Published online Nov 21, 2017. doi: 10.3748/wjg.v23.i43.7657
Peer-review started: April 25, 2017
First decision: June 22, 2017
Revised: September 1, 2017
Accepted: November 1, 2017
Article in press: November 1, 2017
Published online: November 21, 2017
Processing time: 212 Days and 11.1 Hours
Hepatitis B virus (HBV) is a non-cytopathic virus that causes acute and chronic inflammatory liver diseases, often leading to the pathogenesis of hepatocellular carcinoma (HCC). Although many studies for the roles of HBV on pathogenesis of the liver diseases, such as non-alcoholic fatty liver disease (NAFLD), hepatic inflammation, cirrhosis, and HCC, have been reported, the mechanisms are not fully understood. Endoplasmic reticulum (ER) and mitochondria have the protective mechanisms to restore their damaged function by intrinsic or extrinsic stresses, but their chronic dysfunctions are associated with the pathogenesis of the various diseases. Furthermore, HBV can affect intra- or extracellular homeostasis through induction of ER and mitochondrial dysfunctions, leading to liver injury. Therefore, the mechanism by which HBV induces ER or mitochondrial stresses may be a therapeutic target for treatment of liver diseases.
Core tip: Endoplasmic reticulum (ER) is the major site of protein folding and calcium storage. Beside the role of ER in protein homeostasis, it controls the cholesterol production and lipid-membrane biosynthesis as well as surviving and cell death signaling mechanisms in the cell. It is well-documented that abnormal metabolic regulation induces adverse effects in liver disorders, such as non-alcoholic steatosis hepatitis, fibrosis, cirrhosis, and hepatocellular carcinoma which are associated with hepatitis B virus (HBV) infection. Recent animal model and human studies have showed ER stress as an emerging factors involved in the development of metabolic and liver diseases. In this review, we will summarize the crucial effects of ER stress response in the pathogenesis of HBV-induced liver diseases.
- Citation: Kim SY, Kyaw YY, Cheong J. Functional interaction of endoplasmic reticulum stress and hepatitis B virus in the pathogenesis of liver diseases. World J Gastroenterol 2017; 23(43): 7657-7665
- URL: https://www.wjgnet.com/1007-9327/full/v23/i43/7657.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i43.7657
Hepatitis B virus (HBV), a prototype member of the Hepadnavirus family, is a small enveloped DNA virus with a virion diameter of 42 nm. The HBV genome is a relaxed circular, partially double-stranded DNA molecule encoding four overlapping open reading frames (ORFs), named C, S, P, and X coding for core protein, surface proteins (pre-S1, pre-S2, and S), DNA polymerase, and X protein, respectively[1]. Of the HBV-encoded proteins, the function of hepatitis B virus X protein (HBx) is not clearly understood, but it may function as a multifunctional transactivator in HBV replication and host gene transcription through interaction with host proteins[2]. HBV primarily infects hepatocytes and causes acute and chronic liver diseases. In particular, chronic HBV infection can lead to cirrhosis of the liver, liver failure, liver cancer, and even death. According to the report of World Health Organization (WHO), there are more than 350 million people worldwide who have chronic HBV infections and more than 780 thousand people die every year due to the acute or chronic HBV infection[3]. To date, many studies have been reported on the molecular mechanisms for relation between HBV infection and pathogenesis of hepatic diseases, but the mechanisms are still not fully understood.
Cellular organelle is a specialized compartment enclosed by lipid bilayers within a cell and has specific functions. It is classified into major organelle such as endoplasmic reticulum (ER), Golgi apparatus, mitochondria, vacuole, and nucleus and minor organelle such as autophagosome, lysosome, peroxisome, and vesicle. Damage or dysfunction of cellular organelles by intra- or extra-cellular stress is associated with the pathogenesis of various diseases. For examples, mitochondrial dysfunction induces the diseases such as myopathy, diabetes, and multiple endocrinopathy[4] and ER dysfunction induces the diseases such as obesity, diabetes, atherosclerosis, and cancer[5]. Here, we review relation between the HBV-encoded proteins and damage of cellular organelles and the influence on pathogenesis of hepatic diseases.
Mitochondria are the double membrane-bound structure and consist of five compartments with specialized functions including the outer mitochondrial membrane, the inter membrane space, the inner mitochondrial membrane, the cristae space, and matrix. Mitochondria have their own independent genome which is a single circular DNA molecule encoding 37 genes[6]. Division and genome of mitochondria are similar to those of bacterial cell. Mitochondria play critical roles in production of cellular energy, calcium and redox homeostasis, cellular signaling, regulation of cellular metabolism and cell death, and heat production[7-9]. Of the functions of mitochondria, the most prominent function is to synthesize cellular energy, adenosine triphosphate (ATP), which is used as a source of chemical energy for metabolism and a substrate in signaling pathways[10].
Mitochondria are very dynamic and continually fuse and divide in response to physiological conditions. Moreover, mitochondria are fragmented as the consequence of enhanced fission during apoptosis[11] and elongated to maintain ATP production during starvation[12]. Many mitochondrial protein complexes are composed of nuclear or mitochondrial DNA-encoded proteins. Any imbalance in the complex assembly can lead to accumulation or aggregation of unassembled or unfolded proteins[13]. In order to cope with the accumulation of unassembled or unfolded proteins within mitochondria, mitochondria activates the mitochondrial unfolded protein response (UPR) that up-regulates the expression of mitochondrial chaperones and proteases like ER stress response[14]. Although the environmental conditions inducing mitochondrial stress are still not clearly understood, the accumulated evidences suggest that high levels of reactive oxygen species (ROS) or inhibition of mitochondrial genome replication and transcription can induce mitochondrial UPR[15-17]. Unfolded proteins accumulated in matrix activate mitogen-activated protein kinase kinase (MEK)/c-Jun N-terminal protein kinase 2 (JNK2)/c-Jun pathway and protein kinase R (PKR). Activated c-Jun increases the transcription of transcription factors C/EBP homologous protein (CHOP) and CCAAT/enhancer-binding protein β (C/EBPβ), and then the heterodimer of CHOP and C/EBPβ activate the transcription of mitochondrial proteases and chaperons[18]. Activated PKR phosphorylates eukaryotic translational initiation factor 2α (eIF2α), leading to attenuating translation similar with the protein kinase RNA-like ER kinase (PERK) pathway of ER stress response[19]. In addition, unfolded proteins accumulated in the intermembrane space (IMS) activate estrogen receptor and NAD-dependent deacetylase sirtuin-3 (SIRT3). Activated estrogen receptor up-regulates the transcription of nuclear respiratory factor 1 (NRF1) and mitochondrial serine protease, high-temperature requirement A2 (HTRA2), and SIRT3 pathway induces anti-oxidant machinery and mitophagy to alleviate mitochondrial stress[20,21].
Accumulated evidences have suggested that HBx protein is associated with mitochondrial aggregation or damage. HBx protein induces an abnormal aggregation of mitochondrial structures at the periphery of nucleus, which may be eventually connected with cell death[22]. In HBx-expressing cells, the abnormal aggregation of mitochondria is induced by the increase of microtubule-dependent dynein activity through HBx-induced p38 mitogen-activated protein kinase (MAPK) activation[23]. Siddiqui group showed that HBx protein is associated with mitochondrial damage through interaction with voltage-dependent anion channel (HVDAC3) [24], which is known as mitochondrial porins and form pores in the outer membranes of mitochondria[25]. The interaction induces the alteration of mitochondrial transmembrane potential leading to generation of ROS, resulting in activation of transcription factors signal transducer and activator of transcription 3 (STAT3) and nuclear factor kappa B (NFĸB)[24,26]. The ability of HBx protein to transactivate AP-1 and NFB is abolished by positioning HBx protein in the nucleus artificially[27]. These evidences represent the ability of HBx protein as a transactivator, which is to induce gene expression through cytoplasmic factors but not nucleus.
HBx protein can affect mitochondrial functions or cell fate by regulating gene expression or translocating a series of proteins to mitochondria, respectively. HBx down-regulates the expression of mitochondrial encoded subunit proteins of electron transport in oxidative phosphorylation, resulting in a high level of cellular ROS by impairment of electron transport[28]. Besides, HBx down-regulates the expression of nuclear encoded genes involved in mitochondrial β-oxidation of fatty acids, resulting in a low level of cellular ATP by deficiency of energy sources[29]. HBx translocates Raf-1 kinases involved in the Ras-induced MAPK pathway or apoptosis regulator bcl-2-associated X protein (BAX) to mitochondria, leading to hepatic cell proliferation or apoptosis, respectively[30,31]. These evidences suggest that mitochondria dysfunction by HBx contributes to the HBV-induced pathogenesis of hepatocellular carcinoma such as proliferation, metastasis, chemoresistance and other aspects of tumorigenesis.
To date a lot of researches have reported for relation between HBx protein and mitochondrial damage. However, the research for HBx protein and mitochondrial UPR has not yet been reported. Therefore, the research to clarify correlation between HBx protein or HBx-induced mitochondrial damage and mitochondrial UPR needs to be performed in future. In addition, it will be a worthy to research for association between HBx protein and the proteins activated by mitochondrial UPR such as SIRT3, estrogen receptor, CHOP, or C/EBPβ.
The endoplasmic reticulum (ER) is dynamic tubular structure and forms an interconnected network with almost every membrane-bound organelles, including mitochondria, the Golgi apparatus, endosome, peroxisome, and plasma membrane through contact sites[32,33]. The ER acts as a sensor for intra- or extra-cellular stimuli and is essential for cell homeostasis. The ER is classified into rough ER and smooth ER, which are externally distinguished by ribosome, a molecular machine synthesizing biological protein. The rough ER has the ribosome binding sites, named translocon on the ER outer membrane and is involved in synthesis, folding, and glycosylation of secretary or integral membrane proteins[34]. Therefore rough ER is well-developed in specialized secretary cells such as hepatocytes, pancreatic islet cells and immune cells. The smooth ER lacks ribosome and involved in several metabolic processes including synthesis of lipids (phospholipids and steroids), metabolism of carbohydrates, regulation of calcium concentration, detoxification of drugs[35]. The smooth ER plays also a fundamental role in the assembly of very low density lipoprotein (VLDL) particles in liver. In addition to the above-mentioned functions, ER also participate in the following processes through contact sites with other organelles: e.g., ER-mitochondria: mitochondria biogenesis, lipid exchange during biosynthesis, and Ca2+ transfer from ER to mitochondria[32,35]; ER-Golgi: transport of secretory proteins and non-vesicular lipid transport[36]; ER-endosome: regulation of the intracellular distribution of endosomes[37]; ER-peroxisome: non-vesicular lipid transport[38]; ER-plasma membrane: regulation of phosphatidyl inositol metabolism and non-vesicular sterol transfer[39,40].
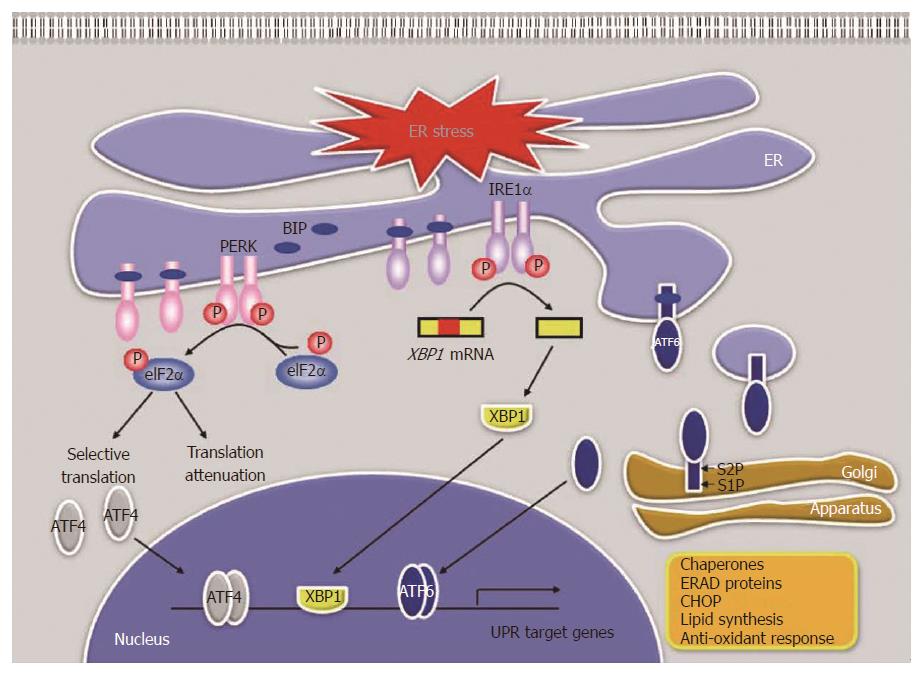
Of the ER functions, the proper folding and modification of proteins are the most important and best characterized function of the ER, and are processed under strict quality-control process (QCR)[41]. QCR means that only correctly matured proteins are exported to the Golgi complex and misfolded proteins are left in the ER to complete the process or to degrade the proteins[42]. Many ER-resident proteins such as chaperones, foldases, and lectins are involved in the QCR. Most ER chaperones are Ca2+ dependent and have ATPase activities. N-linked glycosylation and disulfide bond formation by foldases also play significant roles in protein maturation[43]. Viral infection induces the synthesis of a vast amount of viral proteins, leading to protein overload in ER. Therefore, the QCR is inhibited by the various stimuli such as Ca2+ output, nutrient deficiency or overload, hypoxia, and viral infection. The unfolded or misfolded proteins induced by the stimuli are accumulated and aggregated in ER, leading to ER stress. Fortunately, the ER has four adaptive mechanisms, named UPR, to alleviate ER stress and restore ER to its normal physiological conditions[5,41]: (1) translational attenuation for reduction of protein load, (2) induction of the expression of ER chaperones and foldases for enhancement of folding capacity, (3) induction of the expression of ER associated degradation (ERAD) proteins for clearance of unfolded proteins, (4) induction of apoptosis for removal of cells impaired by ER stress. In mammalian cells, these four responses are regulated by the regulatory pathways as described below (Figure 1).
PERK pathway: PERK is a type I transmembrane protein located in the ER. Under a normal condition, PERK exists as a monomer, an inactive state, by binding with ER chaperone, 78kD glucose-regulated protein (GRP78)/binding immunoglobulin protein (BiP)[44]. In response to ER stress, BiP is dissociated from PERK, which is activated through oligomerization and trans-autophosphorylation[45]. Activated PERK phosphorylates eIF2α, leading to translational attenuation[45]. Interestingly, phosphorylated eIF2α allows the translation of activating transcription factor 4 (ATF4) mRNA[46] and ATF4 induces the expression of UPR target genes involved in amino acid metabolism, antioxidant response, and ER-stress-induced apoptosis[47].
IRE1 pathway: Inositol-requiring protein 1 (IRE1) is a type I transmembrane protein that has a RNase domain. Under a normal condition, IRE1 exists as a monomer by binding with BiP like PERK[44]. In response to ER stress, BiP is dissociated from IRE1, which is activated through oligomerization and trans-autophosphorylation. Activated IRE1 triggers its RNase activity, which catalyses unconventional splicing of x-box binding protein 1 (XBP1) pre-mRNA to synthesize the active transcription factor spliced XBP1 (XBP1s)[48]. XBP1s induces the expression of UPR target genes involved in ERAD and lipid synthesis as well as the expression of ER chaperones[49,50].
ATF6 pathway: Unlike PERK and IRE1, activating transcription factor 6 (ATF6) is a type II transmembrane protein and a basic leucine zipper transcription factor[51]. Under a normal condition, translocation of ATF6 to the Golgi apparatus is inhibited because Golgi-localization signal of ATF6 is covered by BiP[52]. In response to ER stress, ATF6 is released from BiP and translocates to the Golgi apparatus[53]. In the Golgi apparatus, ATF6 is cleaved by site 1 protease (S1P) and S2P, and then a functional fragment of ATF6 is released into the cytosol[54]. This fragment translocates to the nucleus and induces the expression of ER chaperone, ERAD components, and XBP1[55].
Hepatocytes have well-developed ER because liver is a highly active organ for protein and lipid synthesis. The UPR could alleviate and restore ER damaged by extraordinary task as a protein synthesis factory. In addition to protein synthesis, ER has a variety of functions, which also are affected by physiological or pathological stress. The UPR activated by rhythmic or transient physiological conditions (e.g., feeding-fasting cycles) is sufficient to restore ER stress[56]. However, the UPR activated by irreversible or chronic stress (e.g., viral infection and obesity) is not sufficient to restore ER stress[56] and causes hepatic dysfunction, leading to the pathogenesis of the liver diseases including non-alcoholic fatty liver disease (NAFLD), cholestatic liver disease, insulin resistance, diabetes, viral hepatitis, and liver cancer[57-59].
In HBV-infected cells, a vast amount of HBV surface proteins is synthesized and folded in ER during its productive life-cycle, often leading to perturbation of the ER homeostasis, resulting in ER stress. This becomes known by identifying mutant surface proteins (preS1 and preS2 mutants) accumulated in the ER of the cells, termed ground glass hepatocyte (GGH) showing a hypertrophy of ER[60,61]. ER stress activated by HBV can lead to the expression of ER degradation enhancer, mannosidase alpha-like 1 (EDEM1) involved in ERAD pathway through the activation of IRE1/XBP1 pathway[62]. The activated ERAD pathway can limits the amount of surface proteins to alleviate ER stress and to protect the cells. Therefore, ER stress is essential for proper viral protein folding and HBV replication, enabling the chronic HBV infection. In general, autophagy is highly regulated catabolic process that removes the damaged organelles or intracellular microbial pathogens through the formation of double-membrane-bound structure called the autophagosome[63]. However, HBV can induce autophagy via ER stress or HBx protein for viral replication and envelopment[64-66]. Moreover, HBV can strategically protect itself from autophagic degradation through accumulation of immature lysosomes induced by HBx protein[67].
In vivo it seems insufficient to induction of ER stress by only surface proteins except productive life-cycle. ER stress can be induced by the change of intracellular conditions through other viral proteins. For example, HBx protein can generate ROS and decrease mitochondrial membrane potential and cellular ATP/ADP ratio through mitochondrial damage[29]. Although the molecular mechanisms by which HBx induces ER stress are not clearly understood, these changes by HBx protein may synergistically contribute to induction of ER stress in conjunction with surface proteins. In fact, some researchers reported that UPR or ERAD pathway are activated by HBx protein alone[62,68]. We also reported that ER stress is induced by low intracellular glucose or ATP levels as well as HBx protein[29].
As mentioned previously, rhythmic or transient ER stress is a protective mechanism for cell survival. However, chronic ER stress under the pathological conditions such as chronic HBV infection can cause various liver diseases. We showed that HBx up-regulates the expression of cyclo-oxygenase 2 (COX2) and stromal cell-derived factor-1 (SDF1) through PERK-eIF2α-ATF4 pathway and IRE1-XBP1 pathway activated by ER stress, respectively[29,69]. COX2 converts arachidonic acid to prostaglandin (e.g., prostaglandin E2), which is an important mediator of inflammation. SDF1, a small cytokine, is strongly chemotactic for lymphocytes and induces the recruitment of immune cells into liver of HBx transgenic mice[69]. These evidences suggest that chronic ER stress induced by HBx protein may contribute to pathogenesis of hepatic inflammation and fibrosis (Figure 2).
Cyclic AMP responsive element-binding protein H (CREBH) is an ER-resident transmembrane bZIP transcription factor and a member of old astrocyte specifically induced substance (OASIS) family which show cell- or tissue-specific expression pattern[70]. CREBH is abandant expressed in liver and its activation mechanism is similar to that of ATF6[70]. Activated CREBH plays critical roles in iron metabolism, triacylglycerol metabolism, hepatic gluconeogenesis and lipogenesis, and inflammation by regulating the expression of various genes as a master gene[71-75]. Considering the published papers, activated CREBH plays essential roles in various hepatic metabolisms under physiological conditions and in hepatic inflammation and cell proliferation under pathological conditions. We showed that CREBH is activated by HBV and HBx protein as well as ER stress inducer, leading to hepatic cell proliferation by inducing the expression of oncogenic genes in cooperation with HBx protein[75]. These evidences suggest that ER stress may be closely associated with pathogenesis of HCC in patients with chronic hepatitis B.
To date, the various drugs against HBV are developed and used to treat HBV patients, e.g. lamivudine, adefovir, entecavir, and tenofovir[76]. All of the drugs are nucleoside/nucleotide analogues that target reverses-transcriptase (RT) domain of HBV polymerase which play the essential role on viral replication[76,77]. Although the drugs can repress the viral replication efficiently, existing virus and covalently closed circular DNA (cccDNA) are not eliminated by the drugs from the infected cells. Since HBV polymerase lacks a proofreading exonuclease activity, misincorporated bases can’t be removed from newly synthesized viral genome, leading to the mutations in the progeny DNA. Therefore, the resistance for the drugs often occurs in HBV patients during long-term therapy. Besides, there are the adverse effects of the drugs, including myopathy induced by depletion of mitochondrial DNA and myonecrosis, and nephrotoxicity induced by inhibition of kidney function[78].
In addition to nucleoside/nucleotide analogues, the non-nucleoside agents that target viral entry, protein, or replication are developing. The development of agents that target viral entry was available due to identification of sodium taurocholate cotransporting polypeptide (NTCP) known as a HBV entry receptor[79]. Cyclosporin A and Myrcludex-B strongly inhibit HBV infection into hepatocytes by binding to NTCP on plasma membrane[80,81]. Anti-HBV agents that inhibit the secretion of viral proteins or the interaction between core and surface proteins have been reported[82,83]. Anti-HBV agents that inhibit the viral replication by blocking RNA packing or gene expression also have been reported[84,85]. Sorafenib, anti-liver cancer drug, suppresses the HBV gene expression, but it induces cell death at high-dose treatment[86,87]. On the other hand, since surface protein and HBx proteins can induce ER or mitochondria stress which lead to pathogenesis of liver diseases, the development of the agents which inhibit ER or mitochondria stress is necessary in future. The development of host-targeting anti-HBV agents makes patients expect some advantages, including the low frequency of drug resistance, the synergic effect with currently available anti-HBV agents, and supply an alternative therapy.
Under normal physiological conditions, our body has adaptive system to maintain homeostasis from various stresses. Even though faced with pathological conditions, our body can be protected from the conditions by removing and restoring the damaged cells and tissues through innate and adaptive immune system. However, HBV has the abilities to escape from the host’s immune response and even to utilize autophagy for viral replication and envelopment. The abilities facilitate chronic infection of HBV, leading to chronic ER and mitochondrial stress, resulting in the pathogenesis of various liver diseases including NAFLD, cholestatic liver disease, viral hepatitis, and liver cancer. Here we indicated the mechanisms by which HBV proteins induce the dysfunction of cellular organelles and the hepatic diseases developed by the expression of UPR target genes or by disturbance of cellular signaling pathway. From a therapeutic perspective, it will be important to understand how HBV induce ER or mitochondrial dysfunctions and understanding the mechanisms will provide new treatment options to chronic HBV patients.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: South Korea
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chen YJ, Sazci A S- Editor: Gong ZM L- Editor: A E- Editor: Huang Y
| 1. | Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 929] [Cited by in RCA: 993] [Article Influence: 62.1] [Reference Citation Analysis (1)] |
| 2. | Zhang XD, Wang Y, Ye LH. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med. 2014;11:182-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 3. | World Health Organization. [webpage on the internet]. Hepatitis B. http://www.who.int/mediacentre/factsheets/fs204/en/. . |
| 4. | Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 327] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 5. | Yoshida H. ER stress and diseases. FEBS J. 2007;274:630-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 815] [Cited by in RCA: 878] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 6. | Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1412] [Cited by in RCA: 1521] [Article Influence: 80.1] [Reference Citation Analysis (0)] |
| 7. | Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol. 2013;6:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 453] [Cited by in RCA: 558] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 8. | Green DR. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 908] [Cited by in RCA: 909] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 9. | Hajnóczky G, Csordás G, Das S, Garcia-Perez C, Saotome M, Sinha Roy S, Yi M. Mitochondrial calcium signalling and cell death: approaches for assessing the role of mitochondrial Ca2+ uptake in apoptosis. Cell Calcium. 2006;40:553-560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 513] [Cited by in RCA: 476] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 10. | McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551-R560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1308] [Cited by in RCA: 1431] [Article Influence: 75.3] [Reference Citation Analysis (0)] |
| 11. | Dai J, Wu P, Xu S, Li Y, Zhu Y, Wang L, Wang C, Zhou P, Shi H. Changes in mitochondrial ultrastructure in SH-SY5Y cells during apoptosis induced by hemin. Neuroreport. 2017;28:551-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589-598. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1459] [Cited by in RCA: 1371] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 13. | Sasaki K, Yoshida H. Organelle autoregulation-stress responses in the ER, Golgi, mitochondria and lysosome. J Biochem. 2015;157:185-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 14. | Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849-3855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 410] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 15. | Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Høj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem. 1996;240:98-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 273] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 16. | Cochemé HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by paraquat. J Biol Chem. 2008;283:1786-1798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 437] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 17. | Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117:4055-4066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 495] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 18. | Horibe T, Hoogenraad NJ. The chop gene contains an element for the positive regulation of the mitochondrial unfolded protein response. PLoS One. 2007;2:e835. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 179] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 19. | Rath E, Berger E, Messlik A, Nunes T, Liu B, Kim SC, Hoogenraad N, Sans M, Sartor RB, Haller D. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269-1278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 20. | Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. J Cell Sci. 2011;124:1396-1402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 21. | Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34:699-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 229] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 22. | Shirakata Y, Koike K. Hepatitis B virus X protein induces cell death by causing loss of mitochondrial membrane potential. J Biol Chem. 2003;278:22071-22078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 133] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 23. | Kim S, Kim HY, Lee S, Kim SW, Sohn S, Kim K, Cho H. Hepatitis B virus x protein induces perinuclear mitochondrial clustering in microtubule- and Dynein-dependent manners. J Virol. 2007;81:1714-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 24. | Rahmani Z, Huh KW, Lasher R, Siddiqui A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J Virol. 2000;74:2840-2846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 256] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 25. | Sorgato MC, Moran O. Channels in mitochondrial membranes: knowns, unknowns, and prospects for the future. Crit Rev Biochem Mol Biol. 1993;28:127-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 98] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 26. | Waris G, Huh KW, Siddiqui A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol Cell Biol. 2001;21:7721-7730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 278] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 27. | Doria M, Klein N, Lucito R, Schneider RJ. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747-4757. [PubMed] |
| 28. | Lee YI, Hwang JM, Im JH, Lee YI, Kim NS, Kim DG, Yu DY, Moon HB, Park SK. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem. 2004;279:15460-15471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 147] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Cho HK, Cheong KJ, Kim HY, Cheong J. Endoplasmic reticulum stress induced by hepatitis B virus X protein enhances cyclo-oxygenase 2 expression via activating transcription factor 4. Biochem J. 2011;435:431-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 30. | Chen J, Siddiqui A. Hepatitis B virus X protein stimulates the mitochondrial translocation of Raf-1 via oxidative stress. J Virol. 2007;81:6757-6760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 31. | Kim HJ, Kim SY, Kim J, Lee H, Choi M, Kim JK, Ahn JK. Hepatitis B virus X protein induces apoptosis by enhancing translocation of Bax to mitochondria. IUBMB Life. 2008;60:473-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 802] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 33. | Friedman JR, Voeltz GK. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 2011;21:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 235] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 34. | Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 348] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 35. | de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J. 2010;29:2715-2723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 254] [Cited by in RCA: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 36. | Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 273] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 37. | Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209-1225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 474] [Cited by in RCA: 549] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 38. | Raychaudhuri S, Prinz WA. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc Natl Acad Sci USA. 2008;105:15785-15790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 39. | Schulz TA, Prinz WA. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. Biochim Biophys Acta. 2007;1771:769-780. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 40. | Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 389] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 41. | Schröder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1251] [Cited by in RCA: 1339] [Article Influence: 67.0] [Reference Citation Analysis (0)] |
| 42. | Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 932] [Cited by in RCA: 914] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 43. | Schröder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 495] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 44. | Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1987] [Cited by in RCA: 2172] [Article Influence: 86.9] [Reference Citation Analysis (0)] |
| 45. | Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2430] [Cited by in RCA: 2558] [Article Influence: 98.4] [Reference Citation Analysis (0)] |
| 46. | Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099-1108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2332] [Cited by in RCA: 2506] [Article Influence: 100.2] [Reference Citation Analysis (0)] |
| 47. | Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2380] [Cited by in RCA: 2570] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 48. | Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2898] [Cited by in RCA: 3142] [Article Influence: 130.9] [Reference Citation Analysis (0)] |
| 49. | Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448-7459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1694] [Article Influence: 77.0] [Reference Citation Analysis (0)] |
| 50. | Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35-41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 487] [Cited by in RCA: 521] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 51. | Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 969] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 52. | Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1028] [Cited by in RCA: 1095] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 53. | Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045-13052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 371] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 54. | Shen J, Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279:43046-43051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755-6767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 779] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 56. | Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15:623-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 403] [Cited by in RCA: 448] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 57. | Kaplowitz N, Than TA, Shinohara M, Ji C. Endoplasmic reticulum stress and liver injury. Semin Liver Dis. 2007;27:367-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 118] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 58. | Dara L, Ji C, Kaplowitz N. The contribution of endoplasmic reticulum stress to liver diseases. Hepatology. 2011;53:1752-1763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 316] [Cited by in RCA: 307] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 59. | Jo H, Choe SS, Shin KC, Jang H, Lee JH, Seong JK, Back SH, Kim JB. Endoplasmic reticulum stress induces hepatic steatosis via increased expression of the hepatic very low-density lipoprotein receptor. Hepatology. 2013;57:1366-1377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 60. | Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 61. | Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol. 2003;163:2441-2449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 194] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 62. | Lazar C, Macovei A, Petrescu S, Branza-Nichita N. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS One. 2012;7:e34169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 63. | He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2982] [Cited by in RCA: 2878] [Article Influence: 179.9] [Reference Citation Analysis (0)] |
| 64. | Tang H, Da L, Mao Y, Li Y, Li D, Xu Z, Li F, Wang Y, Tiollais P, Li T. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology. 2009;49:60-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 65. | Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci U S A. 2010;107:4383-4388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 254] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 66. | Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319-6333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 231] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 67. | Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy. 2014;10:416-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 68. | Li B, Gao B, Ye L, Han X, Wang W, Kong L, Fang X, Zeng Y, Zheng H, Li S. Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 2007;124:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 112] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 69. | Cho HK, Kim SY, Seong JK, Cheong J. Hepatitis B virus X increases immune cell recruitment by induction of chemokine SDF-1. FEBS Lett. 2014;588:733-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 70. | Asada R, Kanemoto S, Kondo S, Saito A, Imaizumi K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J Biochem. 2011;149:507-518. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 150] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 71. | Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 642] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 72. | Vecchi C, Montosi G, Zhang K, Lamberti I, Duncan SA, Kaufman RJ, Pietrangelo A. ER stress controls iron metabolism through induction of hepcidin. Science. 2009;325:877-880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 246] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 73. | Lee MW, Chanda D, Yang J, Oh H, Kim SS, Yoon YS, Hong S, Park KG, Lee IK, Choi CS. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 2010;11:331-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 157] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 74. | Lee JH, Giannikopoulos P, Duncan SA, Wang J, Johansen CT, Brown JD, Plutzky J, Hegele RA, Glimcher LH, Lee AH. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat Med. 2011;17:812-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 75. | Cho HK, Kim SY, Kyaw YY, Win AA, Koo SH, Kim HH, Cheong J. HBx induces the proliferation of hepatocellular carcinoma cells via AP1 over-expressed as a result of ER stress. Biochem J. 2015;466:115-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Kim KH, Kim ND, Seong BL. Discovery and development of anti-HBV agents and their resistance. Molecules. 2010;15:5878-5908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 77. | Wang GH, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507-6512. [PubMed] |
| 78. | Fleischer RD, Lok AS. Myopathy and neuropathy associated with nucleos(t)ide analog therapy for hepatitis B. J Hepatol. 2009;51:787-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 79. | Watashi K, Urban S, Li W, Wakita T. NTCP and beyond: opening the door to unveil hepatitis B virus entry. Int J Mol Sci. 2014;15:2892-2905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 80. | Petersen J, Dandri M, Mier W, Lütgehetmann M, Volz T, von Weizsäcker F, Haberkorn U, Fischer L, Pollok JM, Erbes B. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol. 2008;26:335-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 335] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 81. | Watashi K, Sluder A, Daito T, Matsunaga S, Ryo A, Nagamori S, Iwamoto M, Nakajima S, Tsukuda S, Borroto-Esoda K. Cyclosporin A and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP). Hepatology. 2014;59:1726-1737. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 205] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 82. | Shin MS, Kang EH, Lee YI. A flavonoid from medicinal plants blocks hepatitis B virus-e antigen secretion in HBV-infected hepatocytes. Antiviral Res. 2005;67:163-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 83. | Asif-Ullah M, Choi KJ, Choi KI, Jeong YJ, Yu YG. Identification of compounds that inhibit the interaction between core and surface protein of hepatitis B virus. Antiviral Res. 2006;70:85-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 84. | Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res. 2007;76:168-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 127] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 85. | Li Y, Fu L, Yeo H, Zhu JL, Chou CK, Kou YH, Yeh SF, Gullen E, Austin D, Cheng YC. Inhibition of hepatitis B virus gene expression and replication by helioxanthin and its derivative. Antivir Chem Chemother. 2005;16:193-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 86. | Rahmani M, Davis EM, Crabtree TR, Habibi JR, Nguyen TK, Dent P, Grant S. The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulum stress. Mol Cell Biol. 2007;27:5499-5513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 199] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 87. | Kim HY, Jung HU, Yoo SH, Yoo KS, Cheong J, Park BS, Yun I, Yoo YH. Sorafenib overcomes the chemoresistance in HBx-expressing hepatocellular carcinoma cells through down-regulation of HBx protein stability and suppresses HBV gene expression. Cancer Lett. 2014;355:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |