Published online Oct 7, 2017. doi: 10.3748/wjg.v23.i37.6845
Peer-review started: May 19, 2017
First decision: June 8, 2017
Revised: August 1, 2017
Accepted: August 15, 2017
Article in press: August 15, 2017
Published online: October 7, 2017
Processing time: 133 Days and 1.3 Hours
To investigate and compare the analytical and clinical performance of TianLong automatic hypersensitive hepatitis B virus (HBV) DNA quantification system and Roche CAP/CTM system.
Two hundred blood samples for HBV DNA testing, HBV-DNA negative samples and high-titer HBV-DNA mixture samples were collected and prepared. National standard materials for serum HBV and a worldwide HBV DNA panel were employed for performance verification. The analytical performance, such as limit of detection, limit of quantification, accuracy, precision, reproducibility, linearity, genotype coverage and cross-contamination, was determined using the TianLong automatic hypersensitive HBV DNA quantification system (TL system). Correlation and Bland-Altman plot analyses were carried out to compare the clinical performance of the TL system assay and the CAP/CTM system.
The detection limit of the TL system was 10 IU/mL, and its limit of quantification was 30 IU/mL. The differences between the expected and tested concentrations of the national standards were less than ± 0.4 Log10 IU/mL, which showed high accuracy of the system. Results of the precision, reproducibility and linearity tests showed that the multiple test coefficient of variation (CV) of the same sample was less than 5% for 102-106 IU/mL; and for 30-108 IU/mL, the linear correlation coefficient r2 = 0.99. The TL system detected HBV DNA (A-H) genotypes and there was no cross-contamination during the “checkerboard” test. When compared with the CAP/CTM assay, the two assays showed 100% consistency in both negative and positive sample results (15 negative samples and 185 positive samples). No statistical differences between the two assays in the HBV DNA quantification values were observed (P > 0.05). Correlation analysis indicated a significant correlation between the two assays, r2 = 0.9774. The Bland-Altman plot analysis showed that 98.9% of the positive data were within the 95% acceptable range, and the maximum difference was -0.49.
The TL system has good analytical performance, and exhibits good agreement with the CAP/CTM system in clinical performance.
Core tip: The TianLong automatic hypersensitive hepatitis B virus DNA quantification system achieved a limit of detection of 10 IU/mL, limit of quantification of 30 IU/mL and good analytical performance in terms of accuracy, precision, reproducibility, linearity, genotype coverage and cross-contamination. In clinical performance, the TL system showed good correlation and agreement with the Roche CAP/CTM system.
- Citation: Li M, Chen L, Liu LM, Li YL, Li BA, Li B, Mao YL, Xia LF, Wang T, Liu YN, Li Z, Guo TS. Performance verification and comparison of TianLong automatic hypersensitive hepatitis B virus DNA quantification system with Roche CAP/CTM system. World J Gastroenterol 2017; 23(37): 6845-6853
- URL: https://www.wjgnet.com/1007-9327/full/v23/i37/6845.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i37.6845
Hepatitis B virus (HBV) infection is a worldwide disease, and over 2 billion people once have been infected with HBV and of these, 240 million have chronic hepatitis B[1,2]. Although there are effective vaccines and anti-viral medicine, HBV infection is still a major condition threatening the human health[3-5]. Globally, 30% of patients with liver cirrhosis and 45% with liver cancer are caused by HBV infection, respectively. Nationally, 500-600 million people in China have been infected with HBV, accounting for nearly 40% of the total national population[6-8]. Currently, China has approximately 93 million chronic HBV infected individuals, and 20 million of these patients have chronic hepatitis B[9,10]. An even higher percentage of patients with liver cirrhosis (60%) and hepatocellular cancer (HCC) (80%) were caused by HBV infection [11,12].
During the last decade, with the advent of new type interferon and antiviral drugs with a high genetic barrier to resistance, the antiviral treatment of chronic HBV infection has achieved significant progress[13-16]. HBV DNA quantification is one of the most important indicators used in HBV antiviral therapy, and can be used to determine whether the patient is suitable for antiviral therapy, monitor antiviral treatment response and virus resistance[17-19]. Use of hypersensitive HBV DNA quantification system to accurately detect HBV DNA in serum is the key factor in determining the curative effect and endpoints of hepatitis B antiviral therapy.
International associations for the study of liver diseases have required HBV DNA quantification detection, with hypersensitivity (a HBV detection kit should reach the sensitivity criterion of 10-15 IU/mL), wide linearity range (1-9 log10 IU/mL), and high specificity and good repeatability[20].
Many automatic HBV DNA detection systems are currently available in the Chinese market, including COBAS TaqMan HBV assay used in combination with COBAS AmpliPrep, abbreviated to CAP/CTM (Roche Molecular Diagnostics, Pleasonton, CA, United States), and Abbott Real Time HBV assay used in combination with Abbott m2000sp (Abbott Molecular, Des Plaines, IL, United States). However, due to their high cost and slow detection speed, these assays are not widely applied.
Most of the real-time HBV DNA qPCR detection kits in China have common shortcomings, such as poor specificity, low detection sensitivity, poor quantitative accuracy, narrow quantitative linear range and difficulty in detecting low or high viral load. To address these problems, the Chinese Food and Drug Administration (CFDA) released “The guideline principles for the technical review of hepatitis B virus DNA quantitative detection reagents registration” in 2013, which clearly put forward the requirements for HBV DNA detection. These principles will also guide the development direction of hypersensitive HBV DNA detection kits and prompt the rapid development of automatic hypersensitive HBV DNA quantitative detection systems.
The TianLong automatic hypersensitive HBV DNA quantification system (TL system) owns proprietary intellectual property rights, and has been approved by the CFDA. This study evaluates the analytical performance of the TL system and its clinical performance compared with the CAP/CTM system.
HBV-DNA positive samples: 200 residual blood samples were obtained from HBV infected patients in the clinical laboratory at 302 Military Hospital of China. These samples included HBV A genotype (4 cases), HBV B genotype (70 cases), HBV C genotype (66 cases), HBV D genotype (28 cases) and unknown genotype (32 cases).
HBV-DNA negative samples: These serum samples were obtained after immunology testing in the clinical laboratory at 302 Military Hospital of China, and were retested using the CAP/CTM system and confirmed to be negative.
High-titer HBV-DNA mixture samples: High-titer HBV-DNA samples were collected from the clinical laboratory at 302 Military Hospital of China. The high-titer HBV-DNA mixture samples were diluted 100 times and measured three times with the CAP/CTM system. The original concentration of the high-titer HBV-DNA mixture samples was calculated to be 2.5 × 108 IU/mL.
The clinical serum samples were tested for co-infections with HIV, HCV and HDV. All results were negative.
The worldwide HBV DNA performance panel (WWHD301, SeraCare, Milford, Massachusetts, United States) which contains HBV DNA (A-H) genotypes was used in this study. National standard materials for HBV serum were used to verify the assay performance including accuracy, limit of detection and limit of quantification. Details of the national standard materials are shown in Table 1.
| Art. No. | Standard Material No. | Batch No. | Standard material name | Reference Conc. |
| HS-0001 | GBW (E) 090137 | 201511003 | HBV DNA serum standard material | 1.41 × 103 IU/mL |
| HS-0002 | GBW (E) 090138 | 201512004 | HBV DNA serum standard material | 5.9 × 104 IU/mL |
| HS-0003 | GBW (E) 090139 | 201511003 | HBV DNA serum standard material | 4.6 × 105 IU/mL |
The TL system consists of a PANA 9600E automatic nucleic acid workstation, Gentier 96E real-time quantitative PCR system (Xi’an TianLong Science and Technology, Xi’an, Shaanxi, China), nucleic acid extraction kit (magnetic bead method) and a HBV DNA quantitative detection kit (fluorescence qPCR method).
The experimental method for the TL system assay followed the manufacturer’s instructions. Serum of 200 μL was used for HBV DNA extraction, and for PCR reaction assessment, 40 μL for each sample was prepared on the PANA 9600E automatic nucleic acid workstation. The Gentier 96E real-time quantitative PCR system was used for quantitative detection as follows: 50 °C for 2 min, 95 °C for 3 min, [94 °C for 15 s, 60 °C for 30 s (read fluorescence)] × 45 cycles. The data were automatically analyzed by software and the results were expressed in international units per milliliter (IU/mL). The TL system detected 96 samples (89 samples, 4 quantitative standards, a negative control, a weak positive control and a strong positive control) within 140 min (80 min for nucleic acid extraction and 60 min for real-time PCR detection).
HBV DNA was extracted from 650 μL of serum by the Cobas AmpliPrep instrument according to the manufacturer’s instructions. The Cobas TaqMan 48 analyzer was used for automated real-time PCR amplification and detection of PCR products according to the manufacturer’s instructions. The data thus generated were analyzed with Amplilink software. HBV DNA levels were expressed in international units per milliliter (IU/mL).
National standard materials for HBV serum [GBW (E) 090137, Batch No. 201511003] at a concentration of 1.41 × 103 IU/mL were diluted with HBV DNA negative serum to nominal concentrations of 30 IU/mL, 15 IU/mL, 10 IU/mL, 5 IU/mL, and 2.5 IU/mL. 25 replicates per run were tested with three different batches of reagents in three runs, and the test results were log transformed to verify whether the limit of detection of the TL system met the standard (10-15 IU/mL) recommended by the European Association for the Study of Liver (EASL) [21].
National standard materials for HBV serum [GBW (E) 090137, Batch No. 201511003], at a concentration of 1.41 × 103 IU/mL were diluted with HBV DNA negative serum to nominal concentrations of 30 IU/mL, 20 IU/mL, and 10 IU/mL. 25 replicates per run were tested with three different batches of reagents in three runs, and the test results were log transformed to verify whether the limit of detection of the TL system met the standard (30 IU/mL) required in “The guideline principles for the technical review of hepatitis B virus DNA quantitative detection reagents registration”.
National standard materials for HBV serum were used as samples and three replicates were tested at each concentration in three runs with three different batches of reagents. The sample results were log transformed to evaluate the accuracy of the TL system assay.
A total of 200 clinical HBV positive samples with a uniform distribution of concentrations were tested with the TL system and the CAP/CTM system. The quantitative results from the two assays were counted and the Statistical Program for Social Sciences (SPSS 13.0 for Windows; SPSS, Chicago, IL, United States) was used to assess the agreement between the positive results obtained with the two assays.
High-titer HBV-DNA mixture samples were diluted with HBV DNA negative serum to nominal concentrations of 1 × 107 IU/mL, 1 × 104 IU/mL and 1 × 102 IU/mL. Twenty replicates were tested at each concentration with the same batch of reagent to assess the intra-assay precision. The inter-assay precision was evaluated by three tests with different batches of reagents, and five replicates at each concentration were tested per run. The inter-assay precision test was repeated over 4 d, in order to validate the reproducibility of the TL system assay. The results were log transformed for analysis.
High-titer HBV-DNA mixture samples were diluted to nominal concentrations of 1 × 108 IU/mL, 1 × 107 IU/mL, 1 × 106 IU/mL, 1 × 105 IU/mL, 1 × 104 IU/mL, 1 × 103 IU/mL, 1 × 102 IU/mL, 60 IU/mL and 30 IU/mL. Triplicate measurements were made at each concentration level, the sample results were log transformed and then analyzed by Pearson’s correlation analysis and linear regression analysis to assess the quantitative linear range of the TL system.
The HBV DNA performance panel (WWHD301, SeraCare, Milford, Massachusetts, United States) which contains all the genotypes of the HBV DNA (A-H) was used, triplicate measurements were continuously carried out for each HBV genotype for 3 days with the TL system to validate its capability of detecting all HBV DNA (A-H) genotypes.
To assess the anti-cross contamination performance of the PANA 9600E automatic nucleic acid workstation, the “checkerboard” test was performed twice, using high-titer mixture samples interspersed with aliquots of HBV negative serum samples during DNA extraction. Each high-titer sample was included with HBV DNA negative plasma samples in the batch of 96 tests (48 positive samples and 48 negative samples) and the positions of positive samples and negative samples in the second round of testing were interchanged.
The test results of the limit of detection are shown in Table 2, and a detection rate ≥ 95% was taken as an acceptable detection limit criterion. According to the test results, the average detection rates of the samples at five concentration levels were 44.0%, 84.0%, 98.7%, 100%, and 100%, respectively. Of these results, the detection rates for the 10 IU/mL samples were above 95% in all three tests. Therefore, the concentration of 10 IU/mL was determined as the detection limit of the TL system, which meets the standard (10-15 IU/mL) recommended by the EASL.
| Sample Conc. | 1st Batch of reagent | 2nd Batch of reagent | 3rd Batch of reagent | Average detection rate | ||||||
| (IU/ml) | NO. | NO. not | Detection | NO. | NO. not | Detection | NO. | NO. not | Detection | |
| detected | detected | rate | detected | detected | Rate | detected | detected | rate | ||
| 2.5 | 10 | 15 | 40% | 12 | 13 | 48% | 11 | 14 | 44% | 44.0% |
| 5 | 21 | 4 | 84% | 20 | 5 | 80% | 22 | 3 | 88% | 84.0% |
| 10 | 25 | 0 | 100% | 24 | 1 | 96% | 25 | 0 | 100% | 98.7% |
| 15 | 25 | 0 | 100% | 25 | 0 | 100% | 25 | 0 | 100% | 100.0% |
| 30 | 25 | 0 | 100% | 25 | 0 | 100% | 25 | 0 | 100% | 100.0% |
The test results of the limit of quantification are shown in Table 3. Log10 IU/mL difference represents the log value difference between the detected and the expected concentrations. Taking ± 0.4 Log10 IU/mL as an acceptable value range in the detection results, this was converted using detected values of traceable national standard materials according to the National Center for Clinical Laboratories (NCCL) in China. When the detection accuracy rate of certain concentration was ≥ 95%, this was considered the system quantification limit. According to the test results, all three test results for the 30 IU/mL samples showed 100% accuracy rate and the Log10 IU/mL difference values were between -0.22 Log10 IU/mL and 0.19 Log10 IU/mL. Therefore, the concentration of 30 IU/mL was determined as the quantification limit of the TL system, which meets the standard (30 IU/mL) required in “The guideline principles for the technical review of hepatitis B virus DNA quantitative detection reagents registration”.
| Sample Conc. | Test batch | Test result (Log10IU/mL) | Accurate rate | |||
| (IU/mL) | Avg. value | CV | No. ≤ ± 0.4 | per batch | Average | |
| 10 | 1 | 0.79 | 40.57% | 19 | 76.00% | 65.33% |
| 2 | 0.59 | 47.58% | 13 | 52.00% | ||
| 3 | 0.71 | 36.53% | 17 | 68.00% | ||
| 20 | 1 | 1.45 | 14.11% | 21 | 84.00% | 82.67% |
| 2 | 1.45 | 16.65% | 20 | 80.00% | ||
| 3 | 1.36 | 20.55% | 21 | 84.00% | ||
| 30 | 1 | 1.49 | 6.09% | 25 | 100.00% | 100.00% |
| 2 | 1.40 | 8.51% | 25 | 100.00% | ||
| 3 | 1.46 | 5.55% | 25 | 100.00% | ||
The accuracy test results are shown in Table 4. The Log10 IU/mL difference represents the log value difference between the detected and the expected concentrations, and when the Log10 IU/mL difference was less than ± 0.4, the test result was considered accurate. According to the test results, for three concentration levels of HBV national standard serum, the absolute values of Log10 IU/mL differences were 0.04, 0.07, and 0.05, respectively; therefore, the test results meet the accuracy requirement.
| National standard materials for Hepatitis B virus serum | Expected | mean ± SD, | Absolute value of Log10 IU/mL difference | |
| Standard material No. | Reference Conc. | Log10 IU/mL | Log10 IU/mL | |
| GBW (E) 090137 | 1.41 × 103 IU/mL | 3.15 | 3.11 ± 0.02 | 0.04 |
| GBW (E) 090138 | 5.9 × 104 IU/mL | 4.77 | 4.80 ± 0.07 | 0.07 |
| GBW (E) 090139 | 4.6 × 105 IU/mL | 5.66 | 5.70 ± 0.05 | 0.05 |
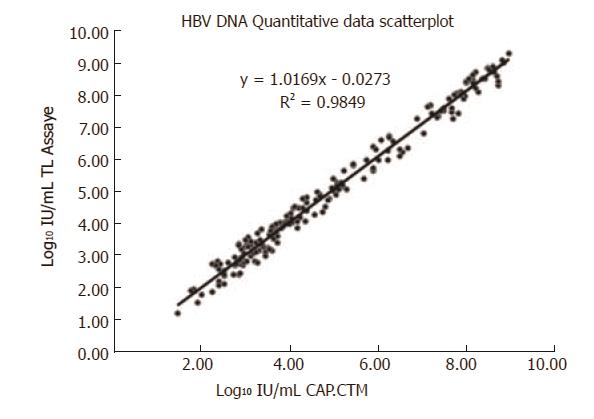
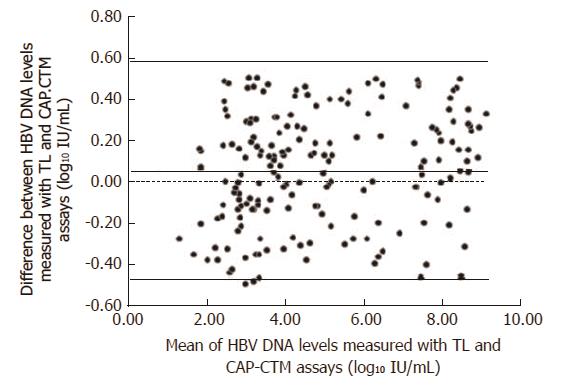
In December 2015, 200 HBV samples were collected and used to compare the results of the TL system assay with the CAP/CTM assay. The results of the two assays showed 100% consistency for both negative and positive results (15 negative samples, 185 positive samples), and the concentrations of positive samples were uniformly distributed between 60 IU/mL and 8.7×109 IU/mL. The Statistical Program for Social Sciences (SPSS 13.0 for Windows; SPSS, Chicago, IL, United States) was used to assess the agreement between the positive results obtained with the two assays. The normality test results indicated that the concentration distribution did not conform to the normal distribution (P = 0.00, P < 0.05). The hypothesis test summary indicated that there were no statistical differences between the TL system and CAP/CTM assays (P = 0.817, P > 0.05). The scatter plot for the two assays is shown in Figure 1, with the linear equation y = 1.0286x - 0.0829 and correlation coefficient r2 = 0.9774, which indicated a significant correlation between the two assays. The Bland-Altman plot analysis was used to assess the difference between the positive results obtained with the two assays (Figure 2). The results showed that 98.9% (183/185) of the positive data were within the 95% acceptable range, the average difference value was 0.06, the maximum difference was -0.49, and 82.3% (153/185) of the difference values were between ± 0.4 Log10 IU/mL.
The precision and reproducibility test results are shown in Table 5. As shown in this Table, in the sample concentration range of 1.0×102 IU/mL to 1.0×107 IU/mL, the CV of intra-assay precision, inter-assay precision and reproducibility was all less than 5%.
| Sample Conc. (IU/mL) | 1.0×107 | 1.0×104 | 1.0×102 | |||||
| Tested Results Log10 (IU/mL) | Mean Value | CV | Mean Value | CV | Mean Value | CV | ||
| Intra assay precision1 | 6.97 | 0.23% | 4.06 | 1.10% | 2.29 | 3.10% | ||
| Inter assay precision | 1st Day2 | 6.94 | 0.91% | 4.19 | 1.82% | 2.26 | 3.79% | |
| 2nd Day2 | 6.95 | 1.04% | 4.31 | 1.49% | 2.41 | 3.25% | ||
| 3rd Day2 | 7.08 | 0.64% | 4.31 | 1.67% | 2.39 | 3.72% | ||
| 4th Day2 | 7.10 | 0.81% | 4.32 | 1.63% | 2.42 | 3.47% | ||
| Reproducibility3 | 7.02 | 1.36% | 4.28 | 2.08% | 2.37 | 4.41% | ||
The linearity test results are shown in Figure 3. The linear regression analysis of eight concentration levels of HBV-DNA samples showed good linearity (the linear equation y=0.9848x - 0.0775 and correlation coefficient r2 = 0.99).
The HBV genotype coverage test results indicated that the TL system was capable of detecting all eight HBV DNA (A, B, C, D, E, F, G, H) genotypes as shown in Table 6. This almost covers all common genotypes worldwide, and meets the clinical requirements.
| Hepatitis B virus DNA | Average tested Ct value | |||
| genotypes | 1st day | 2nd day | 3rd day | Negative |
| A | 25.62 | 25.89 | 25.71 | Undetected |
| B | 25.58 | 25.78 | 25.78 | Undetected |
| C | 25.94 | 25.86 | 25.99 | Undetected |
| D | 22.61 | 22.78 | 22.78 | Undetected |
| E | 22.67 | 22.70 | 22.68 | Undetected |
| F | 22.68 | 22.72 | 22.71 | Undetected |
| G | 24.50 | 24.78 | 24.76 | Undetected |
| H | 24.81 | 24.80 | 24.85 | Undetected |
Following two rounds of “checkerboard” tests with the TL system assay, the detection rate of positive samples was 100%, and the detection rate of negative samples was 0. Therefore, throughout the entire test process, the negative samples were not contaminated.
HBV includes ten genotypes (A-I)[22-24]. According to its distribution in China, genotype B is the most frequent genotype in northern areas, genotype C is commonly seen in southern areas, mixed C/D genotypes are frequent in the Tibetan population in west China and genotype D is prevalent among Uighurs in Xinjiang, where genotype C is rare[25]. Infection with other HBV genotypes has not been reported in China. The 200 clinical HBV positive samples tested in this study were mainly composed of genotype B (70 cases), genotype C (66 cases) and genotype D (28 cases), genotype A (4 cases) and unknown genotypes. In order to verify the HBV genotype coverage using the TL system assay, the worldwide HBV DNA performance panel was used. The test results showed a 100% detection rate, which indicated that the TL system assay was capable of detecting not only the four genotypes in clinical samples, but also the eight HBV DNA (A, B, C, D, E, F, G, H) genotypes included in the worldwide HBV DNA performance panel.
In this study, the quantification limit test criterion adopted the analytical standard, which refers to the acceptable range ≤ ± 0.4 Log10 IU/mL of external quality assessment used by the NCCL of China. The results of three rounds of 30 IU/mL quantification limit tests in this study complied with this criterion. However, there is another international frequently-used criterion that considers CV ≤ 5% as the acceptable quantification limit test criterion. In this study, the CV in three rounds of 30 IU/mL quantification limit tests was 6.09%, 8.51%, and 5.55%, which failed to meet this criterion. However, during the precision and reproducibility tests, the CV values for intra-assay and inter-assay tests of 1.0 × 102 IU/mL samples were both less than 5%. These findings indicate that the TL system assay still requires further optimization for the 30 IU/mL concentration test.
In this study, during the comparison test of 200 clinical samples, the upper limit of linearity range of both the TL assay and CAP/CTM assay was 1 × 108 IU/mL. Of 185 positive clinical samples, the test results of 23 samples obtained using both the TL and CAP/CTM assays exceeded the upper limit of linearity range. Four samples had the test results by the TL assay exceeded the upper limit of linearity range (8.08, 8.03, 8.08, 8.40), while the corresponding test results for the CAP/CTM assay were close to the upper limit of the linearity range (7.88, 7.80, 7.88, 8.00), but the Log10 IU/mL differences between the two assays were all less than ± 0.4. Following elimination of these 27 samples, the correlation analysis results (P = 0.575) showed no significant statistical differences between the two methods and good consistency. However, there were differences between the two assays in the detection of samples at a concentration over or close to the linearity range upper limit. In order to obtain more effective quantitative results, it is recommended that clinical samples are diluted and tested again.
In addition, this study also performed a cross-reaction test to analyze the effect of interfering substances on the TL system performance. However, we were unable to obtain the corresponding drug pharmacokinetic samples; thus, the HBV positive serum samples were adopted as the control group in the cross-reaction test, and these HBV positive serum samples were added to the interfering substances as the test group. The cross-reaction test results showed that the TL system was capable of detecting HBV positive serum samples containing four typical endogenous interfering substances (free hemoglobin, triglyceride, bilirubin and IgG) and four typical exogenous interfering substances (IFNα, lamivudine, adefovir dipivoxil, telbivudine) and the Log10 IU/mL differences between the corresponding samples from the test group and the control group were all less than ± 0.4. Therefore, it can be concluded that the TL system has anti-interference capability for both endogenous and exogenous interfering substances. For more precise evaluation of the anti-interference performance of the TL system, more tests should be carried out with drug pharmacokinetic samples and clinical samples containing interfering substances.
Qiu et al[26] reported a comparison of the Abbott and Da-an real-time PCR for the quantitation of serum HBV DNA. The Abbott assay had a higher sensitivity, shorter assay time, and wider dynamic range compared with the Da-an assay. However, the costs of the Abbott assay limited its routine use in clinical molecular laboratories in China. The clinical performance of the TL system was comparable to the CAP/CTM system, with reasonable costs.
There are some deficiencies in the TL system found by performance verification. First, although no negative samples were detected in the cross-contamination test, two amplification curves for negative samples showed an increased tendency at 43-45 cycles, but did not reach the threshold line. Thus, cleaning and maintenance processes are particularly important in the case of a long run time, and the possibility of accumulated contamination should be validated over this period. Second, the TL system assay adopts pre-packaged nucleic acid extraction kits (24T×4); thus, when the amount of test sample is not a multiple of 24, some nucleic acid extraction reagent is wasted and can increase costs, and this is one of the main deficiencies of the TL system which needs to be improved.
Only 200 μL of sample were required by the TL system, its limit of detection (10 IU/mL) meets the standard (10-15 IU/mL) recommended by the EASL, and its limit of quantification (30 IU/mL) also meets the standard required in “The guideline principles for the technical review of hepatitis B virus DNA quantitative detection reagents registration” issued by the CFDA. The differences between the expected and tested concentration values of national standards were less than ± 0.4 Log10 IU/mL, which demonstrated the high accuracy of the system. In addition, there were no statistically significant differences between the TL system assay and the CAP/CTM assay. This study also assessed the precision, reproducibility, and linearity of the TL system, and the results showed that between 102 IU/mL and 106 IU/mL, the multiple test CV of the same sample was less than 5%; and between 30 IU/mL and 108 IU/mL, the linear correlation coefficient r2 = 0.99. Furthermore, the worldwide HBV DNA performance panel, which contains HBV DNA genotypes (A-H), was used to validate the capability of the TL system to detect all eight genotypes. Finally, two rounds of “checkerboard” tests with 96 samples (48 strongly positive and 48 negative) were tested to validate the anti-cross-contamination performance of the TL system, and the results showed a 100% coincidence rate. In summary, the TL system has good analytical performance, clinical performance and stability.
Use of the hypersensitive hepatitis B virus (HBV) DNA quantification system to accurately detect HBV DNA in serum is important in hepatitis B antiviral therapy. International associations for the study of liver diseases have required HBV DNA quantification detection with hypersensitivity (limit of detection: 10-15 IU/mL), a wide linearity range (1-9 log10 IU/mL), high specificity and good repeatability. The CAP/CTM system is regarded as a worldwide standard for HBV DNA quantification. This study was conducted to validate and compare the automatic hypersensitive HBV DNA quantification system produced in China with the internationally accepted CAP/CTM system.
The sensitivity, reproducibility and system automaticity for HBV DNA quantification have improved in recent years, and are important factors in determining the curative effect and endpoints of hepatitis B antiviral therapy.
This is the first study to evaluate the analytical performance of the TianLong automatic hypersensitive HBV DNA quantification system, which is the first CFDA approved system produced in China, and to compare its clinical performance with the CAP/CTM system. The TL system showed good analytical performance, and good correlation and quantitative agreement with the CAP/CTM system.
This study will be helpful in improving the performance of this system and its clinical applications.
The authors have evaluated the performance characteristics of the domestic TianLong HBV DNA Quantification System by testing 200 clinical HBV samples and compared the results with the international automatic HBV DNA detection systems including Roche CAP/CTM System. There was no statistically significant difference between TianLong and CAP/CTM systems. This study reports valuable results to validate the detection performance of TianLong HBV DNA Quantification System, which can pave the way to reduce cost of HBV DNA quantitative detection in the Chinese market.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Bock CT, Farshadpour F, Jarcuska P, Osna NA, Vaughan G S- Editor: Ma YJ L- Editor: A E- Editor: Huang Y
| 1. | Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212-2219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1328] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 2. | Palumbo E. Hepatitis B genotypes and response to antiviral therapy: a review. Am J Ther. 2007;14:306-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | US Centers for Disease Control and Prevention. The ABCs of Hepatitis. Available from: http://www.cdc.gov/hepatitis/Resources/Professionals/PDFs/ABCTable_BW.pdf. |
| 4. | Beasley RP. Rocks along the road to the control of HBV and HCC. Ann Epidemiol. 2009;19:231-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 5. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1806] [Cited by in RCA: 1994] [Article Influence: 199.4] [Reference Citation Analysis (4)] |
| 6. | Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, Sung JJ. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494-1498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 387] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 7. | Liang X, Bi S, Yang W, Wang L, Cui G, Cui F, Zhang Y, Liu J, Gong X, Chen Y. Epidemiological serosurvey of hepatitis B in China--declining HBV prevalence due to hepatitis B vaccination. Vaccine. 2009;27:6550-6557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 633] [Cited by in RCA: 712] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 8. | He J, Gu D, Wu X, Reynolds K, Duan X, Yao C, Wang J, Chen CS, Chen J, Wildman RP. Major causes of death among men and women in China. N Engl J Med. 2005;353:1124-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 865] [Cited by in RCA: 912] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 9. | Lu FM, Zhuang H. Management of hepatitis B in China. Chin Med J (Engl). 2009;122:3-4. [PubMed] |
| 10. | Liu J, Zhang S, Wang Q, Shen H, Zhang M, Zhang Y, Yan D, Liu M. Seroepidemiology of hepatitis B virus infection in 2 million men aged 21-49 years in rural China: a population-based, cross-sectional study. Lancet Infect Dis. 2016;16:80-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 158] [Article Influence: 17.6] [Reference Citation Analysis (1)] |
| 11. | Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099-2108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 985] [Cited by in RCA: 941] [Article Influence: 85.5] [Reference Citation Analysis (4)] |
| 12. | Goldstein ST, Zhou F, Hadler SC, Bell BP, Mast EE, Margolis HS. A mathematical model to estimate global hepatitis B disease burden and vaccination impact. Int J Epidemiol. 2005;34:1329-1339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 477] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 13. | Bhattacharya D, Thio CL. Review of hepatitis B therapeutics. Clin Infect Dis. 2010;51:1201-1208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 689] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 15. | Lai M, Liaw YF. Chronic hepatitis B: past, present, and future. Clin Liver Dis. 2010;14:531-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Yuen MF, Lai CL. Treatment of chronic hepatitis B: Evolution over two decades. J Gastroenterol Hepatol. 2011;26 Suppl 1:138-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 129] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 17. | McMahon BJ. The natural history of chronic hepatitis B virus infection. Hepatology. 2009;49:S45-S55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 556] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 18. | Saldanha J, Gerlich W, Lelie N, Dawson P, Heermann K, Heath A; WHO Collaborative Study Group. An international collaborative study to establish a World Health Organization international standard for hepatitis B virus DNA nucleic acid amplification techniques. Vox Sang. 2001;80:63-71. [PubMed] |
| 19. | Shyamala V, Cottrell J, Arcangel P, Madriaga D, Linnen J, Phelps B, Chien D. Detection and quantitation of HBV DNA in the WHO International Standard for HIV-1 RNA (NIBSC code: 97/656). J Virol Methods. 2004;118:69-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Caliendo AM, Valsamakis A, Bremer JW, Ferreira-Gonzalez A, Granger S, Sabatini L, Tsongalis GJ, Wang YF, Yen-Lieberman B, Young S. Multilaboratory evaluation of real-time PCR tests for hepatitis B virus DNA quantification. J Clin Microbiol. 2011;49:2854-2858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 21. | European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3745] [Cited by in RCA: 3784] [Article Influence: 473.0] [Reference Citation Analysis (1)] |
| 22. | Chu CM, Liaw YF. Predictive factors for reactivation of hepatitis B following hepatitis B e antigen seroconversion in chronic hepatitis B. Gastroenterology. 2007;133:1458-1465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 135] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 23. | Chu CM, Chen YC, Tai DI, Liaw YF. Level of hepatitis B virus DNA in inactive carriers with persistently normal levels of alanine aminotransferase. Clin Gastroenterol Hepatol. 2010;8:535-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Zehender G, Ebranati E, Gabanelli E, Sorrentino C, Lo Presti A, Tanzi E, Ciccozzi M, Galli M. Enigmatic origin of hepatitis B virus: an ancient travelling companion or a recent encounter? World J Gastroenterol. 2014;20:7622-7634. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 60] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Li GJ, Hue S, Harrison TJ, Yang JY, Chen QY, Wang XY, Fang ZL. Hepatitis B virus candidate subgenotype I1 varies in distribution throughout Guangxi, China and may have originated in Long An county, Guangxi. J Med Virol. 2013;85:799-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Qiu N, Li R, Yu JG, Yang W, Zhang W, An Y, Li T, Liu XE, Zhuang H. Comparison of Abbott and Da-an real-time PCR for quantitating serum HBV DNA. World J Gastroenterol. 2014;20:11762-11769. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |