Published online Feb 14, 2017. doi: 10.3748/wjg.v23.i6.976
Peer-review started: July 1, 2016
First decision: August 19, 2016
Revised: November 27, 2016
Accepted: December 16, 2016
Article in press: December 19, 2016
Published online: February 14, 2017
Processing time: 227 Days and 22.3 Hours
AIM
To explore the mechanism by which microRNA-155 (miR-155) regulates the pathogenesis of experimental colitis.
METHODS
A luciferase assay was performed to confirm the binding of miR-155 to the SHIP-1 3’-UTR. MiR-155 mimics, negative controls and SHIP-1 expression/knockdown vectors were established and then utilized in gain- and loss-of-function studies performed in raw264.7 cells and primary bone marrow-derived macrophages (BMDMs). Thereafter, dextran sulfate sodium (DSS)-induced colitis mouse model with or without antagomiR-155 treatment was established, and the levels of miR-155 and SHIP-1, as well as the pro-inflammatory capabilities, were measured by western blot, quantitative polymerase chain reaction, and immunohistochemistry.
RESULTS
MiR-155 directly bound to the 3’-UTR of SHIP-1 mRNA and induced a significant decrease in SHIP-1 expression in both raw264.7 cells and primary BMDMs. MiR-155 markedly promoted cell proliferation and pro-inflammatory secretions including IL-6, TNF-α, IL-1β, and IFN-γ, whereas these effects could be reversed by the restoration of SHIP-1 expression. In vivo studies showed that antagomiR-155 administration could alleviate DSS-induced intestinal inflammation in Balb/c mice. Moreover, significantly increased SHIP-1 expression, as well as decreased Akt activation and inflammatory response, were observed in the antagomiR-155-treated mice.
CONCLUSION
MiR-155 promotes experimental colitis by repressing SHIP-1 expression. Thus, the inhibition of miR-155 might be a promising strategy for therapy.
Core tip: Our present study identifies SHIP-1 as the functional target of microRNA-155 (miR-155) in macrophages. The up-regulation of miR-155 during colitis led to a significant decrease in SHIP-1 expression as well as a marked enhancement in cell proliferation and pro-inflammatory secretions, whereas the restoration of SHIP-1 expression partly reversed these changes. We further confirmed that antagomiR-155 treatment effectively alleviates dextran sulfate sodium-induced intestinal inflammation in mice, correlated with a significant elevation in SHIP-1 expression levels. Our findings indicate a novel mechanism by which miR-155 influences colitis progression.
- Citation: Lu ZJ, Wu JJ, Jiang WL, Xiao JH, Tao KZ, Ma L, Zheng P, Wan R, Wang XP. MicroRNA-155 promotes the pathogenesis of experimental colitis by repressing SHIP-1 expression. World J Gastroenterol 2017; 23(6): 976-985
- URL: https://www.wjgnet.com/1007-9327/full/v23/i6/976.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i6.976
Inflammatory bowel disease (IBD) is characterized by idiopathic, chronic, recurrent, inflammatory conditions of the human bowel triggered by multi-factorial causes that are not completely understood. IBD predominantly includes ulcerative colitis (UC) and Crohn’s disease (CD)[1,2]. Although its etiology remains unclear, studies have indicated that the combination of the dysfunction of the intestinal mucosal immune system, the imbalanced constitution of the gut flora, genetic susceptibility and environmental factors all contribute to the pathogenesis of IBD[3,4]. Thus far, certain molecular changes in gene and protein expression patterns have been identified during the chronic inflammation process of IBD, and unraveling the molecular events involved in these intracellular signaling transduction pathways may be helpful for IBD diagnosis and treatment.
MicroRNAs are endogenous small non-coding RNAs that regulate gene expression by binding to the 3’-UTR of target messenger RNAs, either targeting the transcripts for degradation or blocking their translation[5,6]. MicroRNA-155 (miR-155), whose expression is induced by inflammatory cytokines and toll-like receptor ligands, has been reported to be involved in tissue development, immune responses, hematopoiesis, and a number of other important physiological functions[7-9]. Because the dysregulation of these same physiological functions is frequently observed in various inflammation or inflammation-induced human diseases[10-12], miR-155 has received a great deal of interest. Recent studies have demonstrated that miR-155 is up-regulated in both UC and CD patients[13,14]; conversely, its deficiency protects mice from experimental colitis[15], although the underlying mechanism stills needs to be elucidated.
Previous studies have proven that the 145-kDa protein Src homology 2 domain-containing inositol 5’-phosphatase-1 (SHIP-1) is a primary target of miR-155[16,17]. The direct repression of SHIP-1 by miR-155 has been demonstrated in many mammalian cell types[18,19]. In fact, the phenotype observed in mice over-expressing miR-155 is closely related to that of SHIP-1 knockout mice[16]. Ubiquitously expressed in hematopoietic cells, SHIP-1 is at the nexus of intracellular signaling pathways in immune cells that mediate the immune response, production of inflammatory and immunosuppressive cytokines, immunoregulatory cell formation, autoimmune diseases, and immune cancers[20-22]. For example, the PI3K-Akt pathway is a crucial intracellular signaling pathway that mediates many biological processes, and SHIP-1 negatively regulates the PI3K-Akt cascade through the dephosphorylation of PIP3[23]. Recent evidence has shown that SHIP-1 is significantly decreased in leukemias and lymphomas[24,25], as well as in some chronic inflammatory diseases such as clinical and experimental arthritis[26]. However, there are few reports on SHIP-1 and IBD, and the currently available studies give inconsistent or even opposing results. In this study, we sought to determine the detailed relationship between miR-155, SHIP-1, and the pathogenesis of IBD by in vitro studies using raw264.7 cells and primary bone marrow-derived macrophages (BMDMs) and by in vivo studies using an experimental colitis mouse model induced by dextran sulfate sodium (DSS).
The raw264.7 cell line was obtained from the American Type Culture Collection and was maintained in low-glucose Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY, United States) supplemented with 10% fetal bovine serum and 1% Pen/Strep. Cells were incubated at 37 °C and in 5% CO2/95% air. BMDMs were isolated by flushing the femurs and tibias of Balb/c mice (female, 6-8 wk, Laboratory Animal Center, Chinese Academy of Sciences, Shanghai, China). Detailed procedures were performed as described previously[27]. BMDM phenotype and purity was determined by FACS analysis for macrophage specific antigen F4/80 (Abcam, United Kingdom). Before function studies, cells were exposed to E. coli lipopolysaccharide (LPS; 1 μg/mL; Sigma, St. Louis, MO, United States) for 24 h.
MiR-155 mimics (UUAAUGCUAAUUGUGAUAGGGGU) and negative controls (CCUACGCCACCAAUUUCGU) were provided by GenePharma (Shanghai, China). To express the murine SHIP-1 gene (Inpp5d), the coding sequence of Inpp5d was amplified from cDNA and was then subcloned into a pcDNA3.1 plasmid (Thermo Fisher Scientific, Waltham, MA), while silencing of SHIP-1 expression was achieved by designing a small-hairpin RNA targeting its coding sequence (shSHIP-1) and inserting this sequence into the vector. Transfection was performed in 6-well plates (5 × 106 cells/well), and the cells were mixed with Lipofectamine2000 reagent (Invitrogen, Carlsbad, CA). Cells were harvested 48 h after transfection for further analyses.
Forty pathogen-free female Balb/c mice were randomly separated into four groups (group 1, group 2, group 3, and group 4). Five mice per cage were maintained in an individual ventilated cage. All protocols concerning laboratory animal usage were submitted and validated by the Animal Care Ethics Committee of Shanghai First People’s Hospital and Nanjing Medical University. Groups 2, 3, and 4 were treated by oral administration of 4.0% (w/v) DSS (MP Biomedicals, Aurora, OH) dissolved in drinking water for 7 d, while group 1 was used as the control group and given normal drinking water. On day 2 and day 5, mice in group 4 were treated with antagomiR-155 (GenePharma) by tail vein injection at doses of 45 mg/kg in 100 μL volumes. Meanwhile, group 3 was treated with a negative control (GenePharma) and group 2 was untreated. The sequences of antagimiR-155 and the negative control were as follows: antagomiR-155: 5’-AsCsCCCUAUCACAAUUAGCAUsUsAsAs-Cholesterol-3’; negative control: 5’-UsUsUGUACUACACAAAAGUAsCs UsGs-Cholesterol-3’.
During the induction phase, weight loss, stool character and bleeding were recorded daily to monitor the disease activity, and the disease activity index (DAI) was determined as previously described[28]. Mice were sacrificed under deep anesthesia at the end of day 7. The colon tissues were stored in 10% buffered formalin or at -80 °C in liquid nitrogen after the colon length was measured and photographed.
Histological examination of the distal colon was performed on paraffin-embedded sections by hematoxylin-eosin (HE) staining. The inflammatory damage score was determined as previously described[29] and was the sum of inflammation infiltrations, depth of lesions, destruction of crypt, and width of lesions. Immunohistochemistry for SHIP-1 was performed using the peroxidase-conjugated avidin-biotin method. Deparaffinized and rehydrated sections were incubated with rabbit polyclonal anti-SHIP-1 (1:300, Santa Cruz, CA, United States) followed by biotinylated secondary antibody (Mai Bio, Shanghai, China). Positive staining was indicated by gray and brown particles. Ten visual fields (× 400 magnification) were chosen randomly in each section for evaluation of stained cells. The final score was the product of the number of stained cells and staining intensities. Detailed counting methods are listed in Supplementary Table 1.
Total RNA was extracted from cells or tissues by the TriPure Reagent (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Reverse transcription was performed using the Transcriptor First Strand cDNA Synthesis kit (Roche). The single-stranded cDNA served as the template for SYBR real-time polymerase chain reaction (PCR) using SYBRGreen PCR Master Mix (Takara Bio, Kyoto, Japan). All reactions were run in triplicate on the MasterCycler Real-Time PCR Detection System (Eppendorf, Hamburg, Germany). Supplementary Table 2 lists all primer sequences used in the study. The fold change of gene expression was calculated using the 2-ΔΔCT method. The expression level of miR-155 was normalized to U6 snRNA, and the expression levels of other genes were normalized to GAPDH.
Cells or colon tissues (stored at -80 °C) were harvested and extracted using the lysis buffer, and an equal amount of protein was separated on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Separated protein bands were transferred into PVDF membranes and then blocked in 5% skim milk powder. The primary antibody against SHIP-1 (Santa Cruz) was diluted according to the manufacturer’s instructions and incubated with the membrane overnight at 4 °C, followed by incubation with secondary antibodies (1:1000 dilution; Mai Bio) at room temperature for 2 h. The immunoreactive bands were visualized using an ECL-PLUS kit (Piscataway, NJ). The relative protein expression levels were normalized to GAPDH.
The murine Inpp5d target site and its mutant version were amplified by primers. The target site was predicted by three databases (miRBase, PicTar and miRanda). The PCR products were cloned downstream of the luciferase gene in psiCHECK-2 luciferase vector (Promega, WI, United States). The constructs were transfected together with miR-155 mimics or the negative controls. Luciferase activity was measured using the Dual-Luciferase Reporter Assay (Promega) 24 h after transfection. Each treatment was performed in triplicate.
Cell proliferation was analyzed using an MTT assay. Briefly, 1 × 103 cells per well were seeded into a 96-well plate and incubated for three days. At the indicated time point, 20 μL of MTT (5 mg/mL) (Sigma-Aldrich) was added into each well and incubated for 4 h. Then, the supernatants were removed and 150 μL of DMSO (Sigma-Aldrich) was added to terminate the reaction. The absorbance value (OD) was measured at 570 nm.
The levels of TNF-α, IL-6, IL-1β and IFN-γ in cell lysate supernatants were measured using corresponding enzyme-linked immunosorbent assay (ELISA) kits (Mai Bio) according to the manufacturer’s instructions.
SPSS 21.0 and GraphPad Prism 5 were used for statistical analyses and the rendering of figures. One-way analysis of variance (ANOVA) was used to analyze the differences between groups. The Student-Newman-Keuls method of multiple comparisons was used when the ANOVA analysis resulted in statistical significance. Data are expressed as the means ± SD. Statistical significance was set at P < 0.05.
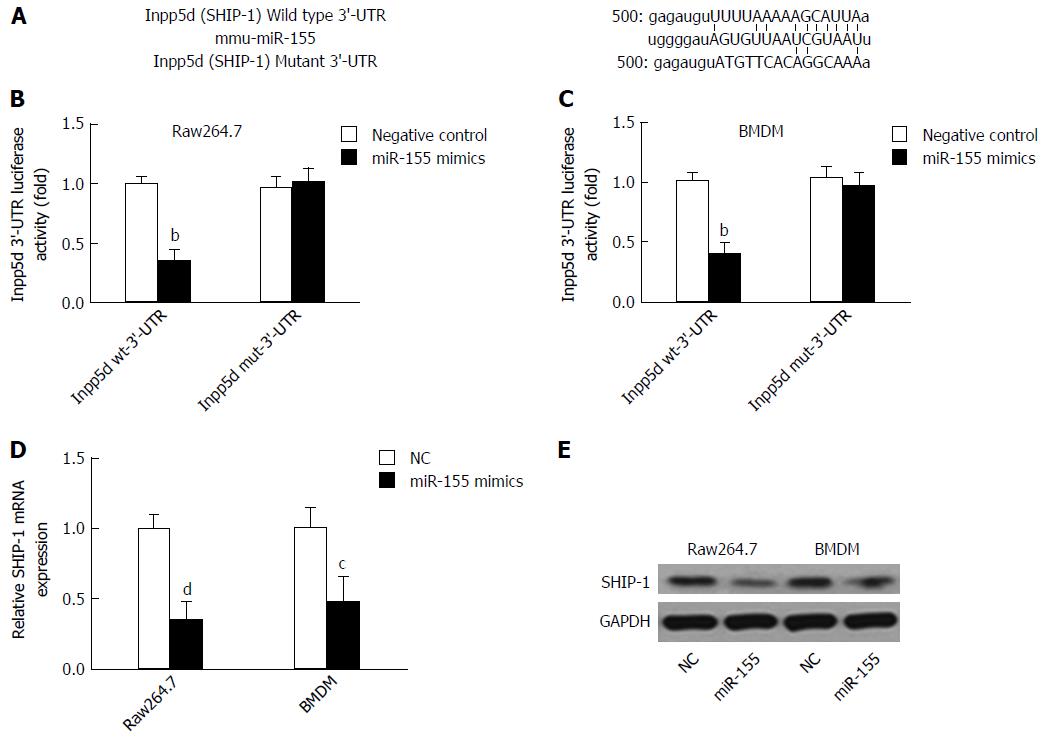
Because SHIP-1 is a well-established target of miR-155, we first performed a dual-luciferase reporter assay by constructing luciferase reporter constructs containing the wild-type or mutant SHIP-1 3’-UTR and co-transfecting them with miR-155 mimics or negative controls into cells (Figure 1A). We found that miR-155 directly bound to the wild-type but not the mutant 3’-UTR of SHIP-1 mRNA and caused the significant reduction of luciferase activities in both the murine macrophage cell line raw264.7 and the primarily isolated BMDM cells (Figure 1B and C). Then, we focused on the expression of SHIP-1 in miR-155 over-expressing raw264.7 cells and BMDMs. As shown in Figure 1D and E, both the mRNA and the protein levels of SHIP-1 were significantly decreased after cells were transfected with 60 nmol/L miR-155 mimics, which confirmed that SHIP-1 is the direct target of miR-155 in murine macrophages.
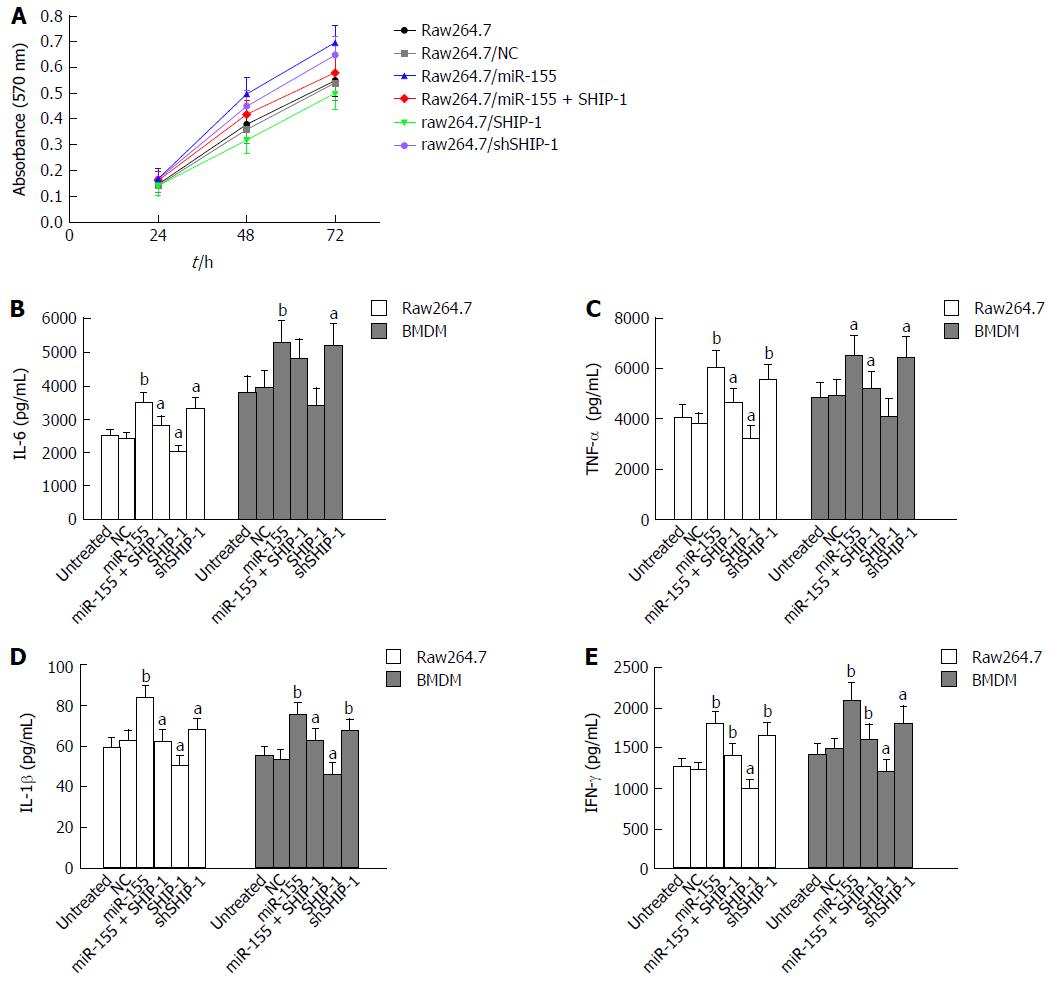
An MTT assay was performed to test the effects of miR-155 and its target SHIP-1 on the proliferation of raw264.7 cells. Cell proliferation was significantly elevated following the over-expression of miR-155 and was decreased after the up-regulation of SHIP-1 (Figure 2A). ELISA analysis was then conducted to determine whether miR-155 and SHIP-1 affected the pro-inflammatory secretions of LPS-stimulated raw264.7 cells and primary BMDMs. After exposure to LPS (1 μg/mL) for 24 h, both the raw264.7 cells and BMDMs showed remarkable secretion levels of IL-6, TNF-α, IL-1β, and IFN-γ, which represent the most important pro-inflammatory cytokines in IBD. The cells that over-expressed miR-155 exhibited the highest levels of secretion of these factors, while SHIP-1 restoration could inhibit the over-production of IL-6, TNF-α, IL-1β, and IFN-γ in these two cell types (Figure 2B-E). These results indicate that miR-155 serves its pro-inflammatory function by repressing SHIP-1 expression in macrophages.
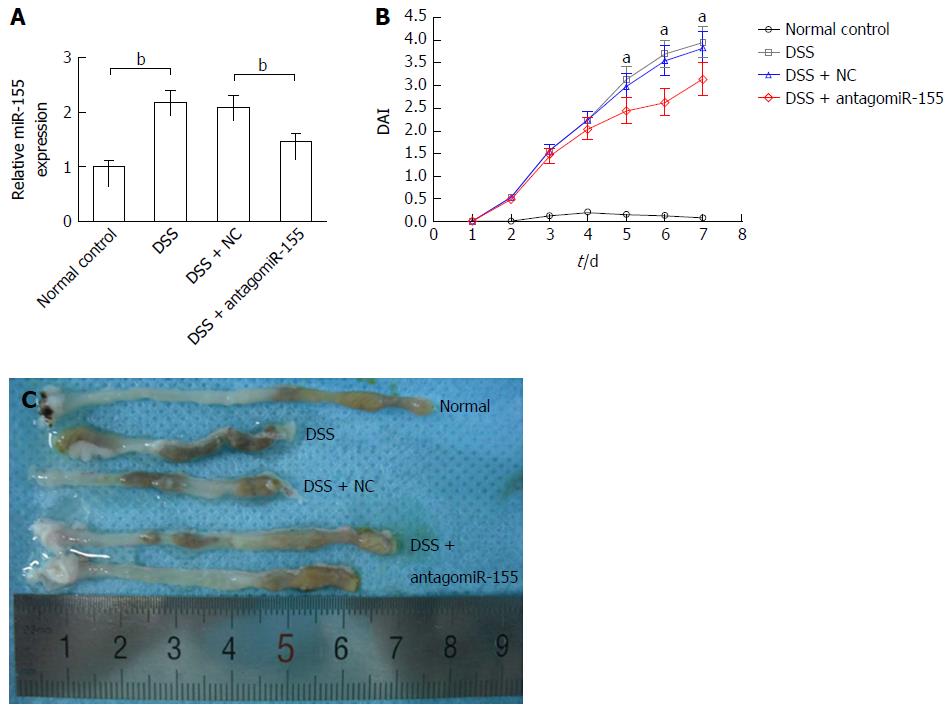
DSS-induced experimental colitis was established in Balb/c mice to determine the role of miR-155 and SHIP-1 in vivo. On day 2 and day 5, one group of mice was injected with 45 mg/kg antagomiR-155 through the tail vein, while another group was treated with the same dose of negative controls. Our PCR analysis confirmed that the level of miR-155 in murine colons was significantly reduced by antagomiR-155 treatment (Figure 3A). During the 7-d DSS induction, changes in body weight, occult blood, and gross bleeding were assessed and scored for the determination of DAI scores. As shown in Figure 3B, the DSS-treated groups exhibited higher DAI scores compared to the normal distilled water-treated group. In the three DSS-treated groups, it was observed that the mice that received antagomiR-155 injection exhibited significantly lower DAI compared with the mice that received random antagomiR treatment and the untreated mice. Additionally, the colon length of DSS-treated mice was markedly shortened compared to controls (8.23 ± 1.35 cm vs 5.45 ± 1.29 cm, P < 0.05), and the inhibition of miR-155 could partly abate such shortening (6.82 ± 1.41 cm vs 8.23 ± 1.35 cm, P > 0.05) (Figure 3C).
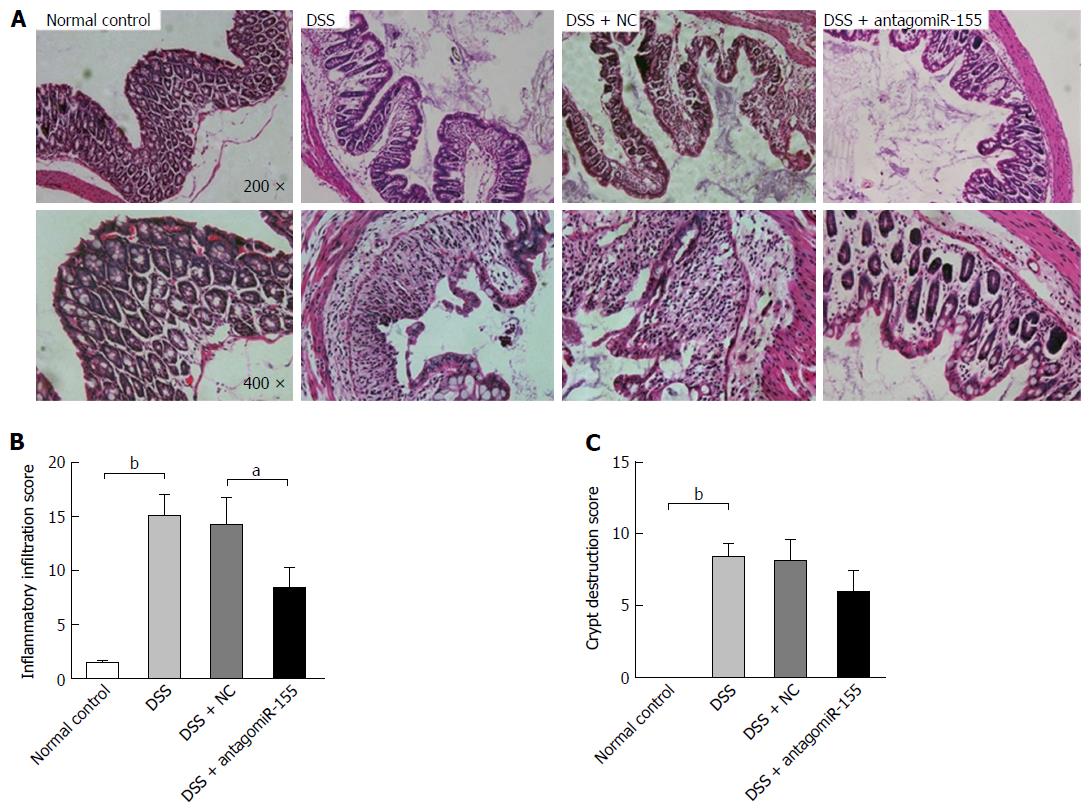
Thereafter, we evaluated the histological changes in colon sections by HE staining and found that the DSS-treated mice exhibited the typical characteristics of intestinal inflammation compared with the normal control mice (Figure 4A). The mice co-treated with antagomiR-155 displayed remarkably reduced levels of colon inflammation, including neutrophil infiltration, epithelial damage, depletion of goblet cells, and distortion of crypt architectures, compared to the mice treated with only DSS or the mice treated with DSS and random antagomiRs (Figure 4B and C).
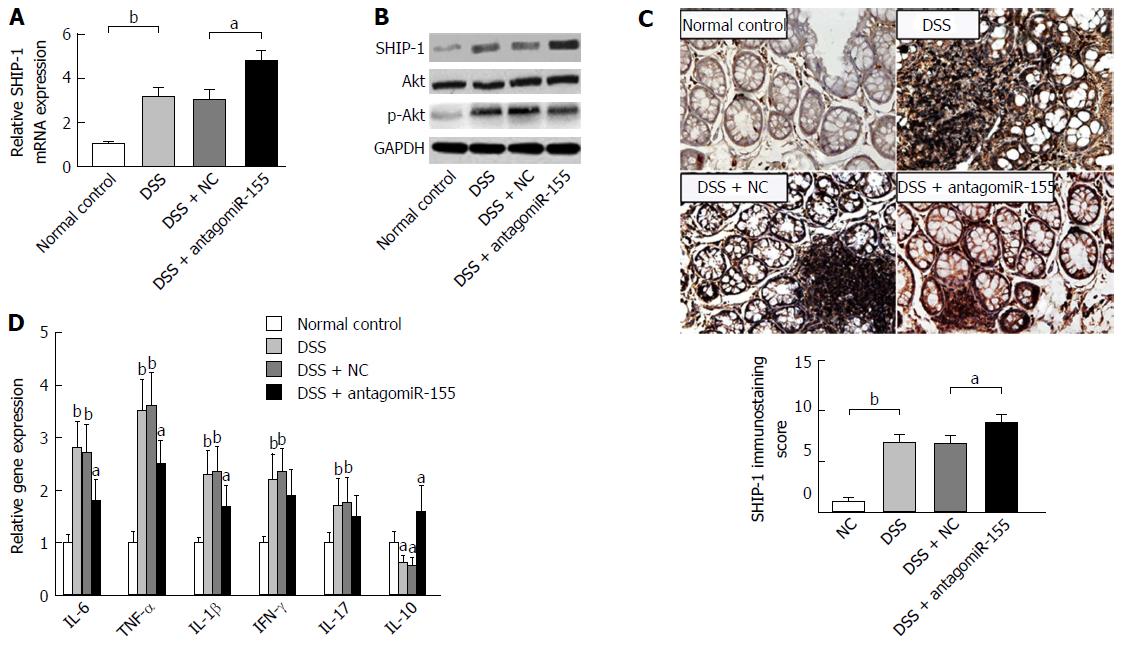
We performed an expression analysis of SHIP-1 in distal colon tissues and found that both the RNA and protein levels of SHIP-1 were significantly increased with antagomiR-155 administration (Figure 5A and B), whereas activity of its major functional target, the Akt signaling pathway, was decreased due to the enhancement of the negative regulation of SHIP-1 upon p-Akt activation (Figure 5B). IHC analysis showed that SHIP-1 was mainly expressed in lymphocytes, neutrophils, and other hematopoietic cells in the inflamed mucosa (Figure 5C). Similarly, the mice co-treated with antagomiR-155 exhibited higher positive staining of SHIP-1 than the mice in other groups. Since the activation of Akt is implicated in cell proliferation, survival, and pro-inflammatory release[30], we then investigated whether the change in SHIP-1 expression and downstream Akt signaling was associated with the inflammatory response in murine colon mucosa. PCR analysis revealed that the main pro-inflammatory mediators in colitis, including IL-6, TNF-α, IL-1β, IFN-γ, and IL-17, were all suppressed to a large degree after antagomiR-155 treatment; however, the paramount anti-inflammatory factor IL-10 demonstrated an opposite trend in expression (Figure 5D), suggesting that the inhibition of miR-155 and the over-expression of SHIP-1 could be an effective strategy to alleviate or suppress the inflammatory cascade in colitis.
Understanding the underlying mechanisms that regulate gene expression and the complex interplay of pathogenic factors is essential to develop novel therapeutics in IBD. Thus far, the ability of microRNAs to target functional genes and intracellular biological signaling pathways has drawn great attention from bench to bedside[31,32]. Over the past few decades, the identification of microRNAs in IBD has made great progress as an initial step in this regard. As a multi-functional microRNA, miR-155 plays an important role in the etiology of autoimmune diseases, and its ectopic up-regulation has been reported in both UC and CD. However, the detailed mechanism by which miR-155 influences the pathogenesis of colitis remains to be elucidated. Since SHIP-1, an important cytoplasmic phosphatase that regulates the number and function of immune cells, has been demonstrated as the direct target of miR-155, we therefore investigated the possible role of miR-155 and SHIP-1 in colitis in the present study.
We first determined that SHIP-1 was directly regulated by miR-155 in murine macrophages including raw264.7 cells and primary BMDMs. As it is well known that macrophages serve as the core regulator of innate immune response during gut inflammation or infection, here we proved that SHIP-1 might play a role in a miR-155-triggered inflammatory cascade during colitis. Singh et al[15] reported that miR-155 deficiency protects mice from experimental colitis by reducing T cell responses, and Min et al[33] found that miR-155 contributes to cytokine secretion in colitis by targeting FOXO3a. In this study, we confirmed the pro-proliferation and pro-inflammation capabilities of miR-155 in murine macrophages. Furthermore, we found that these effects were accompanied by a marked decrease in SHIP-1 expression and that the restoration of SHIP-1 could effectively inhibit or reverse these effects. Since it was first cloned and characterized in 1996, the role of SHIP-1 in immunity and other physiological or pathological processes has gradually emerged from numerous studies[20,21]. Thus far, the dysregulation of SHIP-1 has been described in several chronic inflammation and autoimmune disorders. There have been reports concerning SHIP-1 silencing in immune cells or knockout in animal models leading to the increased release of inflammatory cytokines[34]. Kerr et al[35] in 2011 reported that Ship-1-/- mice develop spontaneous CD-like ileitis, which could be corrected by adoptive transfer of bone marrow from wildtype mice. They further proposed that this type of colitis probably resulted from the imbalance of intestinal immune cells caused by SHIP-1 deprivation. Most recently, Jin et al[36] identified that the miR-155-mediated down-regulation of SHIP-1 promotes gouty arthritis. All of these findings point towards a pivotal role of SHIP-1 in regulation of immune response in the body. Our analysis demonstrated that the anti-inflammation effect of SHIP-1 is possibly via the inhibition of the Akt signaling pathway, both in vitro and in vivo. The pro-inflammatory secretion of cytokines by macrophages was significantly suppressed upon the up-regulation of SHIP-1 expression, indicating a potential for its clinical utility in the future. Although there was a report documenting that the level of SHIP-1 is increased in the intestinal mucosa samples of IBD patients[37], we speculate that this finding was due to the presence of more lymphocytes, monocytes, and neutrophils infiltrating into the colorectal mucosa during colitis.
Previous studies have identified a number of microRNAs as diagnostic biomarkers or potential targets for IBD treatment, such as miR-21 and miR-31[32]. However, to date, no therapeutic manipulation of microRNAs in IBD has been reported in cell lines or animal models. In regards to miR-155, although its aberrant expression in colitis is well established, the prospect of a miR-155-targeted strategy has not been fully investigated. In the present study, we established a DSS-induced colitis model and treated it with antagomiR-155. As expected, the inhibition of miR-155 significantly alleviated the disease activity, the degree of intestinal inflammation, and the release of pro-inflammatory cytokines. We also demonstrated that these curative effects are closely associated with an increase in SHIP-1 expression. These data provide a strong proof-of-concept for miR-155- and SHIP-1-based therapeutic approaches that could modulate inflammation in IBD. Nevertheless, experimental data in a chronic colitis animal model should be provided for further validation.
In conclusion, our current study demonstrated that miR-155 contributes to the pathogenesis of colitis by targeting SHIP-1 expression. Therefore, the inhibition of miR-155 and the restoration of SHIP-1 could effectively alleviate intestinal inflammation and cytokine secretion. Although some other effects of this miR-155 targeting strategy still need to be considered and studied, we cannot help speculating that this promising therapeutic concept may emerge in the near future.
Inflammatory bowel disease (IBD) is one of the major threats to human digestive health and causes a significant increase in the incidence of colorectal cancer. However, thus far, the pathogenesis of IBD remains unclear, highlighting the need for a thorough understanding of its underlying mechanism.
MicroRNAs play important roles in IBD pathogenesis. microRNA-155 (miR-155) has been reported to be upregulated in human IBD samples and animal colitis models, and emerging lines of evidence are unraveling its functional targets, including SHIP-1.
The authors focus on the molecular mechanisms of miR-155 in the immunopathogenesis of IBD using a mouse model of dextran sulfate sodium-induced colitis. This work adds evidence to clarify that the reduction in SHIP-1 levels resulting from increased miR-155 expression is the reason why IBD patients have high levels of miR-155.
This study on the potential role and particularly the mechanisms of miR-155 in IBD is important for the clinical management of the disease and the development of novel therapeutic modalities.
This work offers new insight into the understanding of the inflammatory mechanisms in IBD.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Bian ZX, Chen WX, Zhu YL S- Editor: Qi Y L- Editor: A E- Editor: Zhang FF
| 1. | Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380:1606-1619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1151] [Cited by in RCA: 1521] [Article Influence: 117.0] [Reference Citation Analysis (5)] |
| 2. | Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet. 2012;380:1590-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1544] [Article Influence: 171.6] [Reference Citation Analysis (0)] |
| 4. | de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1100] [Article Influence: 122.2] [Reference Citation Analysis (1)] |
| 5. | Mo YY. MicroRNA regulatory networks and human disease. Cell Mol Life Sci. 2012;69:3529-3531. [PubMed] [DOI] [Full Text] |
| 6. | Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 804] [Cited by in RCA: 842] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 7. | Mashima R. Physiological roles of miR-155. Immunology. 2015;145:323-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 170] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 8. | Vigorito E, Kohlhaas S, Lu D, Leyland R. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253:146-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 248] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 9. | Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497-505. [PubMed] [DOI] [Full Text] |
| 10. | Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015;29:3595-3611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 374] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 11. | Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253:167-184. [PubMed] [DOI] [Full Text] |
| 12. | Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 280] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 13. | Béres NJ, Szabó D, Kocsis D, Szűcs D, Kiss Z, Müller KE, Lendvai G, Kiss A, Arató A, Sziksz E. Role of Altered Expression of miR-146a, miR-155, and miR-122 in Pediatric Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:327-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 14. | Svrcek M, El-Murr N, Wanherdrick K, Dumont S, Beaugerie L, Cosnes J, Colombel JF, Tiret E, Fléjou JF, Lesuffleur T. Overexpression of microRNAs-155 and 21 targeting mismatch repair proteins in inflammatory bowel diseases. Carcinogenesis. 2013;34:828-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Singh UP, Murphy AE, Enos RT, Shamran HA, Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143:478-489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 16. | O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci USA. 2009;106:7113-7118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 674] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 17. | Pedersen IM, Otero D, Kao E, Miletic AV, Hother C, Ralfkiaer E, Rickert RC, Gronbaek K, David M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol Med. 2009;1:288-295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 18. | Wu R, Li Y, Guo Z, Gong J, Zhu W, Li N, Li J. Triptolide ameliorates ileocolonic anastomosis inflammation in IL-10 deficient mice by mechanism involving suppression of miR-155/SHIP-1 signaling pathway. Mol Immunol. 2013;56:340-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 19. | Costinean S, Sandhu SK, Pedersen IM, Tili E, Trotta R, Perrotti D, Ciarlariello D, Neviani P, Harb J, Kauffman LR. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114:1374-1382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 20. | Kerr WG. Inhibitor and activator: dual functions for SHIP in immunity and cancer. Ann N Y Acad Sci. 2011;1217:1-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 21. | Condé C, Gloire G, Piette J. Enzymatic and non-enzymatic activities of SHIP-1 in signal transduction and cancer. Biochem Pharmacol. 2011;82:1320-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Kalesnikoff J, Sly LM, Hughes MR, Büchse T, Rauh MJ, Cao LP, Lam V, Mui A, Huber M, Krystal G. The role of SHIP in cytokine-induced signaling. Rev Physiol Biochem Pharmacol. 2003;149:87-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 23. | Viernes DR, Choi LB, Kerr WG, Chisholm JD. Discovery and development of small molecule SHIP phosphatase modulators. Med Res Rev. 2014;34:795-824. [PubMed] [DOI] [Full Text] |
| 24. | Hamilton MJ, Ho VW, Kuroda E, Ruschmann J, Antignano F, Lam V, Krystal G. Role of SHIP in cancer. Exp Hematol. 2011;39:2-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 25. | Fukuda R, Hayashi A, Utsunomiya A, Nukada Y, Fukui R, Itoh K, Tezuka K, Ohashi K, Mizuno K, Sakamoto M. Alteration of phosphatidylinositol 3-kinase cascade in the multilobulated nuclear formation of adult T cell leukemia/lymphoma (ATLL). Proc Natl Acad Sci USA. 2005;102:15213-15218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, Reilly J, Ierna M, Fraser AR, Stolarski B. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. 2011;108:11193-11198. [PubMed] [DOI] [Full Text] |
| 27. | Lee CM, Hu J. Cell density during differentiation can alter the phenotype of bone marrow-derived macrophages. Cell Biosci. 2013;3:30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 28. | Li X, Xu Y, Zhang C, Deng L, Chang M, Yu Z, Liu D. Protective Effect of Calculus Bovis Sativus on Dextran Sulphate Sodium-Induced Ulcerative Colitis in Mice. Evid Based Complement Alternat Med. 2015;2015:469506. [PubMed] [DOI] [Full Text] |
| 29. | Cui Y, Wei H, Lu F, Liu X, Liu D, Gu L, Ouyang C. Different Effects of Three Selected Lactobacillus Strains in Dextran Sulfate Sodium-Induced Colitis in BALB/c Mice. PLoS One. 2016;11:e0148241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Patel RK, Mohan C. PI3K/AKT signaling and systemic autoimmunity. Immunol Res. 2005;31:47-55. [PubMed] |
| 31. | Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. MicroRNAs: new players in IBD. Gut. 2015;64:504-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 206] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 32. | Fisher K, Lin J. MicroRNA in inflammatory bowel disease: Translational research and clinical implication. World J Gastroenterol. 2015;21:12274-12282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 33. | Min M, Peng L, Yang Y, Guo M, Wang W, Sun G. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm Bowel Dis. 2014;20:652-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Kalesnikoff J, Baur N, Leitges M, Hughes MR, Damen JE, Huber M, Krystal G. SHIP negatively regulates IgE + antigen-induced IL-6 production in mast cells by inhibiting NF-kappa B activity. J Immunol. 2002;168:4737-4746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 108] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 35. | Kerr WG, Park MY, Maubert M, Engelman RW. SHIP deficiency causes Crohn’s disease-like ileitis. Gut. 2011;60:177-188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 36. | Jin HM, Kim TJ, Choi JH, Kim MJ, Cho YN, Nam KI, Kee SJ, Moon JB, Choi SY, Park DJ. MicroRNA-155 as a proinflammatory regulator via SHIP-1 down-regulation in acute gouty arthritis. Arthritis Res Ther. 2014;16:R88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 37. | Arijs I, De Hertogh G, Lemmens B, Van der Goten J, Vermeire S, Schuit F, Rutgeerts P. Intestinal expression of SHIP in inflammatory bowel diseases. Gut. 2012;61:956-957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |