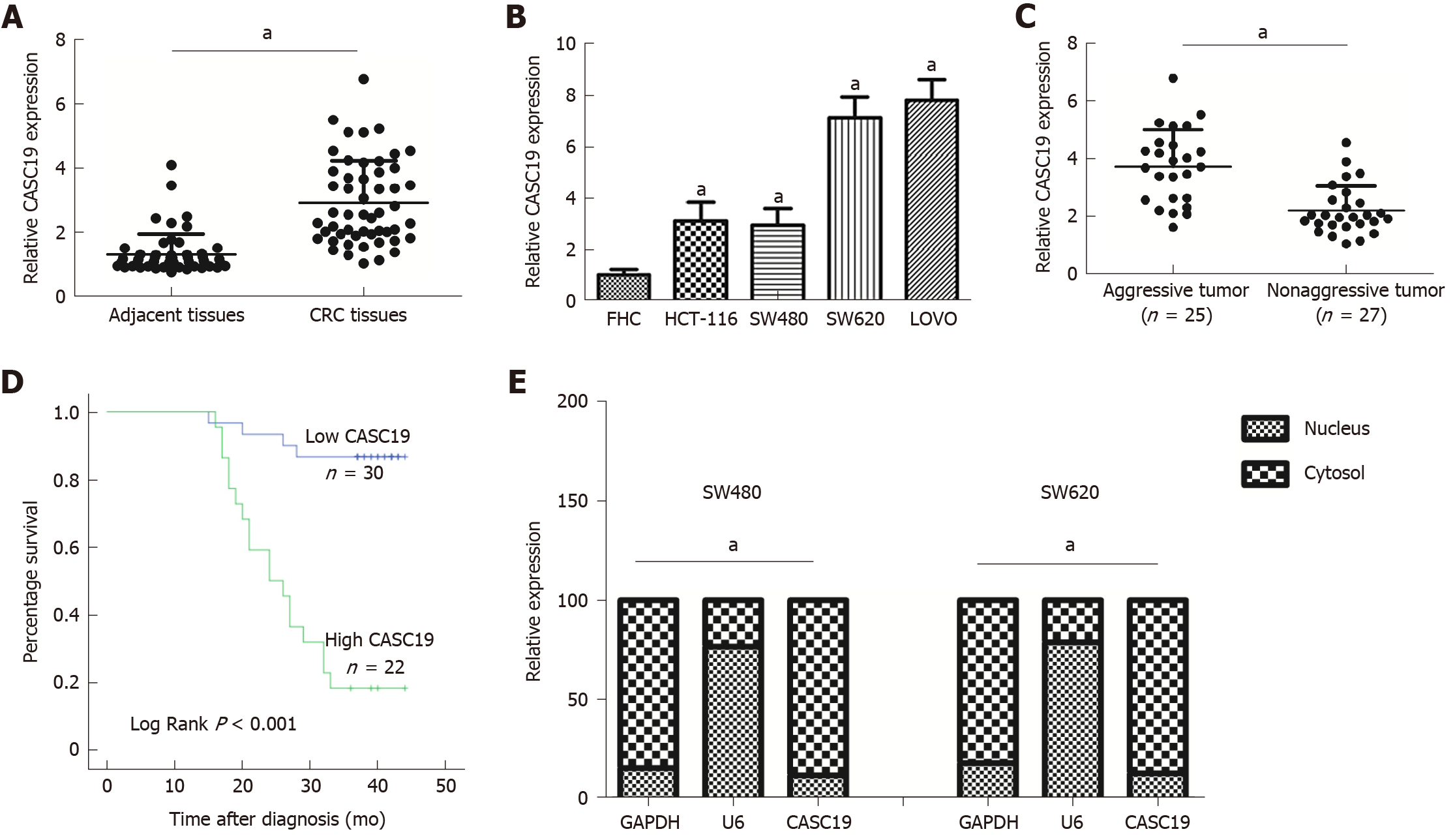
CASC19 expression was analyzed in 52 pairs of CRC tissues and adjacent normal colonic mucosal tissues using qRT-PCR. CASC19 exhibited significantly increased expression in CRC tissues in contrast to adjacent healthy colonic mucosal tissues (P < 0.05) (Figure 1A). Consistently, the CASC19 expression in CRC cell lines was markedly elevated in comparison to FHC cells (P < 0.05) (Figure 1B). For further in vitro studies, HCT-116 and SW480 cell lines were used.
We also focused on determining the association between expression of CASC19 and clinical phenotypes of patients with CRC. Participants were stratified into an aggressive tumor group (n = 25) and a non-aggressive tumor group (n = 27) based on whether the patient had or not lymph node or liver metastasis. Results showed that aggressive CRC tumors had higher CASC19 expression (P < 0.05) (Figure 1C). Next, we calculated the average of relative CASC19 expression levels in tumor tissues, and the average value (2.9310) was determined to be the cutoff point between “High CASC19” and “Low CASC19”. Results showed that 22 samples were included in the high CASC19 group while another 30 were included in the low CASC19 group. Subsequent correlation analysis revealed that CASC19 expression was associated with liver metastasis (P < 0.01), lymphatic metastasis (P < 0.01), and TNM stage (P < 0.05) (Table 1) (Supplementary Figure 1). Significant associations were not present between CASC19 expression and other clinical parameters such as tumor size, local invasion, age, and gender (P > 0.05) (Table 1). Kaplan-Meier survival curves demonstrated that patients who possessed raised expression of CASC19 experienced shorter survival durations in contrast to those who had lower CASC19 expression (log-rank P < 0.001) (Figure 1D). We then carried out multivariable Cox regression analysis to further determine the impact of clinicopathological features and expression of CASC19 on the overall survival times of patients with CRC. The findings indicated that CASC19 expression [the exponent of B (Exp (B) = 8.893, 95% confidence interval (CI): 2.596-30.464, P = 0.001] was an independent prognostic factor for CRC (Table 2).
