Published online Jul 7, 2017. doi: 10.3748/wjg.v23.i25.4548
Peer-review started: February 3, 2017
First decision: March 16, 2017
Revised: April 10, 2017
Accepted: June 1, 2017
Article in press: June 1, 2017
Published online: July 7, 2017
Processing time: 162 Days and 10.9 Hours
To characterize the gut bacterial microbiota of patients with primary sclerosing cholangitis (PSC) and ulcerative colitis (UC).
Stool samples were collected and relevant clinical data obtained from 106 study participants, 43 PSC patients with (n = 32) or without (n = 11) concomitant inflammatory bowel disease, 32 UC patients, and 31 healthy controls. The V3 and V4 regions of the 16S ribosomal RNA gene were sequenced on Illumina MiSeq platform to cover low taxonomic levels. Data were further processed in QIIME employing MaAsLin and LEfSe tools for analysis of the output data.
Microbial profiles in both PSC and UC were characterized by low bacterial diversity and significant change in global microbial composition. Rothia, Enterococcus, Streptococcus, Veillonella, and three other genera were markedly overrepresented in PSC regardless of concomitant inflammatory bowel disease (IBD). Rothia, Veillonella and Streptococcus were tracked to the species level to identify Rothia mucilaginosa, Streptococcus infantus, S. alactolyticus, and S. equi along with Veillonella parvula and V. dispar. PSC was further characterized by decreased abundance of Adlercreutzia equolifaciens and Prevotella copri. Decrease in genus Phascolarctobacterium was linked to presence of colonic inflammation regardless of IBD phenotype. Akkermansia muciniphila, Butyricicoccus pullicaecorum and Clostridium colinum were decreased in UC along with genus Roseburia. Low levels of serum albumin were significantly correlated with enrichment of order Actinomycetales.
PSC is associated with specific gut microbes independently of concomitant IBD and several bacterial taxa clearly distinguish IBD phenotypes (PSC-IBD and UC).
Core tip: This study demonstrates specific microbial patterns associated with PSC and/or concomitant inflammatory bowel disease (PSC-IBD). Several bacterial taxa convincingly distinguish PSC-IBD from ulcerative colitis. Gut microbiota composition also differs in patients with PSC overlap with autoimmune hepatitis. Disease-specific microbial features traceable down to the species level may lead to establishing suitable biomarkers or outlining new research directions in the field of PSC and IBD pathogenesis.
- Citation: Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J, Drastich P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol 2017; 23(25): 4548-4558
- URL: https://www.wjgnet.com/1007-9327/full/v23/i25/4548.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i25.4548
Primary sclerosing cholangitis (PSC) is a chronic liver disorder of unknown etiology, characterized by inflammation and stenosis of the bile ducts[1,2]. The disease may progress to severe liver fibrosis and subsequent liver cirrhosis, and may eventually lead to liver failure and death[3]. Orthotopic liver transplantation (OLT) is the only currently available effective treatment for end-stage liver disease[4]. Inflammatory bowel disease (IBD) is present in 60% to 80% of patients with PSC[5]. Concomitant colonic disease is often classified as ulcerative colitis (UC)[6], but is also considered to be a distinct phenotype and referred to as “PSC-IBD”[7].
The involvement of gut bacteria in IBD is widely accepted, but the etiology and pathogenesis of IBD is still not fully understood. It is generally assumed that the inflammation results from an aberrant immune response to antigens of gut microbiota resident in genetically susceptible individuals[8]. It has been proposed that either an imbalance of the intestinal microbiota (dysbiosis), or presence of commensal bacteria with increased virulence could promote exaggerated local and systemic immune responses by disrupting microbiota-mucosa interactions[9,10].
Evidence from animal models[11] and success of antibiotic treatment in PSC patients[12] suggest that disruption of gut microbiota may play a significant role in pathogenesis of PSC. Recently, several studies described specific changes in the gut microbiota of PSC patients that were not related to dysbiosis with concomitant IBD[13-20]. However, those studies differed in sampling, experimental methods and design, in addition to outcome.
The aim of this study was to identify microbial features specific to patients with PSC (with and without concomitant IBD), in an independent, well-characterized cohort, to compare them with healthy controls (HC) and patients with UC, and to verify previous findings.
PSC, PSC-IBD, and UC patients for this single center, cross-sectional study were recruited at the outpatient department of the Institute for Clinical and Experimental Medicine, Prague. Healthy controls (HCs) were age and sex matched volunteers with insignificant medical history that were randomly selected from the hospital patient database. HCs were eligible if they had no history of malignant or autoimmune disease, major abdominal surgery, gastrointestinal disease or chronic gastrointestinal symptoms. Anyone, healthy or diseased, with antibiotic use within the previous 3 mo, a history of colorectal surgery, or ongoing or recent infectious colitis were excluded. Patients who received liver transplants were excluded because we assumed that OLT and related medications, immunosuppressive therapy in particular, might significantly influence the microbiota composition, and therefore cause substantial bias. To exclude the potential impact of obesity or malnutrition, only patients and controls with a normal BMI were recruited.
Stool samples were freshly collected in standardized, sterile collection tubes by participants and brought to the clinic when they made their routine clinic visits. All samples were delivered within 6 h of collection, immediately frozen at -20 °C and within 2 wk transferred to a -80 °C freezer for long-term storage. Care was taken to prevent thawing until samples were suspended in extraction buffer[21,22]. A MasterPure™ Complete DNA and RNA Purification Kit (Epicentre) was used for DNA extraction using a FastPrep®-24 Instrument homogenizer and Lysing Matrix Y zirconium oxide spheres (both MP Biomedicals). DNA concentration was equalized in each sample after quantitation with a Qubit™ 2.0 Fluorometer and a dsDNA BR Assay Kit (Life Technologies). KAPA 2G Robust Hot Start DNA Polymerase (Kapa Biosystems) was used to amplify segments of the 16S rRNA gene including the V3 and V4 regions using 341F and 806R primers[23]. The PCR protocol was initialized with 94 °C for 3 min, followed by 25 cycles of denaturation (94 °C, 30 s), annealing (54.2 °C, 1 min) and extension (72 °C, 1 min and 15 s), with the final extension (72 °C) for 10 min. The PCR products were purified and normalized for concentration with SequalPrep™ Normalization Plate Kit (ThermoFisher Scientific).
After pooling, sample ligation was performed with a TruSeq DNA PCR-free LT Sample Preparation Kit (Illumina) following the standard protocol provided by the manufacturer. The libraries were validated by a KAPA Library Quantification Kit (Kapa Biosystems) prior to submission to The Genomics Core Facility, Central European Institute of Technology (CEITEC; Brno, Czech Republic) for sequencing on a MiSeq Platform (Illumina) using a Miseq Reagent Kit v3 (Illumina).
Sequencing data were processed using a QIIME package[24]. Briefly, raw reads were demultiplexed and quality filtered, allowing no N characters, a maximum of three consecutive low-quality base calls, a maximum unacceptable Phred quality of Q20, and a maximum of 1.5 barcode errors. Chimeric reads were detected and discarded using USEARCH algorithms[25]. In the final dataset, the median of number of reads per sample reached 42459 sequences. Community statistics and sample comparisons were done on resampled datasets at the level of 16400 sequences. This led to the exclusion of two samples, one from the PSC and one from the HC group because of an insufficient number of reads.
The appropriateness of sequencing depth was checked in rarefaction plots. Alpha diversity statistics were calculated in QIIME[24] using Shannon, Simpson, and observed species diversity indices, and the P value for group comparisons was determined by analysis of variance (ANOVA). Principal coordinate analysis plots were constructed to illustrate the beta diversity of samples based on phylogenetically informed weighted and unweighted Unifrac distance matrices[26]. Significance of clustering patterns was tested by PERMANOVA, Adonis and ANOSIM in the R-vegan package. The multivariate homogeneity of group dispersions was assessed by Permdisp methods. Multivariate association with linear models (MaAsLin) for multivariate analysis[27] applying an additive general linear model was used to assess the association between microbial abundance and patient metadata: Age, sex, gender, presence of PSC/AIH overlap, IBD activity, probiotic use (E. coli Nissle 1917), and use of anti-TNF treatment. A P value < 0.05 was considered statistically significant after adjusting for false discovery rate (FDR). Raw demultiplexed sequencing data, with sample annotations, were submitted to the Short Read Archive (http://www.ncbi.nlm.nih.gov/bioproject/368966).
This study was approved by The Ethics Committee with multicenter competence of the Institute for Clinical and Experimental Medicine and Thomayer Hospital. All patients and healthy controls signed the informed consent form at the time of sample collection.
A total of 106 individuals were enrolled, including 32 patients with PSC-IBD, 11 with PSC (without IBD), 32 with UC and 31 HCs. The diagnosis of PSC or PSC overlap with autoimmune hepatitis (PSC/AIH) syndrome was established following recommended biochemical, immunological, histological, and cholangiographic evaluation[28-30]. The extent of UC was categorized endoscopically[31] and Mayo classification[32] was used to describe disease severity. All other patient characteristics were obtained on the day of sample collection (Table 1).
| PSC-IBD (n = 32) | PSC (n = 11) | UC (n = 32) | Healthy controls (n = 31) | |
| Gender; male/female | 24/8 (75%/25%) | 10/1 (90.9%/9.1%) | 17/15 (53.1%/46.9%) | 13/18 (41.9%/58.1%) |
| Median age (range), yr | 35 (18-60) | 45 (18-69) | 40 (20-71) | 44 (22-72) |
| Overlap syndrome PSC/AIH | 6 (18.8) | 2 (18.2) | N/A | N/A |
| Total bilirubin (μmol/L; mean ± SD) | 47 ± 56.8 | 34.4 ± 32.8 | 13.1 ± 6.7 | 12.8 ± 6.2 |
| AST (μkat/L; mean ± SD) | 1.5 ± 0.8 | 1.1 ± 0.8 | 0.4 ± 0.1 | 0.5 ± 0.3 |
| ALT (μkat/L; mean ± SD) | 1.8 ± 0.9 | 1.3 ± 1.1 | 0.5 ± 0.2 | 0.6 ± 0.4 |
| ALP (μkat/L; mean ± SD) | 7.4 ± 5.7 | 4.3 ± 4.9 | 1.1 ± 0.4 | 1 ± 0.3 |
| GGT (μkat/L; mean ± SD) | 7.5 ± 7.4 | 12.2 ± 25.5 | 0.5 ± 0.5 | 0.7 ± 1.6 |
| IBD extent: | ||||
| Pancolitis | 28 (87.5) | N/A | 25 (78.1) | N/A |
| Left-sided | 0 (0) | N/A | 7 (21.9) | N/A |
| Right sided | 4 (12.5) | N/A | 0 (0) | N/A |
| IBD activity: | ||||
| Mild or remission | 24 (75) | N/A | 20 (62.5) | N/A |
| Moderate | 4 (12.5) | N/A | 3 (9.4) | N/A |
| Severe | 4 (12.5) | N/A | 9 (28.1) | N/A |
| Medication during last month: | ||||
| UDCA | 33 (100) | 11 (100) | 0 (0) | 0 (0) |
| 5-ASA | 26 (81.3) | 0 (0) | 31 (96.9) | 0 (0) |
| Corticosteroids | 12 (37.5) | 2 (18.2) | 3 (9.4) | 0 (0) |
| Azathioprine | 13 (40.6) | 1 (9.1) | 14 (43.8) | 0 (0) |
| Anti-TNFα | 0 (0) | 0 (0) | 10 (31.3) | 0 (0) |
| Probiotics, E. coli Nissle 1917 | 4 (12.5) | 1 (9.1) | 9 (28.1) | 0 (0) |
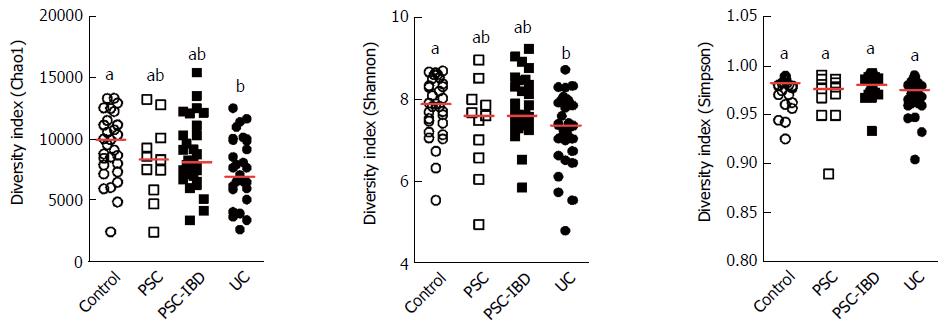
A decrease of alpha diversity in all patients compared with HCs was apparent from differences in the Chao1 and Shannon indices, the decrease was particularly clear in patients with IBD, especially in the UC group. No evident differences in the Simpson indices of the groups were observed (Figure 1).
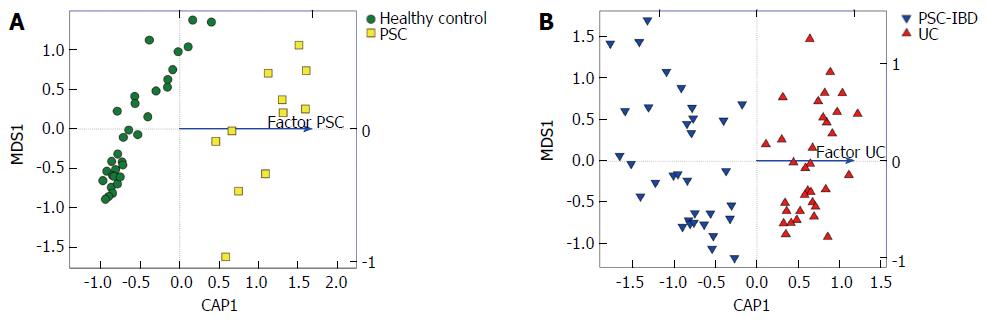
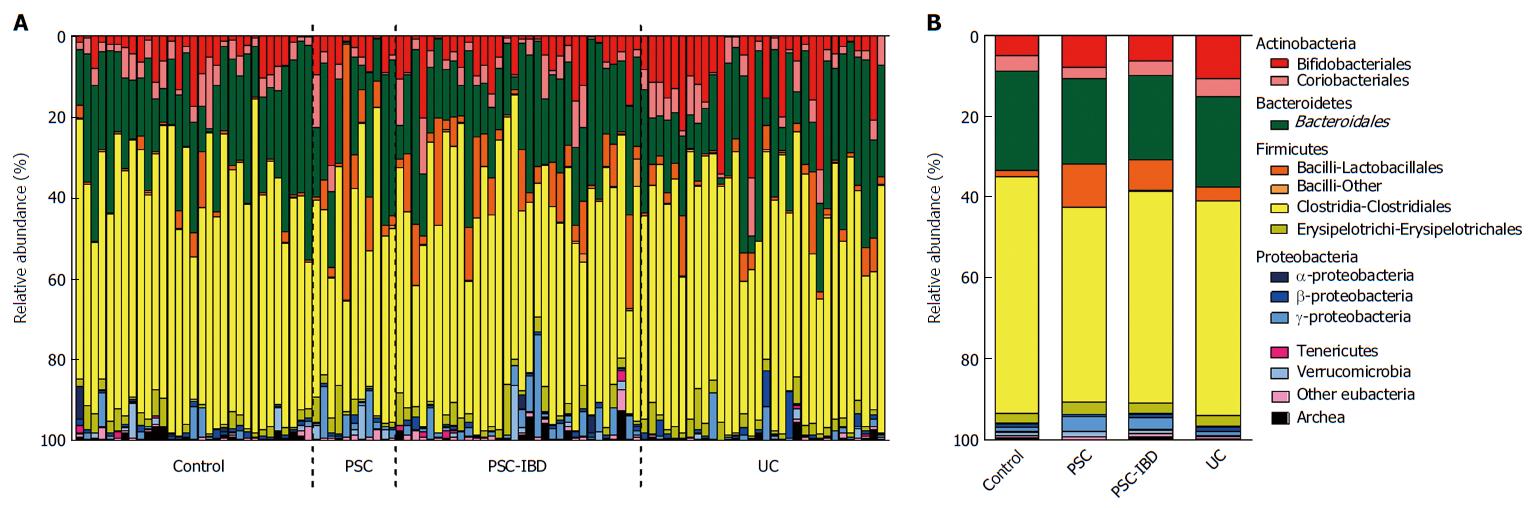
The global microbiota composition was clearly shifted by the presence of liver disease in PSC patients compared with HCs (Adonis R2 = 0.021, P = 0.001; Figure 2A). Similarly, samples from patients with IBD clustered separately, depending significantly on the UC factor (Adonis R2 = 0.021, P < 0.001) (Figure 2B). However, the homogeneity of group dispersion was significantly different when measured with Permdisp (P > 0.1). No such significant shifts were detected when comparing PSC and PSC-IBD groups. The disruption of bacterial beta diversity is shown in stack plots (Figure 3).
We compared relative abundance corrected for FDR, and identified 12 genera that were significantly increased in patients with PSC compared with HCs (Table 2). Seven of these remained relatively more abundant regardless of concomitant IBD. These were Rothia, Enterococcus, Streptococcus, Clostridium, Veillonella and Haemophilus. On the other hand, Coprobacillus, Escherichia, Corynebacterium and Lactobacillus genera were associated with PSC-IBD, but not with isolated PSC. A distinct increase in the relative abundance of family Micrococcaceae (P < 0.001) in PSC and PSC-IBD (P < 0.05) was driven by the strong overrepresentation of Rothia. Similarly, enrichment of genus Lactobacillus and Streptococcus along with increase of family Carnobacteriaceae (P < 0.01) was responsible for a significant increase of order Lactobacillales (P < 0.001) and class Bacilli (P < 0.001) in patients with PSC-IBD. Additionally, genus Coprococcus was significantly reduced (P < 0.01) in all diseased individuals, including those with PSC and UC, along with several other unidentified genera and species belonging to family Lachnospiraceae. A low abundance of genus Phascolarctobacterium positively correlated (P < 0.01) with the presence of IBD, both PSC-IBD and UC.
| Genus | PSC | PSC-IBD | UC |
| Rothia | ↑↑↑ | ↑↑ | NS |
| Enterococcus | ↑↑↑ | ↑↑ | NS |
| Streptococcus | ↑↑ | ↑↑ | NS |
| Clostridium | ↑↑ | ↑ | NS |
| Veillonella | ↑ | ↑ | NS |
| Haemophilus | ↑ | ↑ | NS |
| Staphylococcus | NS | ↑ | NS |
| Coprobacillus | NS | ↑ | NS |
| Escherichia | NS | ↑ | NS |
| Corynebacterium | NS | ↑ | NS |
| Lactobacillus | NS | ↑ | NS |
| Coprococcus | ↓↓ | ↓↓ | ↓↓ |
| Phascolarctobacterium | NS | ↓↓ | ↓↓ |
| Akkermansia | NS | NS | ↓↓↓ |
| Roseburia | NS | NS | ↓ |
At the species level, seven taxa belonging to genera Rothia, Lactobacillus, Streptococcus and Veillonella, were overrepresented, specifically in PSC patients, compared with both HCs and patients with UC (Table 3). Conversely, abundance of Prevotella copri was negatively associated with PSC, particularly in patients with concomitant IBD (P < 0.001). A highly significant decrease of Adlercreutzia equolifaciens (P < 0.001) was detected only in PSC patients without concomitant IBD. Faecalibacterium prausnitzi, Coprococcus catus and Ruminococcus gnavus were decreased in all patients compared with HCs.
| Species | PSC | PSC-IBD | UC |
| Rothia mucilaginosa | ↑↑↑ | ↑↑ | NS |
| Lactobacillus salivarius | NS | ↑↑↑ | NS |
| Streptococcus infantis | ↑↑ | ↑↑ | NS |
| Streptococcus alactolyticus | ↑↑ | ↑↑ | NS |
| Streptococcus equi | ↑↑ | ↑↑ | NS |
| Veillonella dispar | ↑↑ | ↑↑ | NS |
| Veillonella parvula | ↑ | ↑↑ | NS |
| Prevotella copri | ↓ | ↓↓↓ | NS |
| Adlercreutzia equolifaciens | ↓↓↓ | NS | NS |
| Faecalibacterium prausnitzi | ↓ | ↓↓↓ | ↓↓ |
| Coprococcus catus | ↓ | ↓↓ | ↓ |
| Ruminococcus gnavus | ↓↓ | ↓↓ | ↓↓↓ |
| Akkermansia muciniphila | NS | NS | ↓↓↓ |
| Butyricicoccus pullicaecorum | NS | NS | ↓↓↓ |
| Clostridium colinum | NS | NS | ↓↓↓ |
A distinct dysbiotic pattern characterized by the significant underrepresentation of phylum Verrucomicrobia (P < 0.001) distinguished patients in the UC group from HCs. The low signal of this group of bacteria was also evident at the family (Verrucomicrobiaceae, P <0.001) and genus (Akkermansia, P < 0.001) levels and with the extremely low abundance of Akkermansia muciniphila (P < 0.001), reduction of Butyricicoccus pullicaecorum (P < 0.05) and genus Roseburia (P < 0.05). The decrease of genus Akkermansia (P < 0.01) and Akkermansia muciniphila (P < 0.01) remained significant (along with the respective family and phylum signals) when compared with microbiota from PSC-IBD patients. Furthermore, Clostridium colinum was significantly underrepresented when compared with both HCs (P < 0.001) and PSC-IBD patients (P < 0.01). On the other hand, three genera that distinguished PSC patients from HCs (Rothia P < 0.05; Streptococcus, P < 0.05; Veillonella, P < 0.05) were increased in PSC-IBD compared to UC. The pattern continued at the species level with R. mucilaginosa (P < 0.05), V. parvula (P < 0.05), V. dispar (P < 0.05), and unclassified species of genus Streptococcus (P < 0.01) and genus Blautia (P < 0.05). Order Fusobacteriales (P < 0.01) and family Fusobacteriaceae (P < 0.05) were more abundant in UC then in PSC-IBD, but we were not able to track this tendency to lower taxonomic levels (Tables 2 and 3).
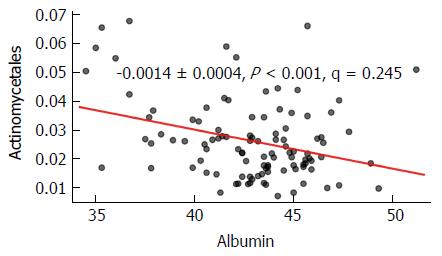
In the multivariate analysis, an OTU (operational taxonomic unit) belonging to genus Actinomycetes was significantly (P < 0.01) enriched, when overlapping AIH was present in patients with PSC (including those with PSC-IBD). Multivariate analysis using MaAsLin found a significant tendency of order Actinomycetales to be increased in study subjects with low serum albumin level (P < 0.01). This finding was no longer significant when adjusting for the specific patient groups and HCs (Figure 4).
The intestinal microbiota is involved in the pathogenesis of various diseases and is, therefore, a potential biomarker or even a therapeutic target[33]. We identified specific signatures of the fecal microbiota that distinguished patients with PSC from those with UC and from HCs. This is the first study of its kind from central Europe, a region characterized by specific dietary habits. Moreover, since recruitment centre may have a serious impact on microbial composition in multi-centric studies[18], we decided to perform this study in a single-centre design with meticulous care for consistency of sample collection, processing and analysis.
We found disruption of gut bacteria homeostasis in patients with UC, which was characterized by decreased microbial diversity. This is in line with previous studies in which low bacterial diversity was associated with IBD[34]. Moreover, significant disease-specific shifts of global microbiota composition were apparent when comparing PSC patients with healthy controls and PSC-IBD with UC patients.
In complex diseases, several distinct environmental factors may influence the composition of the gut microbial community. In our series of PSC and PSC-IBD patients, it was apparent that liver disease was primary factor associated with disease-specific dysbiosis, independent of dysbiotic influences of IBD, as the changes in gut microbiota did not significantly differ in PSC patients with and without established, concomitant IBD. However, stratifying PSC patients into “with or without IBD” subphenotypes may be controversial because PSC-IBD may have a subclinical course that might be easily missed during endoscopic and histological assessment. In addition, there is no agreement on the recommended interval for endoscopic evaluation in patients with PSC and no clinical suspicion of IBD. Furthermore, previous studies evaluated PSC-UC and PSC-CD as subgroups of their study cohorts[13,16]. However, we assumed, that IBD in PSC rarely has macroscopic features typical for CD and that histological findings might be often misinterpreted and the diagnosis is often eventually changed to UC. Therefore, we included only patients with typical features of IBD associated with PSC and universally categorized the phenotype as PSC-IBD[7].
We found a close association of the relative abundance of several genera with PSC, including Rothia, Streptococcus, Enterococcus, Veillonella, Clostridium, and Haemophilus regardless of the presence or absence of concomitant IBD. The association of Rothia with PSC was the most striking, and to our knowledge, this is the first study to describe such a significant relation with PSC.
The increased abundance of genus Rothia, and R. mucilaginosa in particular, in patients with PSC suggests that oral microbiota may be overrepresented in the lower GI populations of patients with advanced liver disease, which is in line with previous reports[35]. In previous studies, infections caused by Rothia species were predominantly found in immunocompromised individuals and patients with indwelling vascular devices[36]. Since Rothia is sensitive to gastric fluid[37], we speculate, that our findings may reflect contamination of the intestinal microbial community by previous endoscopic retrograde cholangio-pancreatography (ERCP), with repeated stenting in particular. Our data alone are insufficient to indicate a direct link to disease pathogenesis. However, recent reports of successful treatment of PSC and recurrent PSC with vancomycin[12,38], suggest that Rothia or other vancomycin-sensitive microbes[39] are reasonable targets of further research in this field.
The increased abundance of genera Enterococcus and Lactobacillus confirms the results of a recent complex study from Belgium[16]. Even though we did not find a significant increase of genus Fusobacterium (one of three significantly increased genera in the Belgian study), we observed the clear enrichment of family Fusobacteriaceae in PSC-IBD patients (P < 0.001).
We could replicate previous results of enrichment of Veillonella, notably V. parvula and V. dispar, as a key feature of PSC-associated dysbiosis[13]. Previously, this genus was associated with several other progressive fibrotic disorders[40]. As in our study, Kummen et al[13] did not evaluate the influence of liver cirrhosis, which might be the condition that predisposes towards Veillonella increase, not necessarily PSC itself. This is further supported by Sabino et al[16] who found that Veillonella abundance in PSC was no longer significantly changed when patients with confirmed liver cirrhosis were excluded from the analysis. Sabino et al[16] diagnosed liver cirrhosis diagnosis biopsy and/or MRI imaging and/or elastography. Confirming cirrhosis in advanced cholestatic liver disease by elastography may be challenging because of imaging distortion, and biopsy can miss focally expressed morphological features[41]. Therefore, it is not certain, whether the alteration in microbiota composition was a PSC-specific characteristic, or more likely reflects homeostatic disruption in advanced chronic liver disease in general.
In our dataset, genus Adlercreutzia was significantly decreased in PSC compared with HCs. Adlercreutzia is a genus that includes only one species (A. equolifaciens) capable of converting ingested isoflavones, which are abundant in legumes and soya beans, into equol[42]. Equol has a high affinity for estrogen receptors (ERs) and may be a selective estrogen receptor modulator[43]. ER expression on cholangiocytes is increased in cholestatic liver diseases but is absent in healthy individuals[44]. Taking into account the fact that the prevalence of PSC is much decreased in women and Asian populations[45] (and presumably in populations with higher than average consumption of isoflavones), we could speculate upon potential pathogenetic pathways with significant impact on disease development that are influenced by dietary habits and endocrine signaling.
We observed a significant decrease of Ruminococcus gnavus in all diseased individuals, Png et al[46] described a manifold increase of this mucin-degrading species in the colonic mucosa of both CD and UC patients. This discrepancy could be, however, caused by the fact that the luminal and mucosal microbiota are significantly different[47]. The low abundance of another mucolytic bacteria, Akkermansia muciniphila (and therefore genus Akkermansia), in UC is consistent with Png et al[46]. The absence of this bacterium from the microbial community could affect the use of mucins as a carbon sources by other bacteria that are important members of microbial community in healthy individuals[46]. In our study, the UC phenotype was characterized by a substantial decrease of Butyricicoccus pullicaecorum sp., which is in line with Eeckhaut et al[48] who found that B. pullicaecorum attenuated trinitrobenzene sulfonic acid (TNBS)-induced colitis in rats, and that supernatants of cultures of B. pullicaecorum increased transepithelial resistance. Decreased abundance of Faecalibacterium prausnitzii has recently been reported in IBD-associated dysbiosis[49,50], which also occurred in this study in both UC and with PSC. The decrease of F. prausnitzii was particularly significant in patients with an inflamed colon, regardless of IBD phenotype. The decreased abundance of B. pullicaecorum, F. prausnitzii, and genus Roseburia demonstrates disruption in butyrate-producing bacteria, which probably have anti-inflammatory activity. Furthermore, it has been proposed recently that the anti-inflammatory properties of F. prausnitzii is associated with the production of 15 kDa protein capable of inhibiting the NF-κB pathway in intestinal epithelial cells[51].
Bile acids and changes in production, circulation, and conversion may be associated with changes of microbiota composition[52]. Microbiota might thus be influenced by long-term use of ursodeoxycholic acid (UDCA) leading to substantial changes in bile composition. Sabino et al[16] reported that only 66.7% of a series of 147 patients with PSC-UC used UDCA. In our series, all patients with an established diagnosis of PSC received UDCA as routine clinical practice at our center follows the recommendations of EASL[53] and ECCO[54], which are contradictory to the AASLD guidelines advocating against routine use of UDCA in PSC[5]. We were thus not able to evaluate the influence of UDCA on microbiota composition. However, recent reports have not reported substantial differences in the microbiota of PSC patients treated or not treated with UDCA[13,16].
The impact of antibiotics on gut microbiota composition is clear, but there is no consensus on how to adjust for the antibiotic use, we included only subjects who had not used antibiotics within the previous 3 mo. Sabino et al[16] enforced only 1 mo cutoff, and Kummen et al[13] observed no substantial differences related to antibiotic use within the previous 12 mo.
The role of biologics for PSC treatment is currently not sufficiently clear. α4β7 integrin antagonists have demonstrated a positive effect on PSC course[55], and the effect of anti-TNF treatment is dubious[56] with potential risk of serious adverse events[57]. In our series, no PSC patients had been treated with biologics. Nearly one-third of the UC patients had received infliximab, adalimumab, or golimumab, but these agents did not result in microbial shifts that distinguished the UC group.
Multivariate analysis revealed a significant negative correlation of serum albumin level and the relative contribution of order Actinomycetales in the total study population. This might reflect a decrease in this microbial subgroup in patients with advanced liver disease and subsequent alteration of proteosynthesis. As the relation could not be assigned to any specific subgroup of patients, the relevance of this observation is not clear.
Due to the advances in sequencing and bioinformatics, we were able to found some of the most striking changes at the low taxonomic level (species). These disease-specific bacteria could be the cornerstone in search for suitable biomarkers for PSC development or in non-invasive distinguishing of different IBD phenotypes. The fact that most of our findings were consistent with those reported in previous studies validates the study design and methodology. The current evidence supports a key role of microbiota in PSC pathogenesis which, however, needs to be further elucidated by future mechanistic studies.
In conclusion, PSC- and UC-associated dysbiosis was characterized by reduced bacterial diversity, significant changes in global bacterial composition and relative abundance of distinct taxa, primarily at the genus and species levels. The most prominent changes related to PSC were in the genera Rothia, Streptococcus, Veillonella, Enterococcus, Clostridium, Haemophilus, and Adlercreutzia. Several microbes including Akkermansia muciniphila, Butyricicoccus pullicaecorum and Clostridium colinum clearly distinguished the UC and PSC-IBD phenotypes. Specific changes in occurring with PSC/AIH overlap involved enrichment of Actinomyces spp.
Primary sclerosing cholangitis (PSC) is a chronic liver disease with particularly high incidence in northern Europe and North America. Concomitant inflammatory bowel disease (IBD) is present in majority of patients with PSC. This IBD phenotype is distinct from Crohn´s disease and ulcerative colitis and is often referred to as PSC-IBD. Gut microbiota composition is most likely involved in pathogenesis of this complex and often unfavorable clinical entity that involves the liver and intestine.
Disruption of microbial ecology has previously been described in several clinical conditions including IBD. The few reports on gut microbiota composition in PSC and PSC-IBD have used diverse methods and yielded inconsistent results.
This study describes specific gut microbial patterns associated with PSC and differ in individual IBD phenotypes (UC and PSC-IBD).
Identifying disease-specific microbial features could be the cornerstone of further research in the field of PSC and IBD pathogenesis as well as the search for suitable biomarkers.
PSC is a severe liver disease characterized by fibrosis and stenosis of both intra- and extrahepatic bile ducts; PSC-IBD is a specific phenotype of inflammatory bowel disease associated with PSC; gut microbiota comprises the ecological community of microorganisms residing in the (human) intestine intestine.
Interesting and complexe article, a very good work and a beautiful writing. It was difficult to read, because includes so many data, but also very useful in the end.
Specialty type: Gastroenterology and hepatology
Country of origin: Czech Republic
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chu J, Lakatos PL, Popp C S- Editor: Qi Y L- Editor: A E- Editor: Zhang FF
| 1. | Gow PJ, Chapman RW. Liver transplantation for primary sclerosing cholangitis. Liver. 2000;20:97-103. [PubMed] |
| 2. | Chapman RW, Arborgh BA, Rhodes JM, Summerfield JA, Dick R, Scheuer PJ, Sherlock S. Primary sclerosing cholangitis: a review of its clinical features, cholangiography, and hepatic histology. Gut. 1980;21:870-877. [PubMed] |
| 3. | Levy C, Lindor KD. Primary sclerosing cholangitis: epidemiology, natural history, and prognosis. Semin Liver Dis. 2006;26:22-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 55] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Karlsen TH, Schrumpf E, Boberg KM. Update on primary sclerosing cholangitis. Dig Liver Dis. 2010;42:390-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 5. | Chapman R, Fevery J, Kalloo A, Nagorney DM, Boberg KM, Shneider B, Gores GJ. Diagnosis and management of primary sclerosing cholangitis. Hepatology. 2010;51:660-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 888] [Cited by in RCA: 835] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 6. | Hirschfield GM, Karlsen TH, Lindor KD, Adams DH. Primary sclerosing cholangitis. Lancet. 2013;382:1587-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 426] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 7. | Loftus EV, Harewood GC, Loftus CG, Tremaine WJ, Harmsen WS, Zinsmeister AR, Jewell DA, Sandborn WJ. PSC-IBD: a unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut. 2005;54:91-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 518] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 8. | Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1339] [Cited by in RCA: 1376] [Article Influence: 80.9] [Reference Citation Analysis (1)] |
| 9. | Bentley RW, Keenan JI, Gearry RB, Kennedy MA, Barclay ML, Roberts RL. Incidence of Mycobacterium avium subspecies paratuberculosis in a population-based cohort of patients with Crohn’s disease and control subjects. Am J Gastroenterol. 2008;103:1168-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Tannock GW. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol. 2008;1 Suppl 1:S15-S18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Lichtman SN, Keku J, Clark RL, Schwab JH, Sartor RB. Biliary tract disease in rats with experimental small bowel bacterial overgrowth. Hepatology. 1991;13:766-772. [PubMed] |
| 12. | Tabibian JH, Weeding E, Jorgensen RA, Petz JL, Keach JC, Talwalkar JA, Lindor KD. Randomised clinical trial: vancomycin or metronidazole in patients with primary sclerosing cholangitis - a pilot study. Aliment Pharmacol Ther. 2013;37:604-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 199] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 13. | Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, Trøseid M, Marschall HU, Schrumpf E, Moum B. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66:611-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 310] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 14. | Kummen M, Hov JR. Response to ‘Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis’ by Rühlemann et al. Gut. 2017;66:755-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Torres J, Bao X, Goel A, Colombel JF, Pekow J, Jabri B, Williams KM, Castillo A, Odin JA, Meckel K. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment Pharmacol Ther. 2016;43:790-801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 119] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 16. | Sabino J, Vieira-Silva S, Machiels K, Joossens M, Falony G, Ballet V, Ferrante M, Van Assche G, Van der Merwe S, Vermeire S. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016;65:1681-1689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 247] [Cited by in RCA: 313] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 17. | Quraishi MN, Sergeant M, Kay G, Iqbal T, Chan J, Constantinidou C, Trivedi P, Ferguson J, Adams DH, Pallen M. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut. 2017;66:386-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (1)] |
| 18. | Kevans D, Tyler AD, Holm K, Jørgensen KK, Vatn MH, Karlsen TH, Kaplan GG, Eksteen B, Gevers D, Hov JR. Characterization of Intestinal Microbiota in Ulcerative Colitis Patients with and without Primary Sclerosing Cholangitis. J Crohns Colitis. 2016;10:330-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 19. | Rossen NG, Fuentes S, Boonstra K, D’Haens GR, Heilig HG, Zoetendal EG, de Vos WM, Ponsioen CY. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J Crohns Colitis. 2015;9:342-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 20. | Rühlemann MC, Heinsen FA, Zenouzi R, Lieb W, Franke A, Schramm C. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut. 2017;66:753-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 21. | Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012;12:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 22. | Tedjo DI, Jonkers DM, Savelkoul PH, Masclee AA, van Best N, Pierik MJ, Penders J. The effect of sampling and storage on the fecal microbiota composition in healthy and diseased subjects. PLoS One. 2015;10:e0126685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 144] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 23. | Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89:670-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1026] [Cited by in RCA: 968] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 24. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [PubMed] |
| 25. | Edgar RC, Flyvbjerg H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics. 2015;31:3476-3482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 704] [Cited by in RCA: 715] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 26. | Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228-8235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5374] [Cited by in RCA: 5560] [Article Influence: 292.6] [Reference Citation Analysis (0)] |
| 27. | Tickle T, Waldron L, Yiren Lu HC. Multivariate association of microbial communities with rich metadata in high-dimensional studies. . |
| 28. | Beuers U. Hepatic overlap syndromes. J Hepatol. 2005;42 Suppl:S93-S99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Beuers U, Rust C. Overlap syndromes. Semin Liver Dis. 2005;25:311-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 30. | Lindor KD, Kowdley KV, Harrison ME. ACG Clinical Guideline: Primary Sclerosing Cholangitis. Am J Gastroenterol. 2015;110:646-659; quiz 660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 338] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 31. | Langan RC, Gotsch PB, Krafczyk MA, Skillinge DD. Ulcerative colitis: diagnosis and treatment. Am Fam Physician. 2007;76:1323-1330. [PubMed] |
| 32. | Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1958] [Cited by in RCA: 2251] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 33. | Kverka M, Tlaskalova-Hogenova H. Intestinal Microbiota: Facts and Fiction. Dig Dis. 2017;35:139-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 34. | Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, Walter J, Mason AL, Wong GK, Dieleman LA. Mucosal Barrier Depletion and Loss of Bacterial Diversity are Primary Abnormalities in Paediatric Ulcerative Colitis. J Crohns Colitis. 2016;10:462-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 180] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 35. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1537] [Article Influence: 139.7] [Reference Citation Analysis (40)] |
| 36. | Ramanan P, Barreto JN, Osmon DR, Tosh PK. Rothia bacteremia: a 10-year experience at Mayo Clinic, Rochester, Minnesota. J Clin Microbiol. 2014;52:3184-3189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 124] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 37. | Rosen R, Amirault J, Liu H, Mitchell P, Hu L, Khatwa U, Onderdonk A. Changes in gastric and lung microflora with acid suppression: acid suppression and bacterial growth. JAMA Pediatr. 2014;168:932-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 38. | Davies YK, Tsay CJ, Caccamo DV, Cox KM, Castillo RO, Cox KL. Successful treatment of recurrent primary sclerosing cholangitis after orthotopic liver transplantation with oral vancomycin. Case Rep Transplant. 2013;2013:314292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | McWhinney PH, Kibbler CC, Gillespie SH, Patel S, Morrison D, Hoffbrand AV, Prentice HG. Stomatococcus mucilaginosus: an emerging pathogen in neutropenic patients. Clin Infect Dis. 1992;14:641-646. [PubMed] |
| 40. | Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675-G685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 429] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 41. | Mjelle AB, Mulabecirovic A, Hausken T, Havre RF, Gilja OH, Vesterhus M. Ultrasound and Point Shear Wave Elastography in Livers of Patients with Primary Sclerosing Cholangitis. Ultrasound Med Biol. 2016;42:2146-2155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 42. | Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 2008;58:1221-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 209] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 43. | Setchell KD, Zhao X, Jha P, Heubi JE, Brown NM. The pharmacokinetic behavior of the soy isoflavone metabolite S-(-)equol and its diastereoisomer R-(+)equol in healthy adults determined by using stable-isotope-labeled tracers. Am J Clin Nutr. 2009;90:1029-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 44. | Alvaro D, Invernizzi P, Onori P, Franchitto A, De Santis A, Crosignani A, Sferra R, Ginanni-Corradini S, Mancino MG, Maggioni M. Estrogen receptors in cholangiocytes and the progression of primary biliary cirrhosis. J Hepatol. 2004;41:905-912. [PubMed] |
| 45. | Tamura S, Sugawara Y, Kokudo N. Primary sclerosing cholangitis as an intractable disease. Intractable Rare Dis Res. 2012;1:13-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 46. | Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420-2428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 863] [Cited by in RCA: 1036] [Article Influence: 69.1] [Reference Citation Analysis (0)] |
| 47. | Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401-3407. [PubMed] |
| 48. | Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B, Steppe M, Haesebrouck F, Sas B, Ducatelle R. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. 2013;62:1745-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 263] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 49. | Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 654] [Article Influence: 54.5] [Reference Citation Analysis (0)] |
| 50. | Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, Fujiyama Y, Andoh A. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J Gastroenterol Hepatol. 2013;28:613-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 220] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 51. | Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65:415-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 571] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 52. | Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1071] [Cited by in RCA: 1223] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 53. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J Hepatol. 2009;51:237-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1203] [Article Influence: 75.2] [Reference Citation Analysis (1)] |
| 54. | Van Assche G, Dignass A, Bokemeyer B, Danese S, Gionchetti P, Moser G, Beaugerie L, Gomollón F, Häuser W, Herrlinger K. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 3: special situations. J Crohns Colitis. 2013;7:1-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 340] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 55. | Halilbasic E, Fuchs C, Hofer H, Paumgartner G, Trauner M. Therapy of Primary Sclerosing Cholangitis--Today and Tomorrow. Dig Dis. 2015;33 Suppl 2:149-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 56. | Hommes DW, Erkelens W, Ponsioen C, Stokkers P, Rauws E, van der Spek M, ten Kate F, van Deventer SJ. A double-blind, placebo-controlled, randomized study of infliximab in primary sclerosing cholangitis. J Clin Gastroenterol. 2008;42:522-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 57. | Franceschet I, Cazzagon N, Del Ross T, D’Incà R, Buja A, Floreani A. Primary sclerosing cholangitis associated with inflammatory bowel disease: an observational study in a Southern Europe population focusing on new therapeutic options. Eur J Gastroenterol Hepatol. 2016;28:508-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |