Published online Aug 14, 2018. doi: 10.3748/wjg.v24.i30.3448
Peer-review started: May 4, 2018
First decision: May 23, 2018
Revised: June 8, 2018
Accepted: June 25, 2018
Article in press: June 25, 2018
Published online: August 14, 2018
To elucidate tongue coating microbiota and metabolic differences in chronic hepatitis B (CHB) patients with yellow or white tongue coatings.
Tongue coating samples were collected from 53 CHB patients (28 CHB yellow tongue coating patients and 25 CHB white tongue coating patients) and 22 healthy controls. Microbial DNA was extracted from the tongue samples, and the bacterial 16S ribosomal RNA gene V3 region was amplified from all samples and sequenced with the Ion Torrent PGM™ sequencing platform according to the standard protocols. The metabolites in the tongue coatings were evaluated using a liquid chromatography-mass spectrometry (LC-MS) platform. Statistical analyses were then performed.
The relative compositions of the tongue coating microbiotas and metabolites in the CHB patients were significantly different from those of the healthy controls, but the tongue coating microbiota abundances and diversity levels were not significantly different. Compared with the CHB white tongue coating patients, the CHB yellow tongue coating patients had higher hepatitis B viral DNA (HBV-DNA) titers (median 21210 vs 500, respectively, P = 0.03) and a significantly lower level of Bacteroidetes (20.14% vs 27.93%, respectively, P = 0.013) and higher level of Proteobacteria (25.99% vs 18.17%, respectively, P = 0.045) in the microbial compositions at the phylum level. The inferred metagenomic pathways enriched in the CHB yellow tongue coating patients were mainly those involved in amino acid metabolism, which was consistent with the metabolic disorder. The abundances of bacteria from Bacteroidales at the order level were higher in the CHB white tongue coating patients (19.2% vs 27.22%, respectively, P = 0.011), whereas Neisseriales were enriched in the yellow tongue coating patients (21.85% vs 13.83%, respectively, P = 0.029). At the family level, the abundance of Neisseriaceae in the yellow tongue patients was positively correlated with the HBV-DNA level but negatively correlated with the S-adenosyl-L-methionine level.
This research illustrates specific clinical features and bacterial structures in CHB patients with different tongue coatings, which facilitates understanding of the traditional tongue diagnosis.
Core tip: Tongue diagnosis has important guiding significance for clinical syndrome differentiation and drug use in traditional Chinese medicine (TCM), but lacks scientific explanations. This study illustrates the existence of specific clinical features and bacterial structure in chronic hepatitis B (CHB) patients with different tongue coatings. Compared with the CHB white tongue coating patients, the yellow tongue coating patients had higher viral titers and a significantly lower level of Bacteroidetes and higher level of Proteobacteria in their microbial compositions at the phylum level. This study explores the micro-features between different tongue coatings, which will promote our understanding of the TCM tongue diagnosis and facilitate therapeutic strategies for individualized treatment.
- Citation: Zhao Y, Mao YF, Tang YS, Ni MZ, Liu QH, Wang Y, Feng Q, Peng JH, Hu YY. Altered oral microbiota in chronic hepatitis B patients with different tongue coatings. World J Gastroenterol 2018; 24(30): 3448-3461
- URL: https://www.wjgnet.com/1007-9327/full/v24/i30/3448.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i30.3448
Tongue diagnosis is a characteristic diagnostic method of traditional Chinese medicine (TCM) with a long history. TCM believes that the tongue is an important window for changes in the body, which are closely related to the zang and fu (internal organs), meridians, qi and xue (blood) and body fluids. Therefore, changes of the tongue can help doctors distinguish syndromes and guide diagnoses, which has great significance for clinical treatment via TCM. Different tongue coating colors in TCM have different treatment principles. Tongue diagnosis has the advantages of convenience and intuitiveness and represents first-hand information for TCM doctors that should not neglected. However, the physiological and pathological changes that contribute to the TCM tongue diagnosis have not been elucidated. According to TCM theory, the formation of tongue coating is closely related to wei-qi (spleen and stomach). Reports have noted that micro-ecological changes in the local flora are important manifestations of pathological tongue images[1]. As part of the human microbiome, the oral microbiota can interfere with the gut microbiota and is associated with multiple diseases[2]. The microbiota can produce metabolites and active ingredients that participate in the regulation of host metabolism and immunity and is closely related to the pathological processes of many diseases. Some reports have found abnormalities in the microbiota or metabolites in pathological tongue coatings[3,4]. However, integrated analyses of the pathological tongue coating microbiotas and metabolites have rarely been reported. As an important diagnostic factor, elucidating the “intension” of the material basis of tongue coatings and providing the scientific basis for individualized treatment and clinical syndrome differentiation have great significance in TCM.
Chronic hepatitis B (CHB) is a major infectious disease that may lead to liver cirrhosis and hepatocellular carcinoma (HCC). In China, CHB is a huge health burden, and 60% and 80% of China’s liver cirrhosis and HCC patients, respectively, are infected with hepatitis B virus (HBV)[5]. Intestinal bacteria are widely involved in the pathogenesis of chronic liver diseases, which can affect liver homeostasis based on energy absorption and storage, interfere with bile metabolism, participate in immune regulation and affect other processes[6]. Some studies have shown that dysbiosis of the oral microbiota may be involved in the development of HBV-induced chronic liver disease. Moreover, key oral-derived phylotypes may invade the gut as opportunistic pathogens and alter the composition of the gut microbiota, and cirrhotic patients have a more limited salivary defenses and worse inflammation than healthy patients[7]. In China, many patients with chronic liver disease are treated with TCM, which has been reported to decrease the incidence of liver cancer in a recent 15-year follow-up study of 21020 newly diagnosed CHB patients[8]. Changes in the tongue are important for TCM diagnoses[9,10], and CHB patients have different tongue coating phenotypes, which most commonly manifested as a yellow or a white tongue coating. These tongue coatings lead to different treatment strategies in TCM. Therefore, we speculate that microbiota differences may exist between CHB patients with different tongue coatings.
In this study, we characterize the microbiota and metabolic compositions of CHB patients with yellow or white tongue coatings and investigate the association between the tongue coating microbiotas, metabolites and host physiological indices to elucidate the differences between CHB patients with yellow and white tongue coatings. These results will contribute to our understanding of the relationship between the tongue coating appearance and the microbiota and metabolic micro-features differences.
All samples from this study were collected at the Shanghai University of TCM-affiliated Shuguang Hospital in Shanghai, the Nanjing Second Hospital and the Huai’an Fourth People’s Hospital in Jiangsu province, People’s Republic of China, from 2013 to 2014. The criteria used for the diagnosis of CHB were from the “The Guidelines for the Prevention and Treatment of Chronic Hepatitis B (2010 version)” issued by the Chinese Society of Liver Diseases and the Chinese Society of Infectious Diseases. Healthy age- and gender-matched control subjects were recruited from the Shanghai University of TCM-affiliated Shuguang Hospital Medical Center. We excluded patients who had been on absorbable antibiotics within the past 3 mo; were undergoing periodontal disease treatment; had other viral liver hepatitises, liver cirrhosis, or HCC; were pregnant or lactating women; or had other severe primary diseases. Informed written consent was obtained before the participants’ enrollment. This project was approved by the IRB of Shuguang Hospital, which was affiliated with the Shanghai University of TCM (Permit Number: 2012-206-22-01) and conformed to the ethical guidelines of the Declaration of Helsinki (2008).
Each subject underwent serum and tongue coating collection on the same day in the morning before breakfast. For tongue coating collection, all participants were required to rinse their mouth with physiological saline and then were photographed with the tongue diagnostic information acquisition system in a stable light source (Daosh Co., Shanghai, China). Sterile spoons scraped the fixed parts of the tongue surface twice to collect tongue coating samples, which were dissolved in sanitized Eppendorf tubes filled with 2 mL of physiological saline[4]. Spare samples were stored in a -80 °C freezer. Blood samples were taken for clinical analyses after an overnight fast of at least 10 h.
Clinical and biochemical indices were determined for each patient, including the following: (1) liver and kidney function, including serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), γ-glutamyl transpeptidase (GGT), total bilirubin (TBIL), direct bilirubin (DBIL), albumin (ALB), globulin (GLB), creatinine (Cr), blood urea nitrogen (BUN) and uric acid (UA); (2) lipid profiles, including triglycerides (TG), total cholesterol (TC), high-density lipoprotein (HDL) and low-density lipoprotein (LDL); (3) clotting function, including the prothrombin time (PT); and (4) virological indicators, including hepatitis B surface antigen (HBsAg), hepatitis B surface antibody (HBsAb), hepatitis Be antigen (HBeAg), hepatitis Be antibody (HBeAb), hepatitis B core antibody (HBcAb) and the HBV-DNA titers were determined for each patient.
DNA extraction and PCR amplification: Microbial DNA was extracted from the tongue samples using a QIAGEN QIAamp DNA Stool Mini Kit (50) according to manufacturer’s protocols. The V3 region of the bacterial 16S ribosomal RNA (rRNA) gene was amplified by PCR (95 °C for 2 min, followed by 25 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s and a final extension at 72 °C for 5 min) using the primers 341F (5’-CCTACGGGAGGCAGCAG-3’) and 518R (5’-ATTACCGCGGCTGCTGG-3’), which included a barcode that was an eight-base sequence unique to each sample. The PCR reactions were performed in triplicate with a 20 μL mixture containing 4 μL of 5 × FastPfu buffer, 2 μL of 2.5 mmol/L dNTPs, 0.8 μL of each primer (5 μmol/L), 0.4 μL of KAPA HiFi Polymerase, and 10 ng of template DNA.
Ion Torrent PGM™ sequencing: Amplicons were extracted from 2% agarose gels, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions and then quantified using the Qubit 2.0 Fluorometer (Invitrogen, United States). Purified amplicons were pooled at equimolar concentrations and sequenced on an Ion Torrent PGM™ platform according to standard protocols. The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: SRP127002).
Processing of sequencing data: Raw FASTQ files were demultiplexed and quality-filtered using the fastx_Toolkit (version 0.0.13.2) with the following criteria. The 200 bp reads were truncated at any site receiving an average quality score < 20, and truncated reads that were shorter than 50 bp were discarded. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UCHIME (version 7.1 http://drive5.com/uparse/), with also identified and removed chimeric sequences. The taxonomy of each 16S rRNA gene sequence was analyzed using the RDP Classifier (http://rdp.cme.msu.edu/) against the SILVA (SSU123)16S rRNA database with a confidence threshold of 70%[11].
Sample preparations: The tongue coating samples were thawed on ice, and quality control (QC) samples were made by mixing and blending equal volumes (20 μL) of each sample. A 100 μL aliquot of the samples was added to 500 μL of 100% methanol, which was vibrated for 30 s and centrifuged at 15000 g for 10 min. The supernatant was dried in a vacuum centrifuge at 30 °C. The dried samples were dissolved in 300 μL of water/methanol (1:1, 4 °C) and filtered through a 0.22 μm membrane. A 3 μL aliquot of tongue coating was injected for LC-MS analysis, and another 100 μL was sampled for subsequent MS/MS identification.
LC-MS experiments: A Waters ACQUITY UPLC system analysis was performed on an ACQUITY UPLC® HSS T3 column (150 mm × 2.1 mm, 1.8 μm particle size; Waters Corporation, Milford, MA, United States) for separation of samples using a binary gradient mode. The sample was eluted with a gradient mobile phase composed of 0.1% (v/v) formic acid/water (A) and 0.1% (v/v) formic acid/acetonitrile (B) as follows: 0-1 min, 2% B; 1-11 min, linear increase from 2 to 50% B; 11-17 min, linear increase from 50 to 98% B; 17-18 min, 98% B; 18-19 min, linear decrease from 98 to 2% B and 19-20 min, 2% B. The flow rate was set to 0.3 mL/min. The column and sample-tray temperatures were maintained at 35 and 8 °C, respectively.
A Thermo Scientific™ LTQ XL™ mass spectrometer (Thermo Scientific, San Jose, CA, United States) was combined with electrospray ionization (ESI) operating in both the negative and positive ion modes. The positive ionization mode was operated using a spray voltage of 4.8 kV, and the negative ionization mode used a spray voltage of 4.50 kV. The capillary temperature was 325 °C, and the sheath gas and auxiliary gas were set at flow rates of 45 and 10 L/min, respectively. The mass scanning range was set at 89-1000 m/z in positive ionization mode and 87-1000 m/z in negative ionization mode.
Data processing and metabolite identification: The raw LC-MS data files were converted into the mzXML format and then analyzed by the XCMS program in the R statistical language (v3.1.1) for peak identification, filtering and alignment. For the subsequent multivariate linear regression analysis, the XCMS results were ultimately converted into two-dimensional data matrices that consisted of retention time (Rt)-m/z pairs and their peak area. The data were normalized by their sum peak areas, and 1043 positive ion mode variables and 540 negative ion mode variables were finally obtained for the subsequent analysis.
Metabolites driving the differences among the 3 groups were filtered with a variable importance (VIP) > 1, a correlation P(corr) > 0.4, and a threshold of 1 using the 7-fold cross-validated partial least squares-discriminant analysis (PLS-DA) model. By using retention time-m/z pairs as the identifiers for each ion, we examined and optimized the parameters individually. The possible identities of the metabolites were first confirmed based on their exact molecular weights (molecular weight error < 15 ppm), followed by the comparison of their exact molecular weight and MS/MS fragmentation patterns to entries in the Human Metabolome Database (HMDB) (http://www.hmdb.ca) and the METLIN (https://metlin.scripps.edu), MassBank (http://www.massbank.jp) and mzCloud (https://http://www.mzcloud.org) databases.
We compared the clinical difference among the 3 groups using the Kruskal-Wallis test and the Nemenyi test was used for post-pairwise comparisons. The HBV-DNA titers between the CHB yellow and white tongue coating patients and the metabolites differences between the CHB and the healthy controls were compared using the standard non-parametric Mann-Whitney U test. Unsupervised principal component analysis (PCA) and supervised PLS-DA analysis were performed using the Simca-P software (version 13.0, Umetrics, Umea, Sweden). The microbiota results were analyzed using Metastats analysis, the Kruskal-Wallis test and the linear discriminant analysis (LDA) effect size (LEfSe) method was implemented in LEfSe v1.0[12]. The functionality of the microbiota was assessed using PiCRUST[13] and compared among the groups. The metabolomics enrichment analysis was performed using MetaboAnalyst 3.0[14] (http://www.metaboanalyst.ca). We correlated the microbiota with clinical index and metabolites and visualized the results with a Spearman’s correlation coefficient> 0.3 and P < 0.05 in Cytoscape (version 3.6.0).
A total of 75 subjects, including 22 healthy subjects and 53 CHB patients, were enrolled in this study. Twenty-eight of the 53 CHB patients exhibited a yellow tongue coating, and 25 CHB patients exhibited a white tongue coating; their physiological characteristics are shown in Table 1. The ages, genders and body mass indices (BMIs) did not significantly differ among the groups. The CHB group had a significantly higher level of liver function, including ALT, AST and GGT activity. The CHB yellow tongue coating patients had a trend toward higher TBIL, DBIL, ALT, AST, ALP, GGT and PT levels. Additionally, the HBV viral titer was significantly higher in yellow tongue coating patients than in the white coating patients (P = 0.03), whereas the ALB level was lower in the yellow tongue coating patients (P = 0.003).
| Healthy | CHB patients | P value | ||
| White tongue coating | Yellow tongue coating | |||
| Age | 34.09 ± 7.16 | 34.46 ± 9.92 | 38.08 ± 9.89 | 0.255 |
| Gender, male/female | 13/9 | 21/7 | 17/8 | 0.488 |
| BMI (kg/m2) | 21.22 (16.61-25.56) | 23.44 (18.42-31.64) | 23.30 (18.37-45.67) | 0.077 |
| Antiviral drugs (%) | / | 78.60% | 64% | 0.24 |
| Chinese herbs (%) | / | 17.90% | 28% | 0.378 |
| TBIL (μmol/L) | 14.87 (8.29-31.62) | 12.85 (7.9-27.7) | 17.27 (6.4-45.6) | 0.27 |
| DBIL (μmol/L) | 3.065 (1.7-6.76) | 3.9 (1.64-9.2) | 4.8 (2.05-34.7)b | 0.006 |
| IDBIL (μmol/L) | 11.6 (6.3-25.7) | 8.95 (5-18.7) | 10.2 (4.2-19.6) | 0.282 |
| ALT (IU/L) | 15 (10-34) | 28.75 (8.5-170)b | 47 (11.1-533)c | < 0.001 |
| AST (IU/L) | 17.5 (11-31) | 27 (12-133)b | 38 (14.8-543)c | < 0.001 |
| ALP(IU/L) | 15.28 (9-52) | 25.8 (10.01-98) | 38 (8.6-274)c | < 0.001 |
| GGT (IU/L) | 70.5 (35-119) | 72.7 (37.1-131) | 76.3 (41.7-202) | 0.62 |
| ALB (g/L) | 44.97 (40.89-50.77) | 48.7 (43.1-56.3)ad | 46 (36.6-50.1) | 0.003 |
| PT (s) | 12.55 (11.3-13.9) | 12.7 (11.1-16.4) | 13.3 (11.3-16) | 0.032 |
| HBV-DNA (IU/mL) | / | 500 (500-44830000)d | 21210 (500-324100000) | 0.03 |
| BUN (mmol/L) | 4.39 (2.97-6.39) | 4.96 (2.96-8.9) | 4.2 (2.4-7.28) | 0.074 |
| Gr (μmol/L) | 72.355 (45.1-106.67) | 70 (27.4-96.7) | 69 (35.9-83) | 0.449 |
| UN (μmol/L) | 305.5 (201-406) | 325 (207-550) | 294 (154-429) | 0.204 |
| FPG (mmol/L) | 5.2 (3.96-5.63) | 5.2 (4.29-7.09) | 5.215 (4.07-15.54) | 0.532 |
| TC (mmol/L) | 4.04 (0.37-6.34) | 4.55 (3.15-6.52) | 4.36 (3.1-6.74) | 0.102 |
| TG (mmol/L) | 0.84 (0.37-1.91) | 1.17 (0.58-3.31)a | 1.11 (0.68-2.62)a | 0.008 |
| HDL (mmol/L) | 1.14 (0.74-1.76) | 1.145 (0.72-1.86) | 1.07 (0.58-2.02) | 0.779 |
| LDL (mmol/L) | 2.205 (1.38-5.04) | 2.27 (1.47-3.8) | 2.44 (1.12-3.73) | 0.647 |
| APOA-A (g/L) | 1.175 (0.85-1.62) | 1.08 (0.76-1.91)c | 1.02 (0.71-1.42)c | 0.013 |
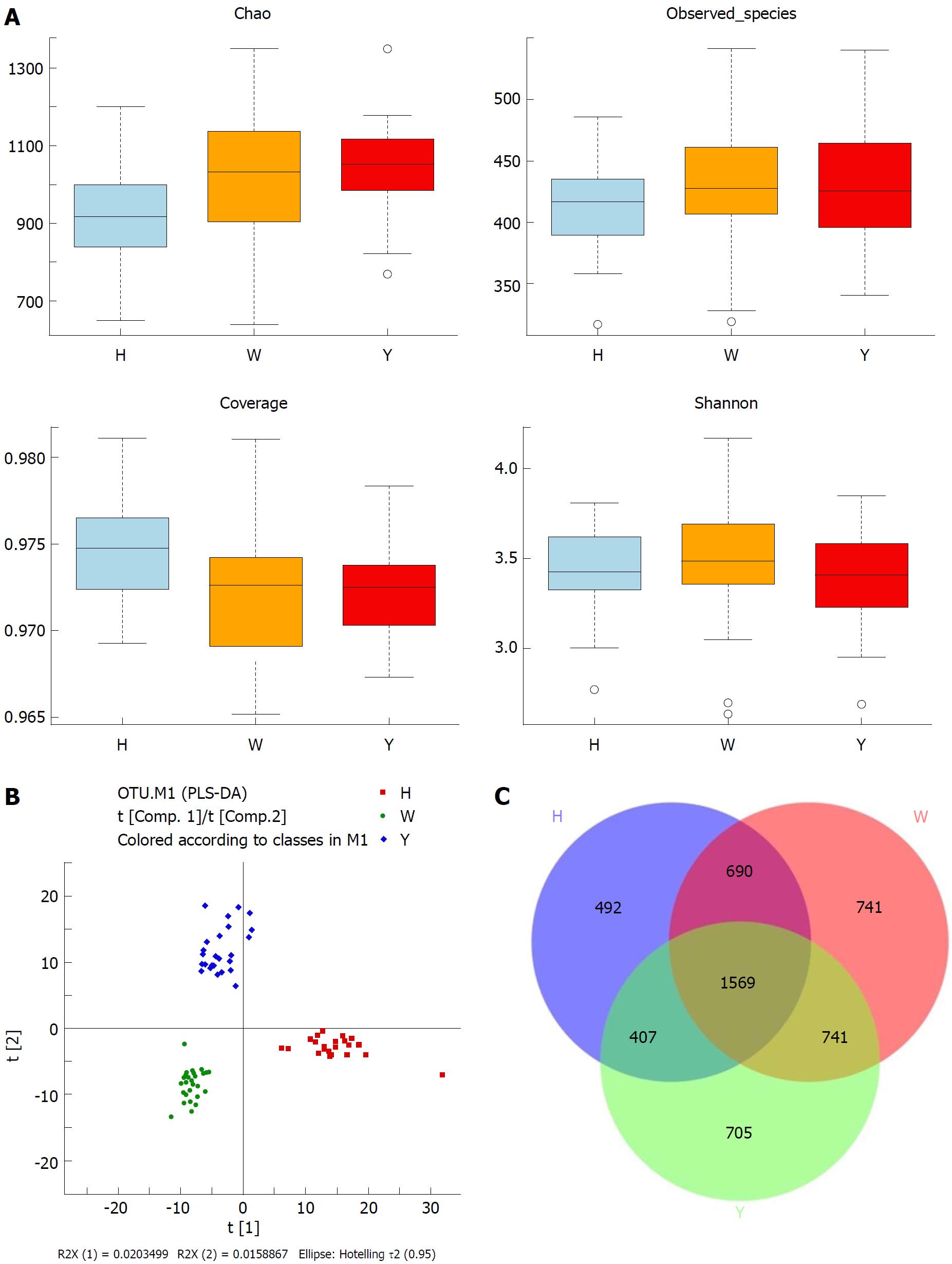
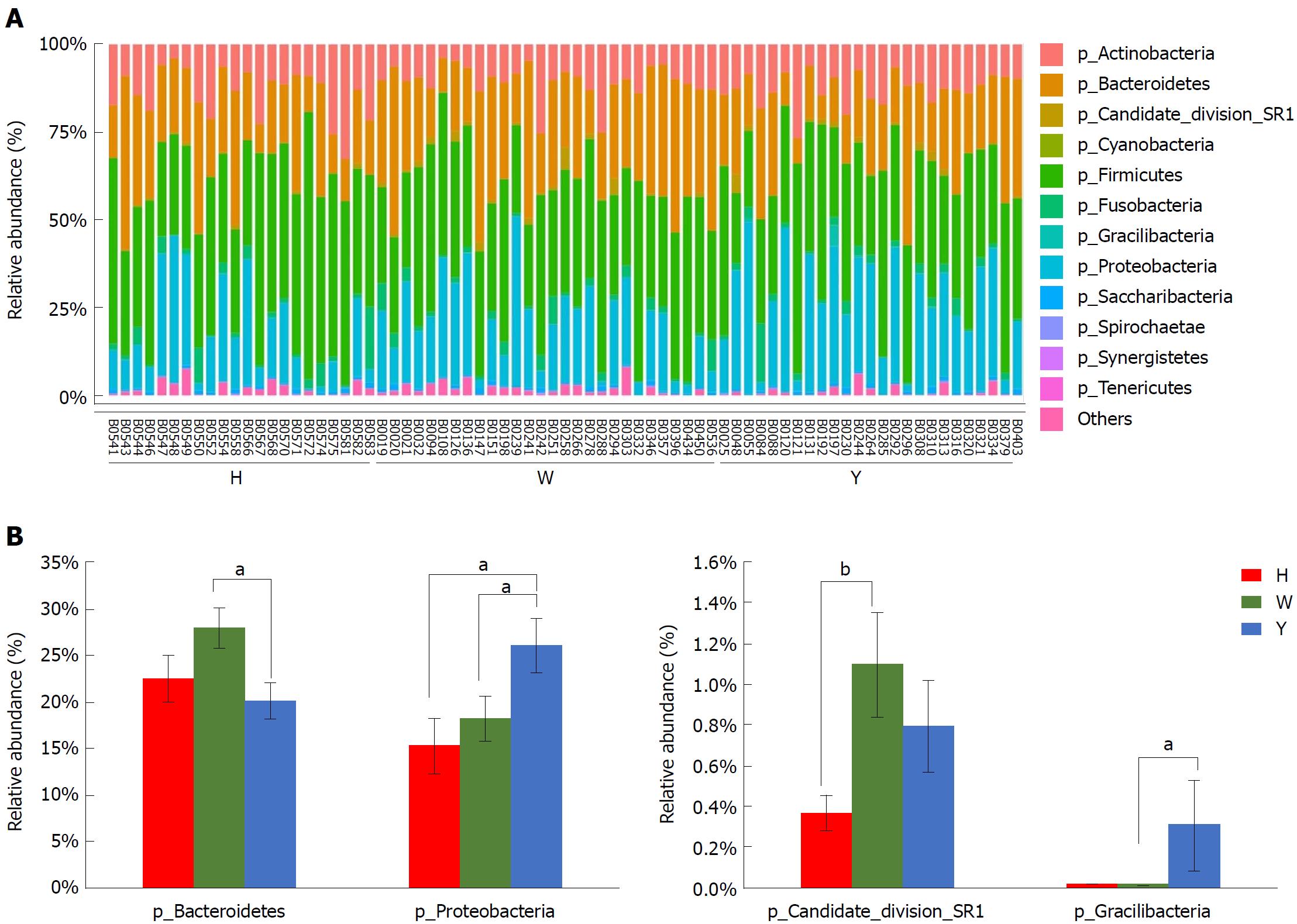
In this study, the qualified sequences were rarefied to 9300 sequences per sample for downstream analysis. A summary is shown in Supplementary Table 1, Good’s coverage values were high in the three groups, indicating that the sequencing depth was sufficient for investigation. As shown in Figure 1A show, the oral microbiota abundance and diversity level did not significantly differ between the CHB patients and the healthy controls. A PCA plot based on the OTU distributions showed that the overall compositions of the tongue coating microbiota were not significantly shifted by age, gender, BMI and antiviral treatment (Supplementary Figure 1). However, the PLS-DA plot showed a clear separation between the CHB patients and the healthy groups, and the CHB yellow tongue coating and white tongue coating patients were also well separated (Figure 1B). A Venn diagram showed that 1569 OTUs were common in all samples and that 705 and 741 OTUs were unique to the CHB yellow tongue coating and white tongue coating patients, respectively (Figure 1C). Most of the tongue coating microbiota were assigned to the Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and Fusobacteria phyla (relative abundance > 1% of the total DNA sequences, Figure 2A). In total, 112 genera were classified, and most of the genera were present in low abundance in the oral microbiota samples.
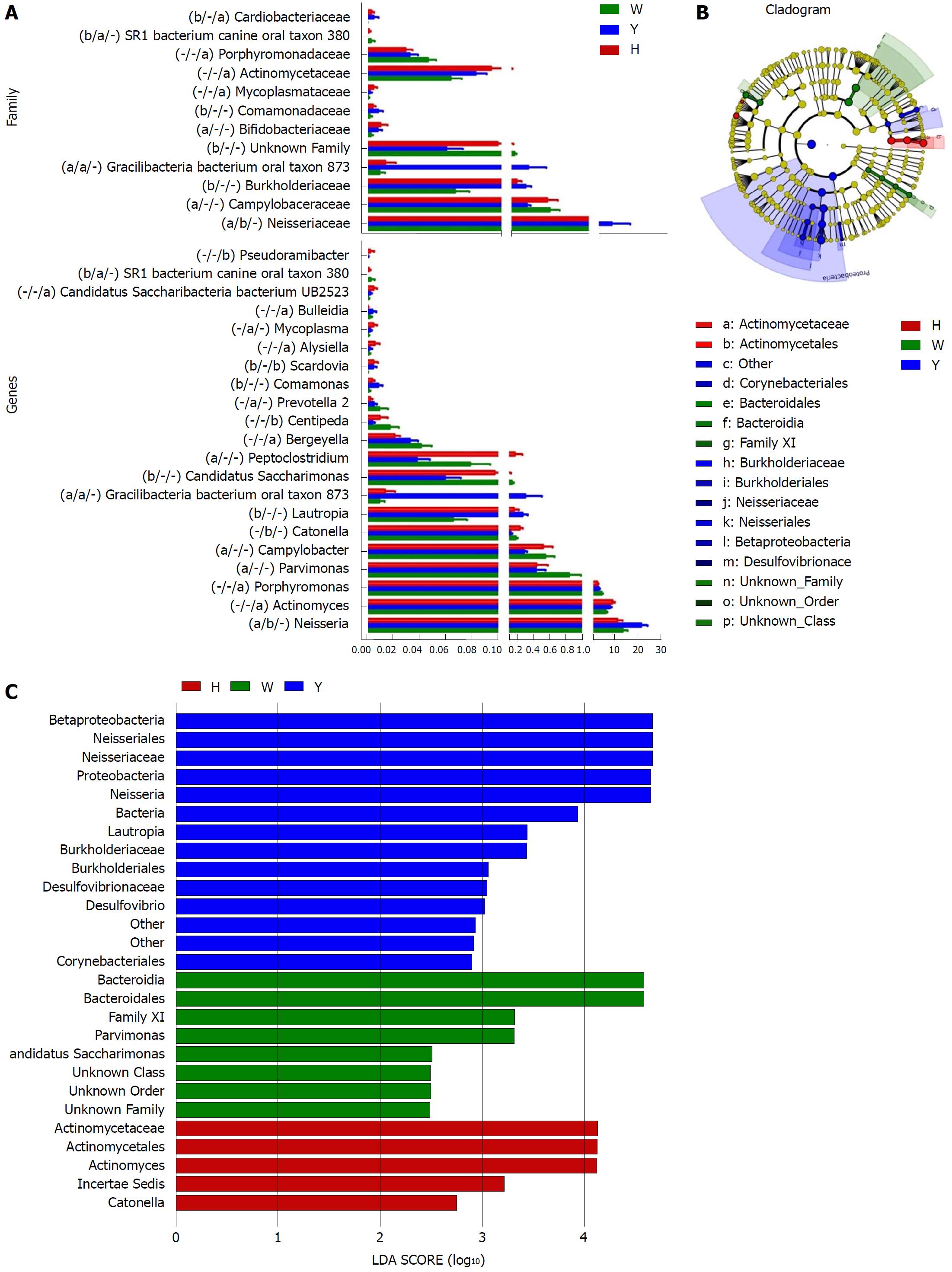
A Metastats analysis was used to detect differentially abundant features at different taxon levels. Compared with the white tongue patients, the relative abundance of Bacteroidetes was decreased in the CHB yellow tongue coating patients, whereas the abundance of Proteobacteria and Gracilibacteria were increased (Figure 2B). At the family level, Neisseriaceae and Gracilibacteria_bacterium_oral_taxon_873 were dominant in the CHB yellow tongue coating and had prevalent differences compared with the abundances in the healthy controls and CHB white tongue coating patients, whereas SR1_bacterium_canine_oral_taxon_380 was decreased. At the genus level, Neisseria and Gracilibacteria_bacterium_oral_taxon_873 were enriched in the CHB yellow tongue coating samples compared with the healthy controls and the CHB white tongue coating patients. The relative abundance of Actinomyces, Porphyromonas, Bergeyella, Centipeda, Alysiella, Bulleidia, Candidatus_Saccharibacteria_bacterium_UB2523 and Pseudoramibacter in the CHB white tongue coating samples were significantly different from those of the healthy controls, but no differences were found compared to the abundances of the CHB yellow tongue coating patients (Figure 3A). We used LEfSe to identify the specific OTUs associated with each group. A cladogram representative of the structure of the tongue coating microbiotas in the CHB yellow tongue coating and CHB white tongue coating patients and the healthy controls is shown in Figure 3B. According to the LDA values (log10LDA score > 4), the tongue coating bacteria with advantages in the CHB yellow tongue coating were Betaproteobacteria, Neisseriales, Neisseriaceae, Proteobacteria and Neisseria, the CHB white tongue coating samples were dominated by Bacteroidia and Bacteroidales, and the healthy controls were dominated by Actinomycetaceae, Actinomycetales and Actinomyces (Figure 3C).
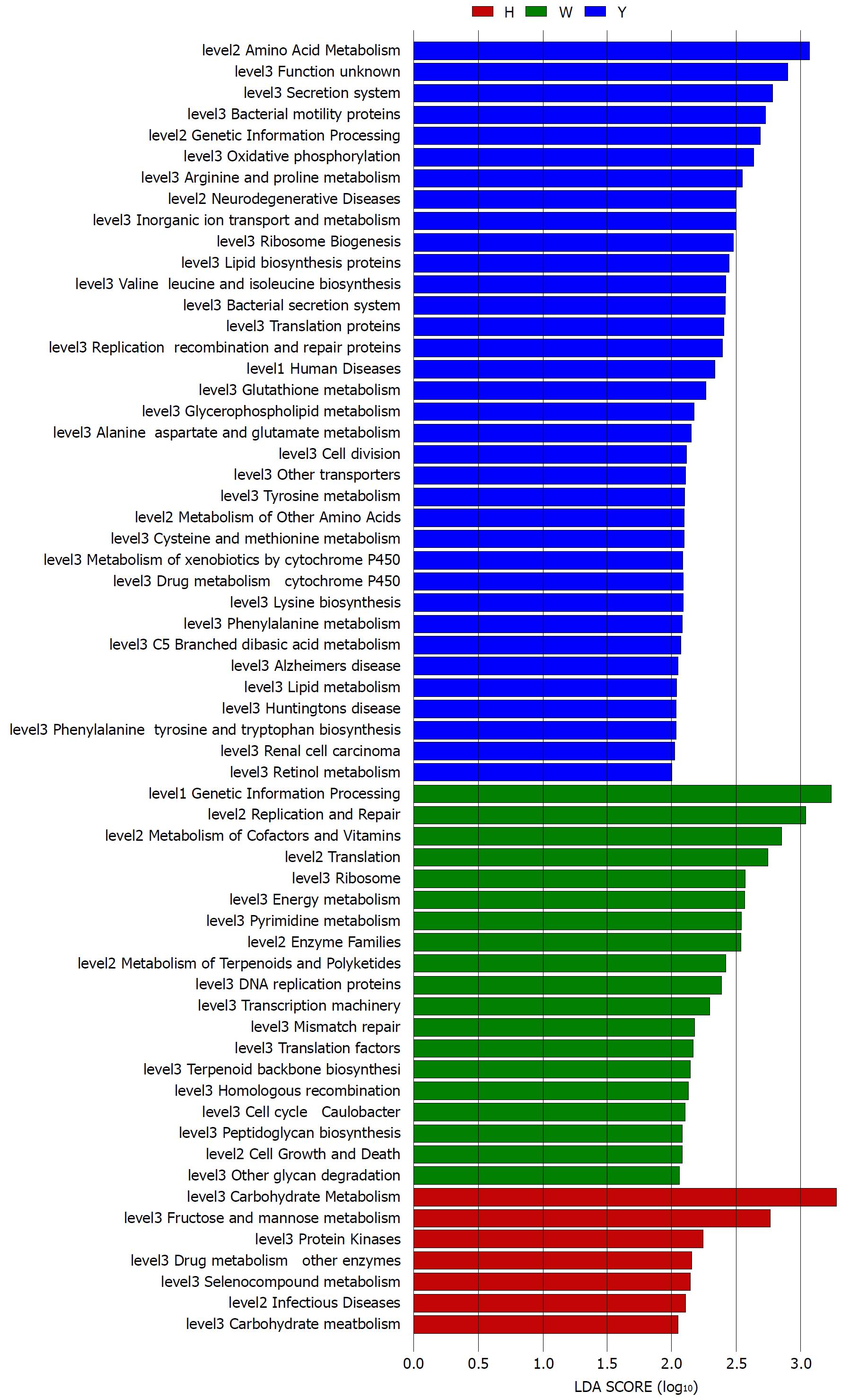
The gene functions in the tongue coating microbiotas were predicted by PICRUSt, and the results were compared between groups (Figure 4). Microbes with greater relative abundances in the healthy controls had functions that were most related to carbohydrate metabolism (fructose and mannose metabolism). The microbiotas in the white tongue coatings were more likely than those in the yellow tongue coatings and healthy controls to have functionality related to renetic information processing, including the level-2 functionality of replication and repair (DNA replication proteins, homologous recombination and mismatch repair) and transcription (transcription machinery, ribosome and translation factors). In contrast, the microbiotas in the yellow tongue coatings were probably related to amino acid metabolism (alanine aspartate and glutamate metabolism, arginine and proline metabolism, cysteine and methionine metabolism, lysine biosynthesis, phenylalanine tyrosine and tryptophan biosynthesis, phenylalanine metabolism, tyrosine metabolism and valine leucine and isoleucine biosynthesis), membrane transport (bacterial secretion system and secretion system) and cell motility (bacterial motility proteins).
The typical total ion chromatograms (TICs) of the samples run in positive ion modes are shown in supplementary Figure 2A. Because the PCA did not distinguish the groups well, the supervised PLS-DA model was used for further analysis and resulted in good separation of the CHB patients and healthy volunteers, however, the CHB yellow tongue coating and the white tongue coating patients were not completely distinguished (Supplementary Figure 2B). A total of 21 endogenous compounds were obtained with a VIP > 1 and P(corr) > 0.4 in the PLS-DA model (Table 2). The presence of 6 compounds differed between the CHB patients and healthy subjects (P < 0.05), and these compounds were mainly involved in histidine metabolism and methylhistidine metabolism (P < 0.05, Supplementary Figure 3). The metabolites were not significantly different between the CHB yellow and white tongue coating patients, but the number of amino acid metabolites was greater in the CHB yellow tongue coating patients than in the white tongue coating patients.
| Metabolites | Formula | Rt (min) | Type | KEGG compounds | VIP | P (corr) | CHB VS H | Healthy | CHBWhite tongue coating | CHBYellow tongue coating | |
| P value | FC | ||||||||||
| L-Proline | C5H9NO2 | 83.3392 | [M+H]+ | C00148 | 1.04 | -0.68 | 0.134 | 1.473 | 1.01 (0.13-12) | 3 (0.16-20.14) | 3.88 (0.09-15.28) |
| L-Valine | C5H11NO2 | 120.7915 | [M+H]+ | C00183 | 1.07 | -0.76 | 0.081 | 1.608 | 2.92 (0.82-17.97) | 3.98 (1.13-38.65) | 7.44 (1.08-31.3) |
| L-Isoleucine | C6H13NO2 | 194.3545 | [M+H]+ | C00123 | 1.15 | -0.81 | 0.069 | 1.641 | 5.55 (0.43-30.81) | 7.23 (1.23-58.36) | 13.28 (0.22-45.18) |
| L-Leucine | C6H13NO2 | 212.576 | [M+H]+ | C00123 | 1.07 | -0.81 | 0.029 | 1.715 | 8.9 (0.02-44.58) | 14.65 (2.89-84.8) | 21.6 (0.44-61.98) |
| Hypoxanthine | C5H4N4O | 154.305 | [M+H]+ | C00262 | 1.07 | -0.80 | 0.039 | 2.218 | 0.42 (0.01-6.93) | 1.31 (0.05-17.87) | 2.46 (0.01-14.33) |
| Urocanic acid | C6H6N2O2 | 132.3225 | [M+H]+ | C00785 | 1.00 | -0.78 | 0.044 | 2.587 | 0.62 (0.07-4.46) | 0.93 (0.08-12.84) | 1.93 (0.08-13.92) |
| L-Lysine | C6H14N2O2 | 64.124 | [M+H]+ | C00047 | 1.01 | -0.72 | 0.683 | 1.510 | 0.08 (0.05-0.28) | 0.08 (0.03-0.81) | 0.08 (0.03-0.65) |
| Glutamate | C5H9NO4 | 76.6581 | [M+H]+ | C00025 | 1.04 | -0.75 | 0.159 | 1.955 | 0.14 (0.02-1.17) | 0.2 (0-2.99) | 0.39 (0-2.52) |
| Methionine | C5H11NO2S | 131.208 | [M+H]+ | C00073 | 1.16 | -0.80 | 0.144 | 1.543 | 1.08 (0.01-7.21) | 1.74 (0.13-10.21) | 2.26 (0-9.75) |
| Xanthine | C5H4N4O2 | 183.102 | [M+H]+ | C00385 | 1.01 | -0.73 | 0.151 | 2.058 | 0.09 (0-2.91) | 0.24 (0-4.93) | 0.47 (0-4.75) |
| L-Phenylalanine | C9H11NO2 | 277.369 | [M+H]+ | C00079 | 1.17 | -0.82 | 0.069 | 1.534 | 6.72 (0.83-24.15) | 11.07 (1.84-51.35) | 13.1 (0.31-36.18) |
| Glycyl-Valine | C7H14N2O3 | 205.364 | [M+H]+ | 1.07 | -0.70 | 0.078 | 1.548 | 0.07 (0-1.1) | 0.15 (0-0.84) | 0.19 (0-1.38) | |
| Citrulline | C6H13N3O3 | 77.3246 | [M+H]+ | C00327 | 1.16 | -0.74 | 0.216 | 1.486 | 0.12 (0-1.28) | 0.14 (0.01-1.7) | 0.3 (0-1.38) |
| L-Tyrosine | C9H11NO3 | 204.441 | [M+H]+ | C00082 | 1.17 | -0.81 | 0.095 | 1.512 | 2.63 (0.33-11.35) | 4.36 (0.71-22.15) | 5.67 (0.1-16.28) |
| Benzophenone | C13H10O | 882.111 | [M+H]+ | C06354 | 1.19 | 0.81 | < 0.001 | 0.752 | 0.25 (0.16-0.3) | 0.19 (0.1-0.3) | 0.18 (0.07-0.25) |
| L-Tryptophan | C11H12N2O2 | 338.856 | [M+H]+ | C00078 | 1.03 | -0.74 | 0.23 | 1.940 | 0.57 (0.09-3.63) | 0.74 (0.07-12.67) | 1.11 (0.1-9.81) |
| 12-Hydroxylauric acid | C12H24O3 | 906.316 | [M+H]+ | C08317 | 1.12 | 0.72 | < 0.001 | 0.697 | 10.81 (8.25-16.18) | 7.3 (0.08-12.49) | 7.61 (0.08-14.08) |
| N-Acetyl-D-glucosamine | C8H15NO6 | 82.3978 | [M+H]+ | C03878 | 1.02 | -0.39 | 0.936 | 0.942 | 0.04 (0-0.46) | 0.07 (0-0.34) | 0.07 (0-0.44) |
| L-cystathionine | C7H14N2O4S | 836.2685 | [M+H]+ | C02291 | 1.10 | 0.67 | 0.24 | 0.919 | 0.48 (0.31-0.79) | 0.48 (0.2-0.81) | 0.43 (0.22-0.66) |
| S-adenosyl-L-methionine | C15H22N6O5S | 66.1963 | [M+H]+ | C00019 | 1.02 | 0.79 | 0.029 | 0.888 | 9.98 (8.09-12.77) | 9.2 (4.71-12.09) | 8.58 (4.98-14.58) |
| Guanosine | C10H13N5O5 | 230.687 | [M+H]- | C00387 | 1.01 | 0.02 | 0.824 | 0.763 | 3.02 (0.55-14.19) | 2.98 (0-10.27) | 3.52 (0-10.89) |
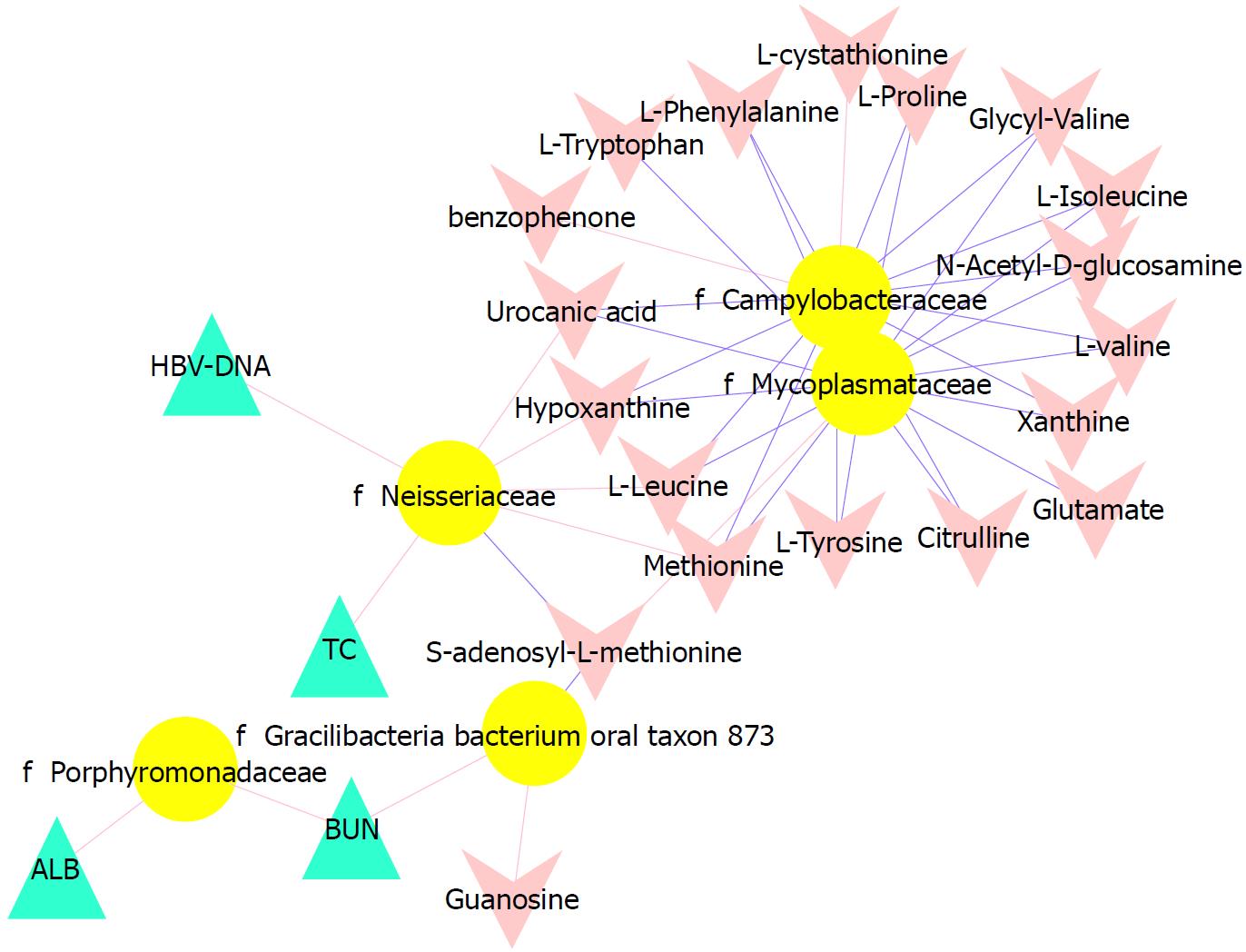
To better understand the relationships among the tongue coating microbiota, metabolites and clinical indicators, we performed a correlation analysis of the differential microbiota at the family level with the metabolites and clinical indicators and constructed the related network (|correlation r| > 0.3, P < 0.05). The abundance of Neisseriaceae, which was the dominant bacterial family in the CHB yellow tongue coating, was positively correlated with the serum HBV-DNA, TC, L-leucine, hypoxanthine, urocanic acid and methionine levels but negatively correlated with thebenzophenone and S-adenosyl-L-methionine levels. Another relatively abundant member of the microbiota in the CHB yellow tongue coating (Gracilibacteria_bacterium_oral_taxon_873) was positively correlated with the guanosine and BUN levels but negatively correlated with the S-adenosyl-L-methionine level (Figure 5).
Chinese medicine theory believes that tongue demonstration is a window of body change and that the oral microbiome is an integral part of the whole body’s microbiome. Indeed, an increasing number of studies have found that many diseases, including atherosclerosis[15], rheumatoid arthritis[16], diabetes[17] and liver cirrhosis[18], are accompanied by disorder of the oral and intestinal microbiotas. Although the oral and gut communities share little taxonomic resemblance, the oral and intestinal microflora have general dynamic characteristics[19], the oral bacterial populations seed the gut[20], and the observed bacterial structures in the oral cavity or intestine can predict each other[21]. Because tongue diagnosis has the advantages of intuition and simplicity, some scholars have recently discovered the importance of the tongue image for the diagnosis and prognostic prediction of diseases[22,23]. The oral microbiota varies according to the different sampling sites with a high diversity[24], but as the tongue texture and tongue color form an important basis for diagnosis and treatment in TCM[25]. The treatments adopted for patients with white and yellow tongue coatings are different, thus, the tongue coating oral microbiotas were selected for analysis in our study, and the samples were grouped by the color of the tongue coating.
The complex and dynamic interactions between microbes and hosts are very important for maintaining homeostasis because microbiome dysbiosis may interact with the host’s immune system and influence the degree of hepatic steatosis, inflammation and fibrosis[26]. Our recent study found that the gut microbiota was significantly altered in CHB patients with low Child-Pugh scores[27]. Evidence is accumulating that the intestinal microbiome has good clinical prospects as a target for the treatment of chronic liver disease. Chinese scholars transplanted the intestinal flora of healthy volunteers into HBeAg-positive CHB patients with long-term antiviral treatment, and this approach showed a good effect on HBeAg seroclearance[28]. However, few studies have been performed on the oral microbiotas in CHB patients. In one of the limited reports, Ling et al[29] noted the decreased diversity of the oral microbiotas in supragingival plaque samples from patients with HBV-induced chronic liver disease. However, in this study, the richness and diversity of the tongue coating did not differ between the CHB patients and healthy controls. Ren et al[30] used 16S rRNA gene pyrosequencing and metagenomic sequencing to examine oral microbial compositions and their functional variations in children with halitosis. They found that the tongue coatings of the subjects with halitosis had greater bacterial richness than those of the healthy subjects, but the number of OTUs and the Chao levels were slightly lower in the saliva samples from the subjects with halitosis than in the healthy subjects. The microbiotas among individuals and habitats are highly personalized in the oral cavity[31]. Therefore, we concluded that the data differences in data might be due to the use of different sampling sites.
Taxonomic and predicted function differences were found between the CHB yellow and white tongue coating patients in the LEfSe analysis. The CHB yellow tongue coating patients had specific clinical and microbiota characteristics that were distinct from those of the white coating patients. First, the CHB patients with yellow tongue coatings had significantly higher viral titers but lower ALB levels than the white coating patients (P < 0.05). The remaining indicators of liver function, including TBIL, DBIL, ALT, AST, ALP, GGT and PT, exhibited elevated trend in the CHB yellow tongue coating patients. Because a yellow tongue is an important indicator for the diagnosis of CHB damp heat syndrome, the results are consistent with studies showing that the CHB damp-heat syndrome patients have higher ALT and AST levels[32], which indicats that these patients have greater liver damage. The correlation analysis between bacteria and metabolites also confirmed this outcome. The relatively dominant bacterial families in the CHB yellow tongue coating (Neisseriaceae and Gracilibacteria_bacterium_oral_taxon_873) were negatively correlated with the S-adenosyl-L-methionine level. S-adenosyl-L-methionine, which is also known as SAM or AdoMet, is a donor in the methylation reactions of all mammalian cells but is found most abundantly in the liver. Patients with chronic liver disease have reduced AdoMet levels, and reduced hepatic AdoMet levels can contribute to the development of oxidative stress, steatohepatitis, and hepatocellular carcinoma (HCC)[33]. In addition, according to TCM, a yellow tongue coating is a manifestation of TCM hot Syndrome, which has been reported to be involved in inflammation or an immune regulation imbalance in the host[34,35]. Jiang et al[3] compared the yellow and white tongue coating microbiomes with those of gastritis patients and found that yellow dense tongue coating bacteria were associated with TCM hot syndrome-related inflammation such as halitosis, tonsil infection and periodontal inflammation. Therefore, we therefore hypothesized that the patients with CHB yellow tongue coatings might have had more inflammation, although this speculation requires further confirmation. Second, the yellow tongue coating subjects showed similarities at the phylum level with a gut disorder in patients with cirrhosis, which was manifested as a reduced abundance of Bacteroidetes and increased Proteobacteria content[18,26,36]. The LEfSe analysis suggested that Neisseriaceae was enriched in the yellow tongue coating patients and was positively correlated with the serum HBV-DNA level. Ling’s research reported that members of Neisseriaceae were significantly more abundant in the oral microbiota of liver cirrhosis patients than in those of CHB patients[29]. Lv et al[37] found that Neisseriaceae, which are opportunistic pathogens, were enriched in primary biliary cirrhosis (PBC) patients. Third, according to the LDA score, the inferred metagenomic pathways that were enriched in the CHB yellow tongue coating patients were mainly those involved in amino acid metabolism, and the detected metabolites were mainly essential amino acids, including L-proline, L-valine, L-isoleucine, L-leucine, L-lysine, glutamate and L-tryptophan, and were generally present in larger amounts trend in the yellow tongue coatings. The relationship between amino acid metabolism and the CHB patients with yellow tongue coatings is unclear, and relevant literature reports are limited. However, one report noted that people with the TCM damp-heat syndrome people with HBV-related chronic liver diseases, whom mainly manifested yellow greasy tongue coatings, had increased leucine, phenylalanine and valine expression in urinary metabolomic studies[38], which indirectly explained the severe disordered amino acid metabolism in CHB patients with the yellow coating. Finally, this study found that membrane transport (secretion system) and cell motility (bacterial motility proteins) were enriched in the yellow coatings, which indicated that the CHB yellow coating patients might be more susceptible to bacterial colonization. The functional analysis of tongue coating flora and metabolites in CHB white tongue coating patients lacked correspondence, the underlying reason might be the microbiotas functionality in the CHB white tongue coating patients were most likely related to genetic information processing, which association with metabolites is relatively little.
Tongue diagnosis is a simple, non-invasive and valuable procedure that has been repeatedly verified by TCM clinical practitioners. We found that the yellow and white tongue coatings of CHB patients had microbiota compositional and functional differences in this study, but the sample sizes were small. In the future, we will expand the sample size to verify the results of this study. In addition, we will focus on the relationship between changes in the oral and intestinal microbiota in different tongue coating patients and explore the effects of oral microbiota variance on immunity and metabolic alterations in the body to further explain the modern theoretical mechanism of TCM tongue diagnosis. These studies will facilitate the development of therapeutic strategies for individualized treatment.
Chronic hepatitis B is a major infectious disease in China and Chinese medicine has a wide range of applications in the treatment of CHB. Tongue diagnosis has important guiding significance for clinical syndrome differentiation and drug use in TCM, but lacks scientific explanations. Some reports have found abnormalities in the microbiota or metabolites in pathological tongue coatings. However, integrated analyses of the pathological tongue coating microbiotas and metabolites have rarely been reported. Elucidating the tongue coatings micro-features differences will promote our understanding of the TCM tongue diagnosis and facilitate therapeutic strategies for individualized treatment.
The motivation of this study was to explore the microfeatures of different tongue coatings, which could promote our understanding of the TCM tongue diagnosis from a modern perspective.
The objective of this research was to elucidate tongue coating microbiota and metabolic differences in CHB patients with yellow or white tongue coatings.
We collected tongue coating samples from 28 CHB yellow tongue coating patients and 25 CHB white tongue coating patients, and an additional 22 samples were collected from healthy controls. The tongue coating bacterial 16S ribosomal RNA gene V3 region was amplified and sequenced with the Ion Torrent PGM™ sequencing platform. The metabolites in the tongue coatings were examined using a LC-MS platform. The microbiota results were analyzed using Metastats analysis, the Kruskal-Wallis test and LEfSe analysis. The functionality of the microbiota was assessed using PiCRUST and compared among groups. The metabolomics enrichment analysis was performed with MetaboAnalyst 3.0 (http://www.metaboanalyst.ca). We correlated the microbiota with the clinical indices and metabolites and visualized the results with Spearman’s correlation coefficients > 0.3 and P < 0.05 in Cytoscape.
This study found taxonomic and predicted function differences between the CHB yellow and white tongue coating patients. Distinct from those of the white coating patients, the CHB yellow tongue coating patients had specific clinical and microbiota characteristics. The microbiota of the CHB patients with yellow tongue coatings had similarities with a gut disorder in patients with cirrhosis at the phylum level, which manifested as a reduced abundance of Bacteroidetes and increased Proteobacteria content. Neisseriaceae, which is a dominant bacterial family enriched in yellow tongue coating patients, was positively correlated with the serum HBV-DNA level. The inferred metagenomic pathways enriched in the CHB yellow tongue coating patients were mainly those involved in amino acid metabolism, whereas the detected metabolites were mainly essential amino acids and generally were present in larger amounts than in the white tongue coatings.
We found that the yellow and white tongue coatings of CHB patients had microbiota compositional and functional differences in this study.
The sample size in this study was small. In the future, we will expand the sample size to further verify the results and focus on the relationship between changes in the oral and intestinal microbiotas to explore the effects of oral microbiota variance on immunity and metabolic alterations in the body. These studies will further explain the modern theoretical mechanism of TCM tongue diagnosis.
The authors thank the doctors from the Shanghai University of TCM-affiliated Shuguang Hospital, the Nanjing Second Hospital and the Huai’an Fourth People’s Hospital for the support with clinical information and samples collection.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Jafari SA, Liu Y, Su SB S- Editor: Gong ZM L- Editor: A E- Editor: Yin SY
| 1. | Ye J, Cai X, Cao P. Problems and prospects of current studies on the microecology of tongue coating. Chin Med. 2014;9:9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 2. | Lira-Junior R, Boström EA. Oral-gut connection: one step closer to an integrated view of the gastrointestinal tract? Mucosal Immunol. 2018;11:316-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 3. | Jiang B, Liang X, Chen Y, Ma T, Liu L, Li J, Jiang R, Chen T, Zhang X, Li S. Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome. Sci Rep. 2012;2:936. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 4. | Sun ZM, Zhao J, Qian P, Wang YQ, Zhang WF, Guo CR, Pang XY, Wang SC, Li FF, Li Q. Metabolic markers and microecological characteristics of tongue coating in patients with chronic gastritis. BMC Complement Altern Med. 2013;13:227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099-2108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 779] [Cited by in F6Publishing: 847] [Article Influence: 84.7] [Reference Citation Analysis (1)] |
| 6. | Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59:328-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 245] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 7. | Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, Unser A, Thacker LR, Sanyal AJ, Kang DJ. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 216] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 8. | Tsai TY, Livneh H, Hung TH, Lin IH, Lu MC, Yeh CC. Associations between prescribed Chinese herbal medicine and risk of hepatocellular carcinoma in patients with chronic hepatitis B: a nationwide population-based cohort study. BMJ Open. 2017;7:e014571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 9. | Kang H, Zhao Y, Li C, Chen Y, Tang K, Yang L, Ma C, Peng J, Zhu R, Liu Q. Integrating clinical indexes into four-diagnostic information contributes to the Traditional Chinese Medicine (TCM) syndrome diagnosis of chronic hepatitis B. Sci Rep. 2015;5:9395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Zhao Y, Kang H, Peng JH, Xu L, Cao ZW, Hu YY. Key symptoms selection for two major syndromes diagnosis of Chinese medicine in chronic hepatitis B. Chin J Integr Med. 2017;23:253-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A, Gaskins HR, Stumpf RM, Yildirim S, Torralba M. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7:1344-1353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 905] [Cited by in F6Publishing: 725] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 12. | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7042] [Cited by in F6Publishing: 8633] [Article Influence: 664.1] [Reference Citation Analysis (0)] |
| 13. | Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5694] [Cited by in F6Publishing: 5765] [Article Influence: 524.1] [Reference Citation Analysis (0)] |
| 14. | Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251-W257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2063] [Cited by in F6Publishing: 2043] [Article Influence: 227.0] [Reference Citation Analysis (0)] |
| 15. | Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V, Behre CJ, Knight R, Fagerberg B, Ley RE. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4592-4598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 687] [Cited by in F6Publishing: 767] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 16. | Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895-905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 903] [Cited by in F6Publishing: 1005] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 17. | Xiao E, Mattos M, Vieira GHA, Chen S, Corrêa JD, Wu Y, Albiero ML, Bittinger K, Graves DT. Diabetes Enhances IL-17 Expression and Alters the Oral Microbiome to Increase Its Pathogenicity. Cell Host Microbe. 2017;22:120-128.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 199] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 18. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1230] [Cited by in F6Publishing: 1330] [Article Influence: 133.0] [Reference Citation Analysis (2)] |
| 19. | Bashan A, Gibson TE, Friedman J, Carey VJ, Weiss ST, Hohmann EL, Liu YY. Universality of human microbial dynamics. Nature. 2016;534:259-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 20. | Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358:359-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 385] [Cited by in F6Publishing: 506] [Article Influence: 84.3] [Reference Citation Analysis (0)] |
| 21. | Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357-360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 551] [Cited by in F6Publishing: 537] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 22. | Han S, Yang X, Qi Q, Pan Y, Chen Y, Shen J, Liao H, Ji Z. Potential screening and early diagnosis method for cancer: Tongue diagnosis. Int J Oncol. 2016;48:2257-2264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | RiYang L, HangYing Y, JunYan Q, YaYu L, YuHui W, YaZhen Y, JiaZhen Y, Jin Y, Jun N, DongRong Y. Association between tongue coating thickness and clinical characteristics among idiopathic membranous nephropathy patients. J Ethnopharmacol. 2015;171:125-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8203] [Cited by in F6Publishing: 7225] [Article Influence: 602.1] [Reference Citation Analysis (2)] |
| 25. | Lee TC, Lo LC, Wu FC. Traditional Chinese Medicine for Metabolic Syndrome via TCM Pattern Differentiation: Tongue Diagnosis for Predictor. Evid Based Complement Alternat Med. 2016;2016:1971295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut. 2016;65:2035-2044. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 296] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 27. | Wang J, Wang Y, Zhang X, Liu J, Zhang Q, Zhao Y, Peng J, Feng Q, Dai J, Sun S. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front Microbiol. 2017;8:2222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 28. | Ren YD, Ye ZS, Yang LZ, Jin LX, Wei WJ, Deng YY, Chen XX, Xiao CX, Yu XF, Xu HZ. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology. 2017;65:1765-1768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 29. | Ling Z, Liu X, Cheng Y, Jiang X, Jiang H, Wang Y, Li L. Decreased Diversity of the Oral Microbiota of Patients with Hepatitis B Virus-Induced Chronic Liver Disease: A Pilot Project. Sci Rep. 2015;5:17098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 30. | Ren W, Xun Z, Wang Z, Zhang Q, Liu X, Zheng H, Zhang Q, Zhang Y, Zhang L, Wu C. Tongue Coating and the Salivary Microbial Communities Vary in Children with Halitosis. Sci Rep. 2016;6:24481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 31. | Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721-5732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1867] [Cited by in F6Publishing: 1895] [Article Influence: 99.7] [Reference Citation Analysis (0)] |
| 32. | Sun YG, Sun YL. Progress of the study on the relationship between TCM Syndromes of chronic hepatitis B and objective indicators. Zhongyiyao Daobao. 2011;17:76-78. [DOI] [Cited in This Article: ] |
| 33. | Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515-1542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 405] [Cited by in F6Publishing: 367] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 34. | Li R, Ma T, Gu J, Liang X, Li S. Imbalanced network biomarkers for traditional Chinese medicine Syndrome in gastritis patients. Sci Rep. 2013;3:1543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 35. | Lu YY, Zhao Y, Song YN, Dong S, Wei B, Chen QL, Hu YY, Su SB. Serum cytokine profiling analysis for zheng differentiation in chronic hepatitis B. Chin Med. 2015;10:24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 674] [Cited by in F6Publishing: 701] [Article Influence: 53.9] [Reference Citation Analysis (1)] |
| 37. | Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, Chen YF, Shao L, Guo FF, Wu WR. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol. 2016;18:2272-2286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 153] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 38. | Guo Z, Wang WY, Dai JY, Fan ZQ, Cao HJ, Sun SJ, Zhang YY, Xu LM, Hu YY, Su SB. Investigation on Urinary Metabolomic in Syndrome Types of Cirrhosis Caused by Hepatitis B. Shijie Kexue Jishu-Zhongyiyao Xiandaihua. 2012;14:1282-1287. [Cited in This Article: ] |