| Feature | Total (n = 52) | CASC19 expression | P-value | |
| High | Low | |||
| Gender | ||||
| Male | 25 | 10 | 15 | 0.483 |
| Female | 27 | 12 | 15 | |
| Age, yr | ||||
| ≤ 60 | 13 | 5 | 8 | 0.503 |
| > 60 | 39 | 17 | 22 | |
| Tumor size | ||||
| ≤ 5 cm | 22 | 9 | 13 | 0.379 |
| > 5 cm | 30 | 13 | 17 | |
| TNM stage | ||||
| I-II | 23 | 6 | 17 | 0.019a |
| III-IV | 29 | 16 | 13 | |
| Local invasion | ||||
| T1 + T2 | 9 | 4 | 5 | 0.585 |
| T3 + T4 | 43 | 18 | 25 | |
| Lymphatic metastasis | ||||
| Yes | 22 | 14 | 8 | 0.008b |
| No | 30 | 8 | 22 | |
| Liver metastasis | ||||
| Yes | 18 | 13 | 5 | 0.002b |
| No | 34 | 9 | 25 | |
| Variable | Univariate analysis | Multivariate analysis | ||
| P-value | Exp(B) | 95%CI | P-value | |
| CASC19 | < 0.001b | 8.893 | 2.596-30.464 | 0.001a |
| Age, yr | 0.202 | - | - | - |
| Gender | 0.719 | - | - | - |
| Tumor size | 0.376 | - | - | - |
| TNM stage | 0.025a | - | - | 0.407 |
| Local invasion | 0.011a | - | - | 0.322 |
| Lymphatic metastasis | < 0.001b | 11.395 | 3.201-40.565 | < 0.001b |
| Local invasion and liver metastasis | < 0.001b | 7.274 | 2.382-22.211 | < 0.001b |
Moreover, the results of subcellular fractionation confirmed that CASC19 was predominantly localized in the cell plasma (Figure 1E). Taken together, our findings demonstrated that CASC19 was a highly expressed lncRNA in CRC and is highly involved in CRC metastasis.
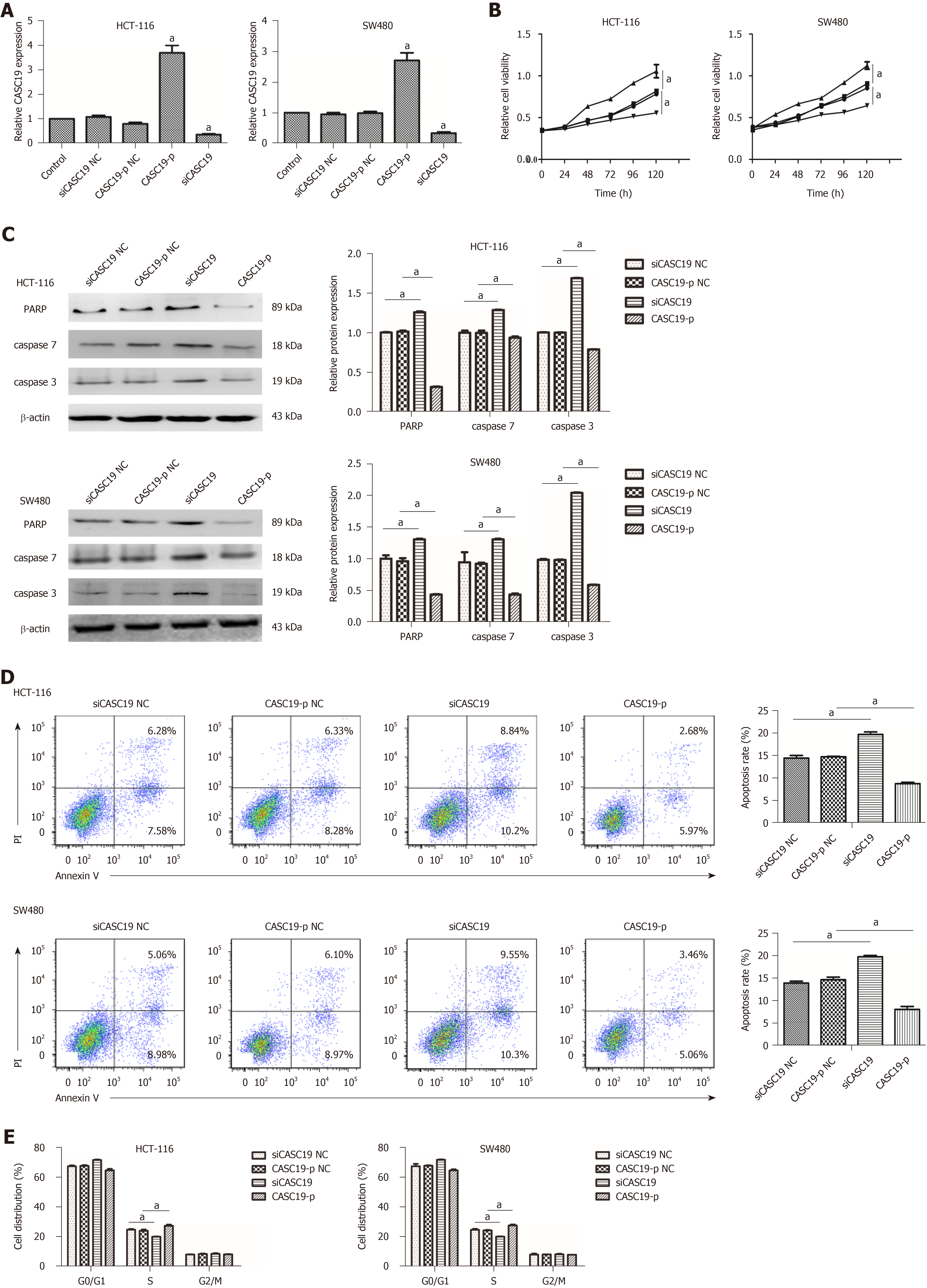
To explore the role of CASC19 in CRC cells, SW480 and HCT-116 cells were transfected with CASC19-p or siCASC19. CASC19 was increased significantly in the CASC19-p group (CASC19 overexpression) in contrast to the control group (P < 0.05) and was substantially attenuated in the siCASC19 group (CASC19 knockdown) (P < 0.05) (Figure 2A). CASC19-p NC or siCASC19 NC did not alter CASC19 expression in CRC cells (Figure 2A).
Additionally, the CCK-8 assay was used to assess the impact of CASC19 on the proliferation of SW480 and HCT-116 cells. Figure 2B demonstrates that HCT-116 and SW480 cells experienced augmented proliferation in the presence of CASC19 overexpression (P < 0.05). Conversely, knockdown of CASC19 inhibited HCT-116 and SW480 cell proliferation (P < 0.05).
Interestingly, high CASC19 expression in CRC cells led to markedly decreased caspase-3, caspase-7, and PARP (P < 0.05 for all) (Figure 2C). Overexpression of CASC19 decreased the apoptotic rate of CRC cells (P < 0.05) (Figure 2D), while CASC19 knockdown enhanced caspase-7, caspase-3, and PARA protein levels. Furthermore, CASC19 knockdown markedly elevated the percentage of apoptotic cells (P < 0.05 for all) (Figure 2D). Cell cycle assay revealed that CASC19 overexpression led to a higher proportion of SW480 and HCT-116 cells that were in the S phase. This proportion was significantly reduced in cells that had suppressed CASC19 levels. It can be concluded that CASC19 promoted CRC cell growth, while knockdown of CASC19 expression could impede CRC cell growth.
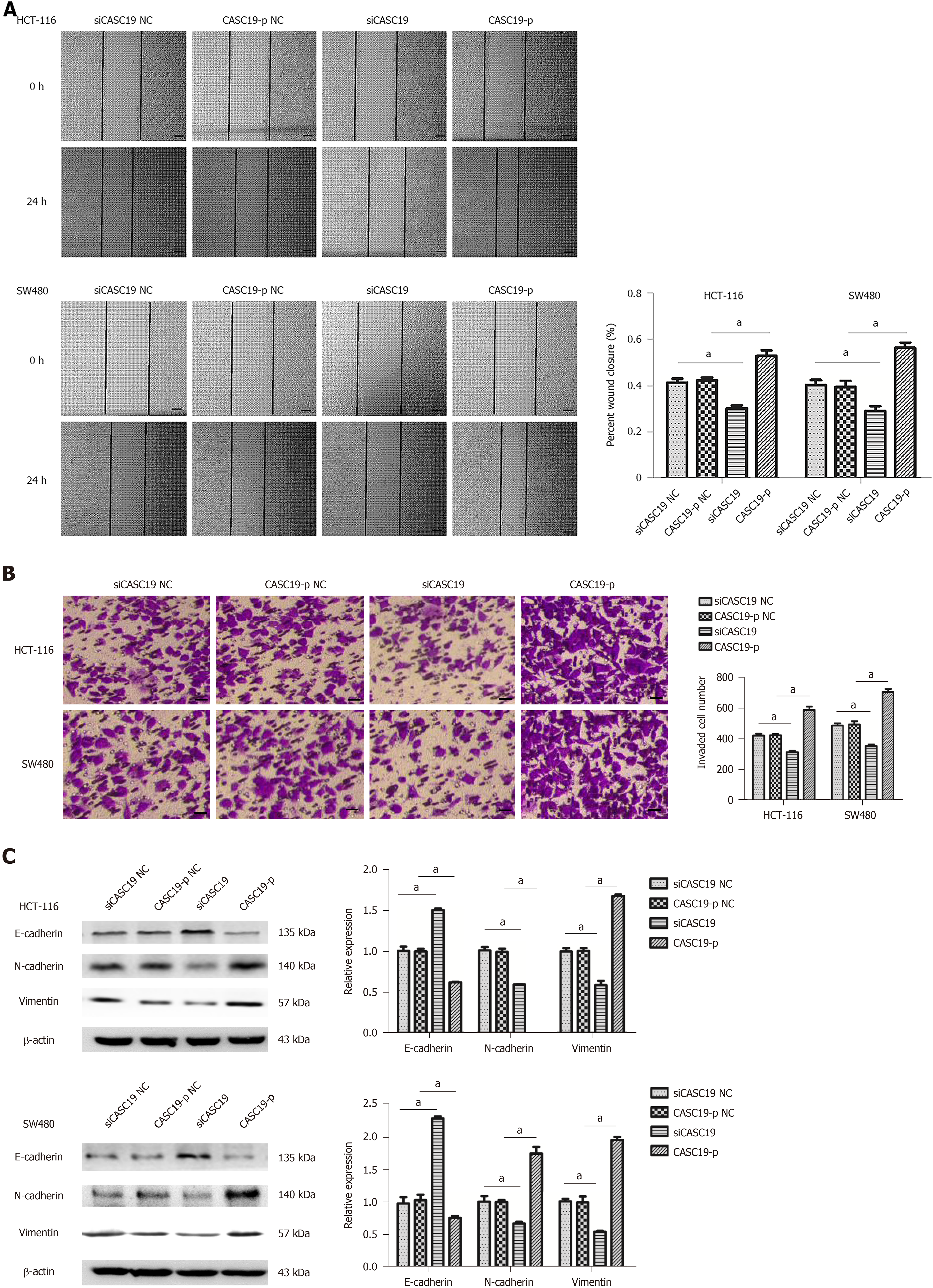
The wound-healing and transwell assays were used to determine the impact of CASC19 expression on CRC cell migration and invasion, respectively. Cells transfected with CASC19-p completed wound healing at a faster rate in comparison to cells that were transfected with CASC19-p NC (P < 0.05) (Figure 3A). HCT-116 and SW480 cells that were induced to overexpress CASC19 were observed to undergo higher invasion rates (Figure 3B). Whereas, the migration and invasion abilities were inhibited in cells that had CASC19 knockdown (Figure 3A and B) (P < 0.05).
Given the crucial function of epithelial-mesenchymal transition (EMT) in the malignant transformation and invasive properties of cancer cells, we sought to clarify the impact of CASC19 on EMT marker expression in CRC cell lines. E-cadherin was downregulated (P < 0.05), while Vimentin and N-cadherin were upregulated (P < 0.05 for all) in HCT-116 and SW480 cell lines that had upregulated CASC19 (Figure 3C). In contrast, E-cadherin expression was increased (P < 0.05), and the expression of Vimentin and N-cadherin was decreased (P < 0.05 for all) in HCT-16 and SW480 cells that had suppressed CASC19 levels (Figure 3C). Together, these data indicate that CASC19 promotes CRC cell metastasis.
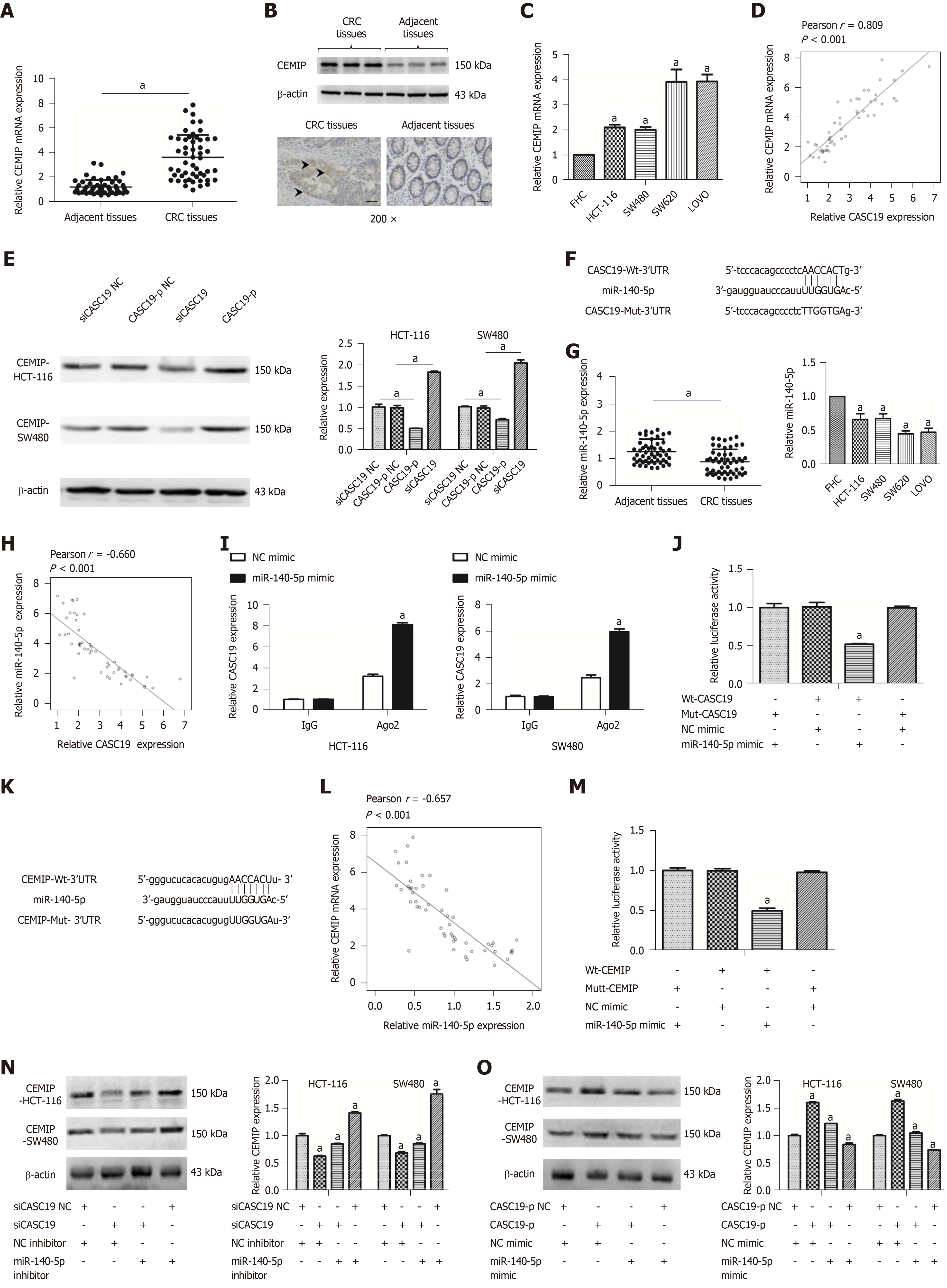
To understand how CASC19 affects the CRC cell migration and proliferation, a ceRNA network analysis was carried out using starBase v 2.0 (http://starbase.sysu.edu.cn/index.php)[38,39]. Interestingly, CASC19 was positively correlated with CEMIT expression. We examined the protein and mRNA levels of CEMIP in CRC cell lines and tissues. CEMIP expression was augmented in CRC tissues in contrast to healthy normal colonic mucosa at both the mRNA and protein levels (P < 0.05 for all) (Figure 4A and B). Similarly, the mRNA levels of CEMIP in CRC cells lines was upregulated in contrast to levels found in FHC cells (P < 0.05) (Figure 4C). Furthermore, Spearman’s correlation analysis revealed CASC19 expression was positively correlated CEMIT mRNA expression in CRC tissues (Pearson r = 0.809, P < 0.001) (Figure 4D). Additionally, we examined the effect of CASC19 on CEMIP expression in SW480 and HCT-116 cell lines. The mRNA and protein levels of CEMIP were upregulated after CASC19 overexpression in both cell lines, while suppression of CASC19 expression decreased CEMIP protein and mRNA levels in the two cell lines (P < 0.05 for all).
It has been demonstrated that lncRNAs are able to bind directly to miRNA and function as ceRNAs[40]. We next predicted miR-140-5p to be a potential target of CASC19 by using starBase v 2.0 program (http://starbase.sysu.edu.cn/index.php)[38,39] (Figure 4F). We went on to examine the expression of miR-140-5p in CRC cell lines and tissues. CRC tissue samples were found to possess a much lower miR-140-5p level in contrast to adjacent healthy colonic mucosal tissues (P < 0.05) (Figure 4G).
Besides, miR-140-5p was suppressed in CRC cell lines in contrast to FHC cells (P < 0.05) (Figure 4G). CASC19 and miR-140-5p expression appeared to negatively correlate in CRC tissues, based on Spearman’s correlation analysis (Pearson r = -0.660, P < 0.001) (Figure 4H). The anti-Ago2 RIP assay was performed to verify the presence of direct binding between CASC19 and miR-140-5p. Endogenous CASC19 was specifically enriched in miR-140-5p mimic transfected CRC cells in contrast to the NC mimic group (P < 0.05) (Figure 4I). A luciferase reporter assay was carried out to determine if CASC19 could directly target miR-140-5p. The luciferase activity was reduced in wild-type CASC19 3’-UTR and miR-140-5p mimic co-transfected 293T cells in comparison to wild-type CASC19 3’-UTR and miR-140-5p mimic NC co-transfected 293T cells (P < 0.05) (Figure 4J).
Interestingly, CEMIP is a potential target gene of miR-140-5p [predicted by TargetScan (http://www.targetscan.org/vert_71/) and miRBase (http://www.mirbase.org/)] (Figure 4K). Spearman’s correlation analysis showed that CEMIP correlated negatively with miR-140-5p in CRC tissues (Pearson r = -0.657, P < 0.001) (Figure 4L). CEMIP is a direct target of miR-140-5p, as revealed by luciferase reporter assays (Figure 4M).
Furthermore, it was suggested that the CEMIP protein expression was downregulated in siCASC19 and NC inhibitor co-transfected CRC cells, a phenomenon which could be reversed with the addition of miR-140-5p inhibitor (P < 0.05) (Figure 4N). CEMIP expression was markedly raised after transfecting CRC cells with CASC19-p and NC mimic. Moreover, the effects of CASC19 were reversed by the addition of miR-140-5p mimic to CRC cell lines (P < 0.05) (Figure 4O). Collectively, these finding indicate that CASC19 is able to function as a ceRNA and indirectly regulate CEMIP expression by sponging miR-140-5p.
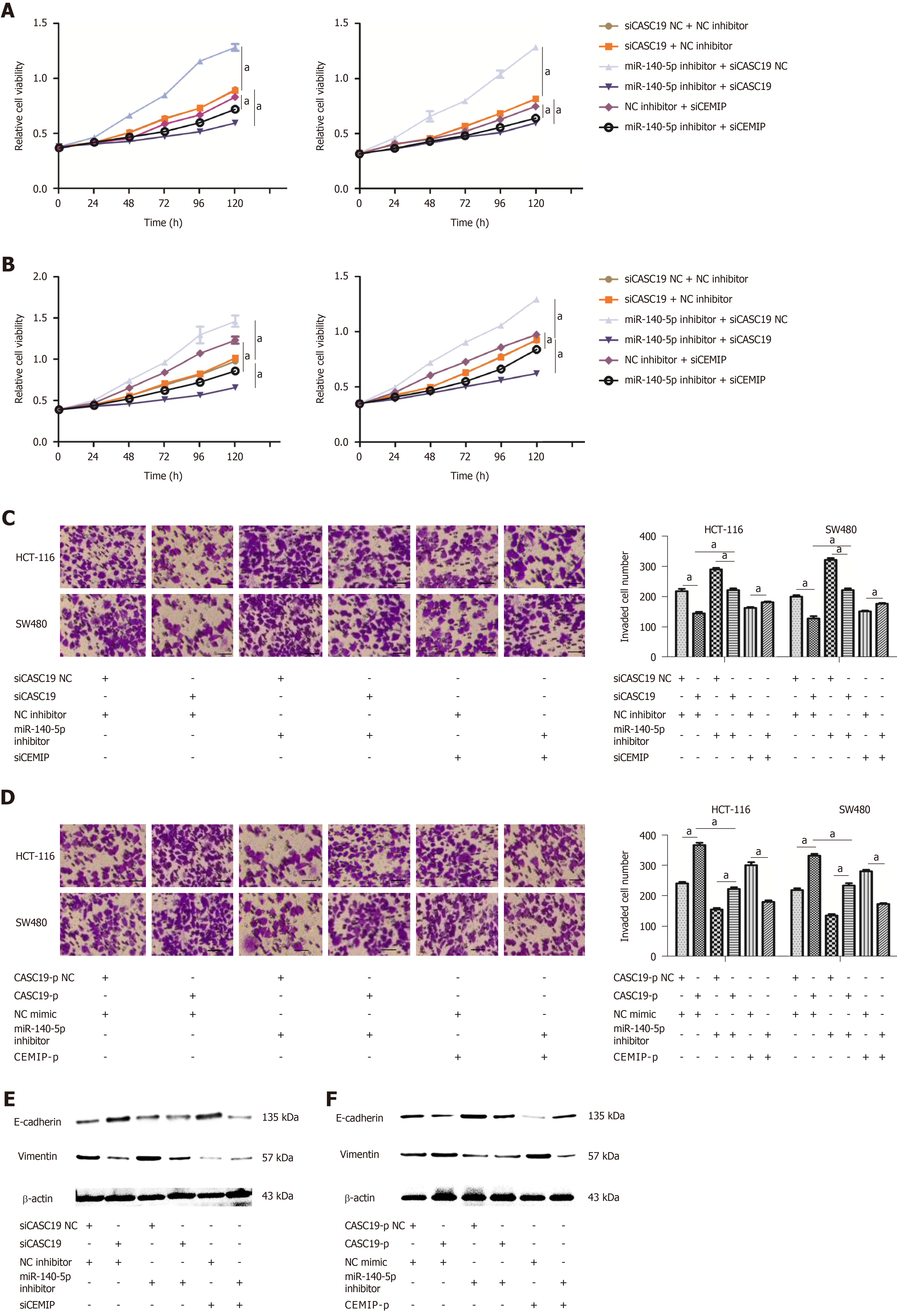
To confirm whether CASC19 exerts its function through sponging miR-140-5p in CRC tissues, CCK-8 and transwell assays were performed. Results demonstrated that knockdown of CASC19 or CEMIP expression significantly inhibited the proliferation and invasion of HCT-116 and SW480 cells (P < 0.05) (Figure 5A-D). The addition of miR-140-5p inhibitor reversed this effect. Moreover, miR-140-5p inhibitor was able to promote the proliferation and invasion abilities of cells. Conversely, overexpression of CASC19 or CEMIP in the cell lines promoted the proliferative and invasive capacities of HCT-116 and SW480 cells, with this effect reversed by miR-140-5p mimic. Furthermore, miR-140-5p alone significantly inhibited the proliferative and invasive capacities of both cell lines (P < 0.05) (Figure 5A-D). Lastly, the expression of E-cadherin and vimentin in transfected CRC cells was analyzed. Consistent with the previous results, the effect of knockdown or overexpression of CASC19 on the expression of the markers was reversed by miR-140-5p inhibitor or miR-140-5p mimic (Figure 5E and F). These data suggested that miR-140-5p is able to counteract the effect of CASC19 on the growth and metastasizing abilities of CRC cells in vitro.
Studies have confirmed that dysregulation of lncRNAs is closely related to the occurrence and development of various tumors. LncRNAs may become potential targets for the development of future chemotherapeutic medications[41,42]. Therefore, studying the clinical significance and function of lncRNA along with its possible mechanisms of action may contribute towards the gene therapy in cancer. Recently, researchers have demonstrated that lncRNAs are involved in the progression of CRC. For instance, the lncRNA SUMO1P3 drives CRC growth and metastasis[16,43,44]. Lymph node metastasis and poor CRC prognosis have been linked to the upregulation of the lncRNA BANCR[43]. The lncRNA HAND2-AS1 sponging miR-1275 suppresses CRC progression by upregulating KLF14[44]. Previous studies demonstrate that the lncRNA CASC19 may promote the progression of several cancers, and it has been suggested that CASC19 may function as a biomarker for CRC poor prognosis[24,45]. Therefore, it is necessary to study the function of CASC19 in CRC. In our study, it was observed that the expression of CASC19 was greatly raised in both CRC cell lines and tissue samples. This result is consistent with previous results, which suggests that CASC19 may be an important lncRNA that regulates CRC heterogeneity. Given that the cellular location of CASC19 was related to lncRNA function, we examined the subcellular distribution of CASC19 using cellular fractionation. Cellular localization showed that CASC19 was mostly located in the cellular plasma, indicating that CASC19 may function to sponge miRNA.
Heterogeneity is a key property that is responsible for malignant processes such as initiation, migration, and metastasis of cancer. One example is the lncRNA HOTAIR that may promote the progression of a variety of cancers by affecting the proliferation, migration, and invasion of various cancer cells[46]. Knockdown of the lncRNA LCPAT1 inhibits autophagy in lung cancer[47]. The lncRNA PVT1 may promote ovarian cancer progression[48]. These data suggest that lncRNAs are emerging as crucial cancer regulators. Correlation analysis showed that high expression of CASC19 was associated with CRC metastasis. Thus, we speculated that CASC19 may be correlated with CRC progression. As we expected, in vitro experiments showed that CASC19 promoted CRC progression.
It has been demonstrated that lncRNAs may function as ceRNAs by sponging miRNAs[49]. For example, HOTAIR promotes renal cell carcinoma progression via absorbing miR-138[50]. The lncRNA SNHG7 sponges miR-216b to upregulate GALNT1, leading ultimately to the promotion of CRC cell proliferation and liver metastasis[51]. Previously, miR-140-5p was observed to be expressed abnormally in several malignancies including CRC[52], breast cancer[53], ovarian cancer[54], glioma[55], and gastric cancer[56]. Similar to previous studies, our study revealed that CASC19 may promote CRC by targeting miR-140-5p. The current investigations revealed that CASC19 directly targets miR-140-5p. Importantly, there was an endogenous interaction between these two molecules in CRC cells. Moreover, inhibiting proliferation and invasion and promoting apoptosis of cancer cells are known to involve miR-140-5p[55,57]. These studies suggest that miR-140-5p may play a tumor suppressive role in different tumors. However, literature is scarce regarding its role in CRC. Our results demonstrated that miR-140-5p was markedly reduced in CRC cell lines and tissue samples. In addition, miR-140-5p reversed the function of CASC19 in CRC cell lines. These results suggest that CASC19 sponging of miR-140-5p may be a mechanism regulating the heterogeneity of CRC. Given that miRNAs are known to simultaneously control the expression of a broad range of genes, our working hypothesis was that miR-140-5p may target different genes in different cell lines, resulting in varying phenotypes. Therefore, we examined the target genes predicted and found out that CEMIP may be a target gene of miR-140-5p in CRC.
Tumor malignancy is driven by its almost limitless capability for proliferation and metastasis. CEMIP was initially known as an enzyme to depolymerize hyaluronic acid. Further studies reveal CEMIP overexpression to be a common phenomenon in several tumors. For instance, CEMIP drives breast cancer progression and predicts poor prognosis in pancreatic ductal adenocarcinoma[31,32]. In CRC, tumor metastasis and invasion are able to be suppressed through the miR-216a-mediated attenuation of CEMIP[58]. CEMIP therefore may be associated with the progression of CRC. Previous studies have demonstrated that CEMIP may interact with the glycogen phosphorylase kinase β-subunit (PHKB) to promote cancer cell survival by breakdown of glycogen[59]. In CRC tissues, the tumor cells are exposed to a nutrient deprivation and hypoxia environment[60]. Energy deprivation is one of the main courses of cell death. It has been demonstrated that glycogen accumulation contributes to cell survival when cancer cells expose to hypoxia[61-63]. At the same time, glycogen lysis is an important process for cell survival. CEMIP overexpressing tumor cells have relatively low glycogen content, while the survival rate of these cells is relatively high. CEMIP may play a vital role in CRC survival and metastasis. Many differing miR-140-5p targets, such as YES1[56], Wnt1[64], and VEGFA[65], have been documented to be associated with cell invasion and migration. In our study, we observed that CASC19 functions as a ceRNA, controlling the availability of miR-140-5p that can be acted on by the CEMIP gene. The current investigations focused on CEMIP, which was found to have strong associations with CASC19 and cancer cell survival. Our results strongly suggest that CASC19 may regulate CEMIP expression via sponging of miR-140-5p.
In conclusion, CASC19 overexpression augmented CRC cell proliferation and metastasizing abilities in vitro. Our study is the first to report a novel mechanism for CASC19 in CRC. CASC19 exerted its effects partially through the CASC19/miR-140-5p/CEMIP axis. Investigation of this CASC19/miR-140-5p/CEMIP pathway allows a deeper understanding of the pathogenesis of CRC, with the members of this pathway serving as useful targets for future preventive and therapeutic innovations in the management of CRC.
There is increasing evidence that long non-coding RNAs (lncRNAs) play an important role in tumor progression. The lncRNA cancer susceptibility 19 (CASC19) is highly expressed in colorectal cancer (CRC) tissues, and may be associated with colon cancer progression. However, there has been no experimental evidence regarding the function of CASC19 in CRC.
Investigation of the functions of the lncRNA CASC19 may suggest potential molecular mechanisms of CRC carcinogenesis and progression, and may further offer the potential for future preventive and therapeutic innovations in the management of CRC.
We measured the lncRNA CASC19 expression in CRC tissues and pair-matched adjacent non-tumor tissues, and investigated biological functions and the possible molecular mechanisms of CASC19 in CRC.
We detected CASC19 expression in CRC tissues and pair-matched adjacent non-tumor tissues using quantitative real-time PCR. The biological behavior of CASC19 in vitro was then assessed by overexpression and knockdown of the expression of CASC19. In further molecular mechanism studies, we confirmed the possible molecular mechanisms by which CASC19 plays a role by overexpression and knockdown of the expression of downstream molecules of CASC19.
The lncRNA CASC19 was upregulated in CRC tissue, which was related to tumor stage and metastasis. The lncRNA CASC19 had a role of promoting proliferation, metastasizing ability, and epithelial-mesenchymal transition, and inhibiting apoptosis by regulating microRNA 140-5p/cell migration inducing hyaluronidase 1 (miR-140-5p/CEMIP) expression in CRC cells.
The lncRNA CASC 19 is overexpressed in CRC tissues and cell lines, which could promote CRC progression through regulating miR-140-5p/CEMIP.
Members of the CASC19/miR-140-5p/CEMIP axis may serve as useful targets for future preventive and therapeutic innovations in the management of CRC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Leon J, Linnebacher M, Serafino A S-Editor: Ma RY L-Editor: Wang TQ E-Editor: Song H
| 1. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11573] [Cited by in RCA: 13112] [Article Influence: 1873.1] [Reference Citation Analysis (4)] |
| 2. | Garden OJ, Rees M, Poston GJ, Mirza D, Saunders M, Ledermann J, Primrose JN, Parks RW. Guidelines for resection of colorectal cancer liver metastases. Gut. 2006;55 Suppl 3:iii1-iii8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 205] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 3. | Gallagher DJ, Kemeny N. Metastatic colorectal cancer: from improved survival to potential cure. Oncology. 2010;78:237-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 4. | Liang C, Sun W, He H, Zhang B, Ling C, Wang B, Huang T, Hou B, Guo Y. Antitumor effect of a new nano-vector with miRNA-135a on malignant glioma. Int J Nanomedicine. 2017;13:209-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 5. | Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 544] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 6. | Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435-1439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 925] [Cited by in RCA: 997] [Article Influence: 76.7] [Reference Citation Analysis (0)] |
| 7. | Li S, Wang Q, Qiang Q, Shan H, Shi M, Chen B, Zhao S, Yuan L. Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol. 2015;141:1909-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Xue WH, Fan ZR, Li LF, Lu JL, Ma BJ, Kan QC, Zhao J. Construction of an oesophageal cancer-specific ceRNA network based on miRNA, lncRNA, and mRNA expression data. World J Gastroenterol. 2018;24:23-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:205-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 10. | Xiao JN, Yan TH, Yu RM, Gao Y, Zeng WL, Lu SW, Que HX, Liu ZP, Jiang JH. Long non-coding RNA UCA1 regulates the expression of Snail2 by miR-203 to promote hepatocellular carcinoma progression. J Cancer Res Clin Oncol. 2017;143:981-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 11. | Wang BG, Xu Q, Lv Z, Fang XX, Ding HX, Wen J, Yuan Y. Association of twelve polymorphisms in three onco-lncRNA genes with hepatocellular cancer risk and prognosis: A case-control study. World J Gastroenterol. 2018;24:2482-2490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Zheng S, Zheng D, Dong C, Jiang J, Xie J, Sun Y, Chen H. Development of a novel prognostic signature of long non-coding RNAs in lung adenocarcinoma. J Cancer Res Clin Oncol. 2017;143:1649-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W, Zhang P, Qian H, Jiang PC, Xu WR, Zhang X. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 262] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 14. | Lu M, Liu Z, Li B, Wang G, Li D, Zhu Y. The high expression of long non-coding RNA PANDAR indicates a poor prognosis for colorectal cancer and promotes metastasis by EMT pathway. J Cancer Res Clin Oncol. 2017;143:71-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 15. | Ping G, Xiong W, Zhang L, Li Y, Zhang Y, Zhao Y. Silencing long noncoding RNA PVT1 inhibits tumorigenesis and cisplatin resistance of colorectal cancer. Am J Transl Res. 2018;10:138-149. [PubMed] |
| 16. | Zhang LM, Wang P, Liu XM, Zhang YJ. LncRNA SUMO1P3 drives colon cancer growth, metastasis and angiogenesis. Am J Transl Res. 2017;9:5461-5472. [PubMed] |
| 17. | Yang S, Sun Z, Zhou Q, Wang W, Wang G, Song J, Li Z, Zhang Z, Chang Y, Xia K, Liu J, Yuan W. MicroRNAs, long noncoding RNAs, and circular RNAs: potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res. 2018;10:2249-2257. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 18. | Liang Y, Zhang C, Ma MH, Dai DQ. Identification and prediction of novel non-coding and coding RNA-associated competing endogenous RNA networks in colorectal cancer. World J Gastroenterol. 2018;24:5259-5270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Fan H, Lv P, Mu T, Zhao X, Liu Y, Feng Y, Lv J, Liu M, Tang H. LncRNA n335586/miR-924/CKMT1A axis contributes to cell migration and invasion in hepatocellular carcinoma cells. Cancer Lett. 2018;429:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Zhou Y, Dai W, Wang H, Pan H, Wang Q. Long non-coding RNA CASP5 promotes the malignant phenotypes of human glioblastoma multiforme. Biochem Biophys Res Commun. 2018;500:966-972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 21. | He Z, You C, Zhao D. Long non-coding RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated stromal cells-mediated glycolysis and invasion of glioma cells. Biochem Biophys Res Commun. 2018;500:569-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 22. | Yu X, Zhao J, He Y. Long non-coding RNA PVT1 functions as an oncogene in human colon cancer through miR-30d-5p/RUNX2 axis. J BUON. 2018;23:48-54. [PubMed] |
| 23. | Wang JJ, Li XM, He L, Zhong SZ, Peng YX, Ji N. Expression and Function of Long Non-coding RNA CASC19 in Colorectal Cancer. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2017;39:756-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 24. | Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G, Goel A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 'gene desert', serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28:1882-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 144] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 25. | Yamada A, Yu P, Lin W, Okugawa Y, Boland CR, Goel A. A RNA-Sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci Rep. 2018;8:575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 26. | Abe S, Usami S, Nakamura Y. Mutations in the gene encoding KIAA1199 protein, an inner-ear protein expressed in Deiters' cells and the fibrocytes, as the cause of nonsyndromic hearing loss. J Hum Genet. 2003;48:564-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Yoshida H, Nagaoka A, Kusaka-Kikushima A, Tobiishi M, Kawabata K, Sayo T, Sakai S, Sugiyama Y, Enomoto H, Okada Y, Inoue S. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci USA. 2013;110:5612-5617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 217] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 28. | Zhang P, Song Y, Sun Y, Li X, Chen L, Yang L, Xing Y. AMPK/GSK3β/β-catenin cascade-triggered overexpression of CEMIP promotes migration and invasion in anoikis-resistant prostate cancer cells by enhancing metabolic reprogramming. FASEB J. 2018;32:3924-3935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 29. | Matsuzaki S, Tanaka F, Mimori K, Tahara K, Inoue H, Mori M. Clinicopathologic significance of KIAA1199 overexpression in human gastric cancer. Ann Surg Oncol. 2009;16:2042-2051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | Fink SP, Myeroff LL, Kariv R, Platzer P, Xin B, Mikkola D, Lawrence E, Morris N, Nosrati A, Willson JK, Willis J, Veigl M, Barnholtz-Sloan JS, Wang Z, Markowitz SD. Induction of KIAA1199/CEMIP is associated with colon cancer phenotype and poor patient survival. Oncotarget. 2015;6:30500-30515. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 31. | Koga A, Sato N, Kohi S, Yabuki K, Cheng XB, Hisaoka M, Hirata K. KIAA1199/CEMIP/HYBID overexpression predicts poor prognosis in pancreatic ductal adenocarcinoma. Pancreatology. 2017;17:115-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 32. | Shostak K, Zhang X, Hubert P, Göktuna SI, Jiang Z, Klevernic I, Hildebrand J, Roncarati P, Hennuy B, Ladang A, Somja J, Gothot A, Close P, Delvenne P, Chariot A. NF-κB-induced KIAA1199 promotes survival through EGFR signalling. Nat Commun. 2014;5:5232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 33. | Evensen NA, Kuscu C, Nguyen HL, Zarrabi K, Dufour A, Kadam P, Hu YJ, Pulkoski-Gross A, Bahou WF, Zucker S, Cao J. Unraveling the role of KIAA1199, a novel endoplasmic reticulum protein, in cancer cell migration. J Natl Cancer Inst. 2013;105:1402-1416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 34. | Liang G, Fang X, Yang Y, Song Y. Knockdown of CEMIP suppresses proliferation and induces apoptosis in colorectal cancer cells: downregulation of GRP78 and attenuation of unfolded protein response. Biochem Cell Biol. 2018;96:332-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 35. | Ding M, Bowman L, Castranova V. Luciferase reporter system for studying the effect of nanoparticles on gene expression. Methods Mol Biol. 2012;906:403-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 36. | Mattheolabakis G, Wang R, Rigas B, Mackenzie GG. Phospho-valproic acid inhibits pancreatic cancer growth in mice: enhanced efficacy by its formulation in poly-(L)-lactic acid-poly(ethylene glycol) nanoparticles. Int J Oncol. 2017;51:1035-1044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 37. | Wang X, Lu J, Cao J, Ma B, Gao C, Qi F. MicroRNA-18a promotes hepatocellular carcinoma proliferation, migration, and invasion by targeting Bcl2L10. Onco Targets Ther. 2018;11:7919-7934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 38. | Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92-D97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2607] [Cited by in RCA: 4037] [Article Influence: 336.4] [Reference Citation Analysis (0)] |
| 39. | Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011;39:D202-D209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 565] [Cited by in RCA: 674] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 40. | Zhou X, Gao Q, Wang J, Zhang X, Liu K, Duan Z. Linc-RNA-RoR acts as a "sponge" against mediation of the differentiation of endometrial cancer stem cells by microRNA-145. Gynecol Oncol. 2014;133:333-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 143] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 41. | Li Y, Duo Y, Bi J, Zeng X, Mei L, Bao S, He L, Shan A, Zhang Y, Yu X. Targeted delivery of anti-miR-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy. Int J Nanomedicine. 2018;13:1241-1256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 42. | Küçüktürkmen B, Bozkır A. Development and characterization of cationic solid lipid nanoparticles for co-delivery of pemetrexed and miR-21 antisense oligonucleotide to glioblastoma cells. Drug Dev Ind Pharm. 2018;44:306-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 43. | Shen X, Bai Y, Luo B, Zhou X. Upregulation of lncRNA BANCR associated with the lymph node metastasis and poor prognosis in colorectal cancer. Biol Res. 2017;50:32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 44. | Zhou J, Lin J, Zhang H, Zhu F, Xie R. LncRNA HAND2-AS1 sponging miR-1275 suppresses colorectal cancer progression by upregulating KLF14. Biochem Biophys Res Commun. 2018;503:1848-1853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 80] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 45. | Teerlink CC, Leongamornlert D, Dadaev T, Thomas A, Farnham J, Stephenson RA, Riska S, McDonnell SK, Schaid DJ, Catalona WJ, Zheng SL, Cooney KA, Ray AM, Zuhlke KA, Lange EM, Giles GG, Southey MC, Fitzgerald LM, Rinckleb A, Luedeke M, Maier C, Stanford JL, Ostrander EA, Kaikkonen EM, Sipeky C, Tammela T, Schleutker J, Wiley KE, Isaacs SD, Walsh PC, Isaacs WB, Xu J, Cancel-Tassin G, Cussenot O, Mandal D, Laurie C, Laurie C, Thibodeau SN, Eeles RA, Kote-Jarai Z, Cannon-Albright L; PRACTICAL consortium; International Consortium for Prostate Cancer Genetics. Genome-wide association of familial prostate cancer cases identifies evidence for a rare segregating haplotype at 8q24.21. Hum Genet. 2016;135:923-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 46. | Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1-9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 236] [Reference Citation Analysis (0)] |
| 47. | Yu X, Ye X, Lin H, Feng N, Gao S, Zhang X, Wang Y, Yu H, Deng X, Qian B. Knockdown of long non-coding RNA LCPAT1 inhibits autophagy in lung cancer. Cancer Biol Med. 2018;15:228-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 48. | Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. 2018;15:238-250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 49. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5512] [Article Influence: 393.7] [Reference Citation Analysis (0)] |
| 50. | Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, Chang C, Qi J, Yeh S. Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37:5037-5053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 51. | Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges miR-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9:722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 52. | Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6:19735-19746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 109] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 53. | Güllü G, Peker I, Haholu A, Eren F, Küçükodaci Z, Güleç B, Baloglu H, Erzik C, Özer A, Akkiprik M. Clinical significance of miR-140-5p and miR-193b expression in patients with breast cancer and relationship to IGFBP5. Genet Mol Biol. 2015;38:21-29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Lan H, Chen W, He G, Yang S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother. 2015;75:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 55. | Cui Y, Yi L, Zhang MM, Zhou Y, Tan ZG, Huang T, Jiang YG. WITHDRAWN: Knockdown of LncRNA HOXA11 Antisense Promotes Glioma Cell Apoptosis Via Sponging MiR-140-5p. Oncol Res. 2017;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 56. | Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y, Zhang T, Khaliq J, Li Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 119] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 57. | Sun Y, Qin B. Long noncoding RNA MALAT1 regulates HDAC4-mediated proliferation and apoptosis via decoying of miR-140-5p in osteosarcoma cells. Cancer Med. 2018;7:4584-4597. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 58. | Zhang D, Zhao L, Shen Q, Lv Q, Jin M, Ma H, Nie X, Zheng X, Huang S, Zhou P, Wu G, Zhang T. Down-regulation of KIAA1199/CEMIP by miR-216a suppresses tumor invasion and metastasis in colorectal cancer. Int J Cancer. 2017;140:2298-2309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 74] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 59. | Terashima M, Fujita Y, Togashi Y, Sakai K, De Velasco MA, Tomida S, Nishio K. KIAA1199 interacts with glycogen phosphorylase kinase β-subunit (PHKB) to promote glycogen breakdown and cancer cell survival. Oncotarget. 2014;5:7040-7050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918-4925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 772] [Article Influence: 48.3] [Reference Citation Analysis (1)] |
| 61. | Pelletier J, Bellot G, Gounon P, Lacas-Gervais S, Pouysségur J, Mazure NM. Glycogen Synthesis is Induced in Hypoxia by the Hypoxia-Inducible Factor and Promotes Cancer Cell Survival. Front Oncol. 2012;2:18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 151] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 62. | Pescador N, Villar D, Cifuentes D, Garcia-Rocha M, Ortiz-Barahona A, Vazquez S, Ordoñez A, Cuevas Y, Saez-Morales D, Garcia-Bermejo ML, Landazuri MO, Guinovart J, del Peso L. Hypoxia promotes glycogen accumulation through hypoxia inducible factor (HIF)-mediated induction of glycogen synthase 1. PLoS One. 2010;5:e9644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 195] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 63. | Shen GM, Zhang FL, Liu XL, Zhang JW. Hypoxia-inducible factor 1-mediated regulation of PPP1R3C promotes glycogen accumulation in human MCF-7 cells under hypoxia. FEBS Lett. 2010;584:4366-4372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 64. | Wu D, Zhang J, Lu Y, Bo S, Li L, Wang L, Zhang Q, Mao J. miR-140-5p inhibits the proliferation and enhances the efficacy of doxorubicin to breast cancer stem cells by targeting Wnt1. Cancer Gene Ther. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 65. | Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G, Hou Y, Jiang P. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37:1123-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |