Published online Feb 7, 2017. doi: 10.3748/wjg.v23.i5.810
Peer-review started: August 30, 2016
First decision: November 9, 2016
Revised: November 21, 2016
Accepted: December 8, 2016
Article in press: December 8, 2016
Published online: February 7, 2017
Processing time: 145 Days and 20.4 Hours
To investigate gut microbial diversity and the interventional effect of Xiaoyaosan (XYS) in a rat model of functional dyspepsia (FD) with liver depression-spleen deficiency syndrome.
The FD with liver depression-spleen deficiency syndrome rat model was established through classic chronic mild unpredictable stimulation every day. XYS group rats received XYS 1 h before the stimulation. The models were assessed by parameters including state of the rat, weight, sucrose test result and open-field test result. After 3 wk, the stools of rats were collected and genomic DNA was extracted. PCR products of the V4 region of 16S rDNA were sequenced using a barcoded Illumina paired-end sequencing technique. The primary composition of the microbiome in the stool samples was determined and analyzed by cluster analysis.
Rat models were successfully established, per data from rat state, weight and open-field test. The microbiomes contained 20 phyla from all samples. Firmicutes, Bacteroidetes, Proteobacteria, Cyanobacteria and Tenericutes were the most abundant taxonomic groups. The relative abundance of Firmicutes, Proteobacteria and Cyanobacteria in the model group was higher than that in the normal group. On the contrary, the relative abundance of Bacteroidetes in the model group was lower than that in the normal group. Upon XYS treatment, the relative abundance of all dysregulated phyla was restored to levels similar to those observed in the normal group. Abundance clustering heat map of phyla corroborated the taxonomic distribution.
The microbiome relative abundance of FD rats with liver depression-spleen deficiency syndrome was significantly different from the normal cohort. XYS intervention may effectively adjust the gut dysbacteriosis in FD.
Core tip: Gut microflora has been found to associate with promotion of health and occurrence and development of different gastrointestinal/non-gastrointestinal disease. Some traditional Chinese drugs exert effects on diseases by regulating the equilibrium of intestinal microflora. In the present study, we described gut microbial diversity in functional dyspepsia with liver depression-spleen deficiency syndrome in rat models and the effect of Xiaoyaosan intervention.
- Citation: Qiu JJ, Liu Z, Zhao P, Wang XJ, Li YC, Sui H, Owusu L, Guo HS, Cai ZX. Gut microbial diversity analysis using Illumina sequencing for functional dyspepsia with liver depression-spleen deficiency syndrome and the interventional Xiaoyaosan in a rat model. World J Gastroenterol 2017; 23(5): 810-816
- URL: https://www.wjgnet.com/1007-9327/full/v23/i5/810.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i5.810
Functional dyspepsia (FD) is a common gastrointestinal disorder[1]. FD is not life-threatening but adversely impacts the patient’s quality of life. The pathogenesis of FD is uncertain because of complicated multiple factors. Clinical observation has shown that patients with FD suffer from obvious gastrointestinal motility obstacles, visceral hypersensitivity, Helicobacter pylori infection, neuropsychological factors and so on[2]. The occurrence and development of FD is closely related with mood disorders, such as anxiety and depression. Traditional Chinese Medicine (TCM) holds that gastrointestinal function abatement is usually due to spleen deficiency (pixu) and mood disorders with associated liver depression (ganyu). Thus, liver depression-spleen deficiency is regarded as one of the main pathogenesis of FD. In all syndrome types, FD with liver depression-spleen deficiency syndrome is the most common FD in traditional Chinese medical syndrome typing[3].
Xiaoyaosan (XYS) is a well-known Chinese herbal formula and is prescribed to sooth liver, tonify spleen, and nourish blood[4,5]. The Song Dynasty (960-1127 AD) book of “Taiping Huimin Heji Jufang” recorded that XYS comprises the following eight Chinese herbs: Bupleurum root, Chinese angelica root, white peony root, Perenniporia, bighead Atractylodes rhizome, roasted ginger, prepared licorice root, menthol and peppermint. It has long been used for the treatment of FD associated with the syndrome of “liver depression” and “spleen deficiency” in China.
Gastrointestinal microflora plays an important role in the host’s life activities[6,7]. When the host’s balance of microflora in the gastrointestinal tract is disrupted, the host may become ill. In recent years, the gastrointestinal microflora has been shown to be associated with promotion of health and occurrence and development of different gastrointestinal/non-gastrointestinal diseases[8,9]. Therefore, more experimental research has focused on the relationship between disease and gastrointestinal microflora. Besides, research has shown that some traditional Chinese drugs exert their effects on disease by regulating the equilibrium of intestinal microflora[10,11]. Many molecular biology techniques have been used to illustrate gut microbial diversity, including denaturing/temperature gradient gel electrophoresis, terminal restriction fragment length polymorphism, and high-throughput sequencing[12]. High-throughput sequencing has the advantage of providing more and detailed information on microbial diversity with high accuracy, and thus has been widely used in the study of gut microbial diversity. Here, we used the Illumina sequencing to analyze gut microbial diversity of FD. The goal of the current study was to describe gut microbial diversity on FD with liver depression-spleen deficiency syndrome in a rat model and to assess the effect of XYS on microflora.
Male Sprague-Dawley rats of clean grade were supplied by the Experimental Animal Center of Dalian Medical University, Dalian, China. Rats were kept at a temperature of 23 ± 2 °C and a relative humidity of 55% ± 2% on a 12 h light-dark cycle for 7 d before experimentations. Rats were randomly divided into three groups: normal group (FD1), FD with liver depression-spleen deficiency syndrome group (FD2) and XYS-treated group (FD3). The FD with liver depression-spleen deficiency syndrome rat model was established by including classic chronic mild unpredictable stimulation (bondage, swim-induced fatigue, electrical stimulation, fasting and concussion) every day[13]. The normal and FD model groups were given normal saline intragastrically before initiating stimulation. The XYS group was given XYS intragastrically 1 h before initiating stimulation. The dose of XYS was 15.3 g/kg per day (gavage volume 2 mL).
The models were assessed by state of the rat, weight, sucrose test result and open-field test result. All the procedures of animal experiments were approved by the local Animal Care Committee and in accordance with the Guide for the Care and Use of Laboratory Animals published by the Science and Technology Commission of P.R.C. (STCC Publication No. 2, revised 1988).
Stool samples were collected from rats of FD1, FD2 and FD3 groups. The stools were suspended, respectively, in TE buffer with lysozyme. Total bacterial DNAs were extracted using the Stool DNA Isolation Kit (FORE GENE, China) according to the manufacturer’s instruction. The total DNAs were determined using electrophoresis on 1% agarose gel containing ethidium bromide. The concentration of DNAs was measured using NanoVue plus (GE, United States).
The variable V4 region of 16S rRNA was amplified by PCR using the following barcode primers: upstream primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and downstream primer 806R (5′-GGACTACHVGGGTWTCT AAT-3′). The PCR reaction mixture contained 12.5 μL of Phusion Master Mix (Phusion® High-Fidelity PCR Master Mix with GC Buffer; New England Biolabs, United States), 1 μL of template DNA, 1.25 μL of 10 μmol primer 515F, 1.25 μL of 10 μmol primer 806R and 10 μL of double-distilled H2O. The amplification of PCR was performed as follows: 98 °C for 30 s; 30 cycles of 98 °C for 10 s, 54 °C for 30 s, and 72 °C for 30 s; and 72 °C for 7 min. The amplified products were identified by electrophoresis on a 2% agarose gel containing ethidium bromide. The PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen, Germany).
Library construction was performed using the TruSeq® PCR-Free DNA Sample Preparation Kit and quantified by Qubit and Q-PCR. After library accreditation, purified PCR products of 16S rDNA were sequenced using HiSeq2500 sequencer PE250. Sequencing work was performed in collaboration with Novogene Bioinformatics Technology Co., Ltd (Beijing, China).
The animal protocol was designed to minimize pain or discomfort to the animals. The animals were acclimatized to laboratory conditions (23 ± 2 °C, 12 h/12 h light/dark, 55% ± 2% humidity, ad libitum access to food and water) for 1 wk prior to experimentation. All animals were euthanized by ethyl ether for tissue collection.
After amputation of the barcode and primer sequences, Raw Tags were obtained by joining reads of each sample using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/)[14]. Clean Tags were obtained through the strict filtering process[15]. Effective Tags of all samples were clustered using Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/)[16]. Operational Taxonomic Units (OTUs) were defined by clustering 97% identity sequences. Based on the principles of the algorithm, the highest frequency OTUs were screened as the representative OTUs. The microbiomes species at phylum level were annotated by analyzing the representative OTUs with RDP Classifier (version 2.2, http://sourceforge.net/projects/rdp-classifier/) method[17] and GreenGene database (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi)[18]. The phylogenetic relationships of representative OTUs were identified by multiple sequence alignment using PyNAST software (version 1.2)[19] and “Core Set” of GreenGene database.
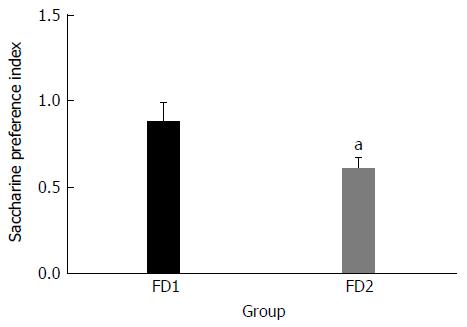
The FD with liver depression-spleen deficiency syndrome rat model was established successfully using chronic mild unpredictable stimulation. Compared with the good mental state of the normal group rats, the model group rats were restless and exhibited high alertness. The fur of the model group rats was slightly rough. It was difficult for the model group rats to grasp. The weight of the model group rats significantly decreased, compared to that of the normal group rats (Table 1). The saccharine preference index of the model group rats was significantly lower than that of the normal group rats (Figure 1). At the end of the modeling process, the numbers of crossed-grids, standing and grooming times were calculated. There were significant differences between the model group and the normal group in all these parameters (Table 2).
| Model before | Model after | |
| FD1 | 220.33 ± 1.20 | 264.17 ± 1.17 |
| FD2 | 220.50 ± 1.06 | 236.00 ± 1.17a |
Amplified PCR products were examined by agarose gel electrophoresis (Figure 2). The V4 region PCR products of 16S rDNA were obtained.
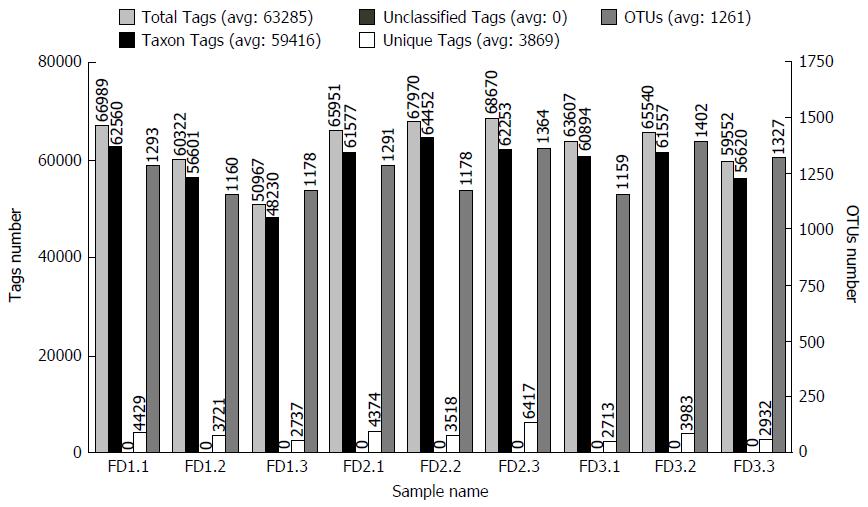
The number of Effective Tags and OTUs observed after sequencing are shown in Figure 3. The Effective Tags of all samples were enough to cluster to OTUs. The representative OTUs were used for annotating.
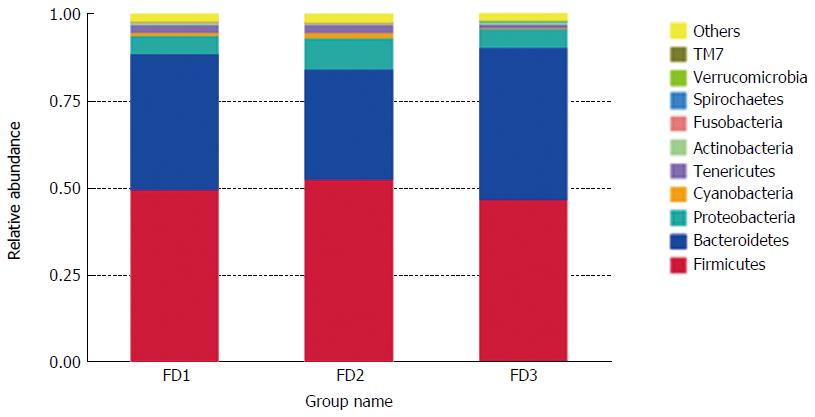
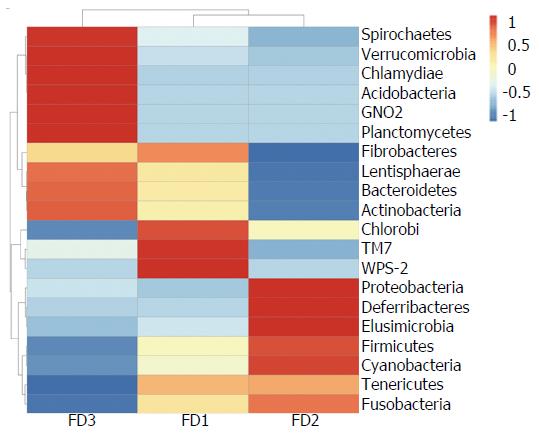
The microbiomes contained 20 phyla and 156 genera in all rat stool samples (Figure 4). The composition microbiome of all samples was similar at the phyla level. Firmicutes, Bacteroidetes, Proteobacteria, Cyanobacteria and Tenericutes were the most abundant taxonomic groups. The relative abundance of Firmicutes, Proteobacteria and Cyanobacteria in the FD model group was individually higher than in the normal group. On the contrary, the relative abundance of Bacteroidetes in the FD model group was lower than observed in the normal group. After XYS treatment, the relative abundance of all altered phyla readjusted to levels comparable to those observed in the normal rats.
The abundance clustering heat map of phyla was consistent with results of phylum level taxonomic distribution (Figure 5).
In this study, we modeled FD with liver depression-spleen deficiency syndrome in rats and observed changes in gut microbial diversity with or without XYS intervention.
FD is a common clinical digestive disease with epigastric discomfort, postprandial full bilge, abdominal bloating, belching, anorexia, nausea, vomiting, heartburn and other symptoms that occur persistently or recurrently[20]. FD with liver depression-spleen deficiency syndrome is one of the most common FD subtypes in TCM. Modern medicinal knowledge has indicated psychiatric factors as the most important etiologic factors. We used unpredictable stimulations to mimic psychiatric stress to induce FD in rats[21]. These stimulations led to successful FD with liver depression-spleen deficiency syndrome through making rats nervous and depressive for a long time.
Some studies have indicated the influence that gut microbial diversity may have on different diseases, including gastrointestinal disease, incretion disease and mental disease[22]. Therefore, understanding the interaction between the microbiota and the host’s physiology may facilitate the exploration of new therapeutic targets for certain diseases. FD with liver depression-spleen deficiency syndrome belongs to a kind of gastrointestinal disease caused by psychiatric factors. While XYS was the TCM prescription used for the treatment of FD with liver depression-spleen deficiency syndrome[23], it has become widely accepted in microbial diversity analysis by employing the sequencing technique in this postmetagenomic era.
In our study, the microbiomes of the normal group, the FD model group and the XYS-treated FD group rats were classified at the phylum level. Firmicutes, Bacteroidetes and Proteobacteria were the three most abundant taxonomic groups in all group samples. These groups of microorganisms have, in fact, been indicated as the three predominant phyla in the gut of animals and humans[24]. The experimental groups had similar microbiome distributions but with varied phyla abundances within each microbiome. Firmicutes was the most favored microflora and the highest in proportion in the gut of mammals. Many bacteria of the phyla Firmicutes can produce butyrate. Butyrate is an important source of energy for colonic epithelial cells[25]. Bacteroidetes and Proteobacteria were the second and third favored microflora, respectively. Many Bacteroidetes are involved in the metabolic processing of polysaccharides, steroids and bile acids, whereas Proteobacteria are pathogenic[26,27]. The increased relative abundance of Firmicutes and Proteobacteria coupled with the decreased relative abundance of Bacteroidetes in the FD microbiome as compared with the microbiome of normal rats suggests an association between increased Proteobacteria with decreased Bacteroidetes and the occurrence of FD with liver depression-spleen deficiency syndrome. Although the relative abundance of Fusobacteria was generally low, it indicated a plausible positive association with Proteobacteria. Fusobacteria is a common resident pathogen in gut mucosa and has been linked to gastrointestinal disease[28]. Upon treatment of FD rats with XYS, the relative abundances of Firmicutes, Proteobacteria and Bacteroidetes were adjusted to similar levels, as identified in the microbiome of normal rats, with enhanced decrease of Fusobacteria. We infer that the therapeutic function of XYS on FD may, at least partly, be due to its ability to restore gut microbial homeostasis. Notwithstanding, further studies are needed to define the molecular factors and downstream mechanisms that are chiefly influenced during the dysbiosis-microbiome restoration activities in FD.
In summary, this study investigated the gut microbial diversity of FD in a rat model and indicated that the microbiome composition of FD with liver depression-spleen deficiency syndrome was significantly different from the normal cohort. Intervention with XYS restored the gut dysbiosis in FD to normalcy. The data thus shed light on a plausible means by which XYS may achieve its therapeutic function in FD.
Functional dyspepsia (FD) with liver depression-spleen deficiency syndrome is not life-threatening but adversely impacts the patient’s quality of life. Xiaoyaosan (XYS) is a common Chinese herbal formula and has long been used for the treatment of FD associated with the syndrome of “liver depression” and “spleen deficiency” in China. Gastrointestinal microflora plays an important role in the host’s life activities. Descriptions of the gut microbial diversity of FD rat models and those intervened with XYS will facilitate the understanding of FD pathogenesis and clarify the mechanism of XYS.
Gastrointestinal microflora has been found to associate with promotion of health and occurrence and development of different gastrointestinal/non -gastrointestinal diseases in recent years. Besides, research has shown that some traditional Chinese drugs exert their effects on disease by regulating the equilibrium of intestinal microflora.
Gut microbial diversity is related to different diseases, such as gastrointestinal disease, incretion disease and mental disease. Some traditional Chinese drugs exert their effects on disease by regulating the equilibrium of intestinal microflora. There is no report to date on the relationship between gut microbial diversity, FD with liver depression-spleen deficiency syndrome and XYS.
These data will facilitate the exploration of new therapeutic targets of disease by increasing the understanding of the interaction between the microbiota and the host’s physiology. Clarification of the XYS intervention will provide a theoretical basis for promotion and application of Chinese traditional medicine.
Illumina sequencing, also known as next-generation sequencing, describes sequencing of millions of gene fragments by synthesis with reversible terminators.
The manuscript is interesting and worthy of publication.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Pessi T, Schuller HM S- Editor: Yu J L- Editor: Filipodia E- Editor: Zhang FF
| 1. | Talley NJ, Walker MM, Holtmann G. Functional dyspepsia. Curr Opin Gastroenterol. 2016;32:467-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Talley NJ. Systemic review: the prevalence and clinical course of functional dyspepsia. Aliment Pharmacol Ther. 2004;19:643-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 219] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 3. | Yang F, Jin XJ. TCM treatment for functional dyspepsia. Jilin Zhongyiyao. 2014;34:4. |
| 4. | Zhang Y, Han M, Liu Z, Wang J, He Q, Liu J. Chinese herbal formula xiao yao san for treatment of depression: a systematic review of randomized controlled trials. Evid Based Complement Alternat Med. 2012;2012:931636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 5. | Du HG, Ming L, Chen SJ, Li CD. Xiaoyao pill for treatment of functional dyspepsia in perimenopausal women with depression. World J Gastroenterol. 2014;20:16739-16744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5700] [Cited by in RCA: 5584] [Article Influence: 279.2] [Reference Citation Analysis (2)] |
| 7. | Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2779] [Cited by in RCA: 3077] [Article Influence: 236.7] [Reference Citation Analysis (0)] |
| 8. | Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330-339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1341] [Cited by in RCA: 1555] [Article Influence: 172.8] [Reference Citation Analysis (0)] |
| 9. | Barbara G, Feinle-Bisset C, Ghoshal UC, Quigley EM, Santos J, Vanner S, Vergnolle N, Zoetendal EG. The Intestinal Microenvironment and Functional Gastrointestinal Disorders. Gastroenterology. 2016;pii:S0016-5085(16)00219-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 231] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 10. | Peng Y, Wang Z, Lu Y, Wu CF, Yang JY, Li XB. Intestinal microflora molecular markers of spleen-deficient rats and evaluation of traditional Chinese drugs. World J Gastroenterol. 2009;15:2220-2227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Peng Y, Wu C, Yang J, Li X. Gut microbial diversity in rat model induced by rhubarb. Exp Anim. 2014;63:415-422. [PubMed] |
| 12. | Simrén M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 655] [Cited by in RCA: 649] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 13. | Deng N, Lin XY, Cai GX. Research survey of the functional dyspepsia TCM animal models. Huanqiu Zhongyiyao. 2013;6:3. |
| 14. | Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957-2963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7679] [Cited by in RCA: 9206] [Article Influence: 657.6] [Reference Citation Analysis (0)] |
| 15. | Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3022] [Cited by in RCA: 2667] [Article Influence: 222.3] [Reference Citation Analysis (0)] |
| 16. | Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8951] [Cited by in RCA: 9957] [Article Influence: 829.8] [Reference Citation Analysis (0)] |
| 17. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12842] [Cited by in RCA: 13174] [Article Influence: 731.9] [Reference Citation Analysis (0)] |
| 18. | DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069-5072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8383] [Cited by in RCA: 7550] [Article Influence: 397.4] [Reference Citation Analysis (0)] |
| 19. | Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2767] [Cited by in RCA: 2584] [Article Influence: 161.5] [Reference Citation Analysis (0)] |
| 20. | Talley NJ, Holtmann G, Walker MM. Therapeutic strategies for functional dyspepsia and irritable bowel syndrome based on pathophysiology. J Gastroenterol. 2015;50:601-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 21. | Yue LF, Ding J, Chen JX, Yue GX, Liang Y, Huo Sk, Li JJ. Establishment and review of rat model of syndrome of liver depression with spleen in sufficiency. Beijing Zhongyiyao Daxue Xuebao. 2008;31:5. |
| 22. | Yoshikawa K, Shimada M, Kuwahara T, Hirakawa H, Kurita N, Sato H, Utsunomiya T, Iwata T, Miyatani T, Higashijima J. Effect of Kampo medicine “Dai-kenchu-to” on microbiome in the intestine of the rats with fast stress. J Med Invest. 2013;60:221-227. [PubMed] |
| 23. | Zhao J. 76 Cases with Functional Indigestion Treated by Modified Xiaoyao Powder. Henan Zhongyi. 2015;35:2. |
| 24. | Lu Y, Chen J, Zheng J, Hu G, Wang J, Huang C, Lou L, Wang X, Zeng Y. Mucosal adherent bacterial dysbiosis in patients with colorectal adenomas. Sci Rep. 2016;6:26337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 25. | den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325-2340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2408] [Cited by in RCA: 3268] [Article Influence: 272.3] [Reference Citation Analysis (3)] |
| 26. | Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1476] [Cited by in RCA: 2431] [Article Influence: 243.1] [Reference Citation Analysis (0)] |
| 27. | Rajilić-Stojanović M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 685] [Cited by in RCA: 770] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 28. | Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 550] [Article Influence: 55.0] [Reference Citation Analysis (0)] |