Published online Jun 7, 2017. doi: 10.3748/wjg.v23.i21.3864
Peer-review started: December 1, 2016
First decision: December 28, 2016
Revised: January 18, 2017
Accepted: March 2, 2017
Article in press: March 2, 2017
Published online: June 7, 2017
Processing time: 189 Days and 11.6 Hours
To investigate the prospective importance of serum micro (mi)RNAs (miR-125b, miR-138b, miR-1269, miR-214-5p, miR-494, miR375 and miR-145) as early biomarkers for the diagnosis of hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC).
Two-hundred and fifty HCV4a patients, 224 HCV4a-HCC patients, and 84 healthy controls were enrolled in the study. Expression levels of miR214-5p, miR-125b, miR-1269 and miR-375 were quantified using quantitative real-time PCR.
Expression of the selected miRNAs in serum was significantly lower in HCC patients than in the healthy controls, except for miR-1269 and miR-494. There was a significant difference between HCC and HCV patients, in particular for HCC and late stage fibrosis, rather than HCV patients and early fibrosis. It is obvious that miR-1269 was significantly upregulated in HCC cases compared to hepatic fibrosis cases. Each miRNA can show HCC progression. Multivariate logistic regression analysis indicated that the tested panel of miRNAs (miR214-5p, miR-125b, miR-1269 and miR-375) represent accurate and specific indictors of HCC development.
This study presents a panel of miRNAs with strong power as putative diagnostic and prognostic biomarkers for HCV-induced HCC. Moreover, miR-214-5p and miR-1269 could be considered as early biomarkers for tracking the progress of liver fibrosis to HCC.
Core tip: Lack of compelling methods for early diagnosis of hepatitis C virus (HCV)-related hepatocellular carcinoma (HCC) disease leads to poor prognosis. The identification of more reliable markers for diagnosis of HCC with easy methodologies for HCC screening and detection in early stage is desperately required. A collection of small non-coding circulating RNAs associated with HCC related-HCV has been found to be differentially communicated and included in the pathogenesis of the disease. Thus, we can use these molecules as prospective biomarkers for HCC, with some of the miRNAs representing biomarkers for liver fibrosis progression.
- Citation: Elemeery MN, Badr AN, Mohamed MA, Ghareeb DA. Validation of a serum microRNA panel as biomarkers for early diagnosis of hepatocellular carcinoma post-hepatitis C infection in Egyptian patients. World J Gastroenterol 2017; 23(21): 3864-3875
- URL: https://www.wjgnet.com/1007-9327/full/v23/i21/3864.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i21.3864
Hepatocellular carcinoma (HCC) is one of the first five cancers represented in the hierarchy of cancer mortality rates, causing around 500000 deaths per annum[1]. Patients with HCC have the least survival compared to patients with other types of cancer, and most die within a year from developing tumor[2]. In Egypt, HCC is the 2nd leading cause of cancer-related death in males and the 5th in females[3].
HCC incidence is on the rise. Several etiologies are the main determinants underlying the sharp increase in HCC rate, with chronic hepatitis infection (either B or C) and fatty liver disease being the most important predisposing factors. Hepatitis C virus (HCV) increases the risk of HCC development by almost 17-fold[4,5]. During the last 10 years, Egypt showed around 2-fold increment increase in HCC development among chronic liver disease patients, mainly due to infection with HCV[6-8].
Multiple studies have suggested that early detection of HCC will increase the lifetime of HCC patients[9]. Current strategies for the diagnosis of HCC fall into two main categories: biomarker tests and imaging. Unfortunately, the diagnostic performance of these strategies is not satisfactory, especially at the early stages of disease. Currently, only 30% to 40% of patients with HCC are qualify for potential curative intervention at diagnosis, because of the lack of clinical presentation and absence of potential early identification biomarkers. Thus, the identification of new reliable markers for HCC, with high sensitivity and specificity, is of great importance[9].
Micro (mi)RNAs are a group of non-coding small RNAs that regulate the expression of other protein-coding RNAs, at both the transcriptional and post-transcriptional levels. It is well known that miRNAs manage every aspect of cell activities, including cell differentiation and development, metabolism, cell cycle regulation, apoptosis, immune response, and oncogenesis[10]. Multiple studies have demonstrated that hepatic cells express high levels of certain miRNAs to control different cell functions, with their expression levels correlating to different pathological states[11]. Characterized by their stability and resistance to RNase degradation, miRNAs are considered as promising biomarkers for many diseases[12,13]. The use of miRNAs as noninvasive biomarkers is of particular interest for the diagnosis of liver diseases[14-16].
miRNAs profiling studies in HCC have revealed significant dysregulation for a number of miRNAs[17-19]. In this regard, there are several examples: miR-1269 showed elevated levels in liver of HCC/HCV patients versus healthy livers[19]. miR-618 and miR-650 showed elevated levels in the urine of HCV/HCC patients, with enhanced specificity reaching 75% upon use of both miRNAs in concert[20]; these findings suggest the promise of urinary miRNAs as biomarkers for early detection of HCC. Moreover, a recent study identified a panel of serum miRNAs that have considerable clinical value in the diagnosis of HCC following hepatitis B virus (HBV) infection[20].
The identification of non-invasive biomarkers for predicting patients at high risk of HCC development allows early intervention and increases the probability of successful treatment, consequently reducing mortality and disease burden. Our study aims to investigate the prospective importance of some serum miRNAs as early biomarkers for the diagnosis of HCV-related HCC.
This study included 474 HCV serum-positive Egyptian patients (224 HCC patients and 250 with chronic liver hepatitis), in addition to 84 controls (HCV-negative patients). Study subjects were recruited from the Clinical Hepatology Department, Theodor Bilhariz Research Institute. Blood samples were collected according to the ethical standards for donor approval required by the national regulatory bodies under supervision of a physician and after having signed informed consent for the use of their blood in this study. All patients were pre-diagnosed HCV antibody-positive by enzyme-linked immunosorbent assay. All samples were subjected to HCV antigen detection in serum. HCC was detected by ultrasound scan and confirmed using computed tomography (CT). Fibrosis level was calculated using Metavir scoring, from F0 representing absence of fibrosis to F4 representing cirrhosis[21]. All HCV cases were reported as fibrotic from F1 to F4, and routine lab tests were performed for each sample. Patients with any hepatitis other than C, or malignancy other than caused by HCV, were excluded from the study. Sex distribution among healthy controls was 56 males and 28 females, with mean age of 44 and standard deviation of 12.5 years. Control subjects had normal levels of liver enzymes and alpha-fetoprotein (AFP)[22].
It is well known that the levels of the selected serum miRNAs are dysregulated in chronically infected patients, simply because all of the aforementioned miRNAs are cell cycle and interferon (IFN)-related, and mostly affect response to IFN therapy[23]. Potentiality of clinical non-invasive serum biomarkers of these miRNAs in HCC-related HCV progression is unknown till now.
RNA was extracted using the miRNeasy Kit (Qiagen, Germany), according to the manufacturer’s protocol, and quantified using the Nanoquant Infinite M200 Microplate Reader (Tecan, Switzerland). A total of 100 ng RNA was reverse transcribed using the mi-Script RT-II Kit (Qiagen) in a reaction volume of 20 μL and subjected to 60 min incubation at room temperature, followed by 5 min incubation at 95 °C. Serum levels of different miRNAs (hsa-miR-1269, hsa-miR-125b, hsa-miR-138, hsa-miR-214-5p, hsa-miR-494, hsa-miR-375 and hsa-miR-145) were assessed using the mi-Script miRNA PCR Array (Qiagen), according to the manufacturer’s procedure. The housekeeping gene miRNA SNORD68 was used as an internal control[24,25]. All the miRNA primers, including for the housekeeping gene, were purchased from Qiagen. QuantiTect SYBR PCR reagent with miScript universal primer was used according to the manufacturer’s protocol (Qiagen). Quantitative PCR amplification and analysis were achieved by Rotor-gene (Qiagen), using the following thermal cycling program: 30 min at 95 °C, then 15 s at 94 °C and 30 s at 55 °C (for 40 cycles), and finally 30 s at 70 °C. The change in miRNA expression level was calculated using the 2-ΔΔCT approach and relative to healthy subjects serving as the calibrator control[26].
Statistical analyses were performed by SPSS-15 (SPSS Inc, United States) and GraphPad Prism-5.0 (GraphPad, United States). Mean ± SD, or median was used to express all the data in this study. Clinical data from the three independent groups were compared using one-way ANOVA test in addition to Tukey Kramer’s for multiple comparisons test and Mann-Whitney U-test. Log transformation was applied to AFP data to facilitate the use of parametric statistical analysis. The χ2 test was used to compare categorical data. Analysis for receiver operating characteristic (ROC) with the area under the curve (AUC) was used to evaluate the diagnostic potential of each studied miRNA. The end-point was categorizing HCC out of late fibrosis (F4) using the miRNAs in the study panel[23].
Patients showed a significant progression from moderate fibrosis to carcinoma with older age (P < 0.0001). HCC patients’ serum levels of alkaline phosphatase, glutamic oxaloacetic transaminase, glutamic pyruvic transaminase, bilirubin and AFP were significantly higher (P < 0.0001) than those of fibrotic groups and was associated with depletion in hemoglobin, prothrombin and albumin levels, as well as platelet count (P < 0.0001). Moreover, leucocyte count did not show any significant variation among these groups, as shown in Table 1.
| Non-malignant chronic HCV (n = 250) | HCC (n = 224) | P value | ||
| Moderate fibrosis (n = 150) | Late fibrosis (n = 100) | |||
| Age (years) | 40 ± 8.6a | 45 ± 6.5c | 55 ± 9.3e | < 0.0001 |
| ALP (IU/L) | 102.3 ± 36.9a | 119 ± 72.1a | 182.5 ± 58c | < 0.0001 |
| SGOT (IU/L) | 58 ± 18a | 62.5 ± 28.2a,b | 85.4 ± 45.2c | 0.001 |
| SGPT (IU/L | 52.1 ± 18a | 68.46 ± 24.8a | 120.4 ± 53.4c | < 0.0001 |
| Bilirubin, total (mg/dL) | 0.40 ± 0.17a | 0.43 ± 0.16a | 0.78 ± 0.69c | < 0.0009 |
| Bilirubin, direct (mg/dL) | 0.68 ± 0.28a | 0.85 ± 0.37a | 1.9 ± 1.2c | < 0.0001 |
| Log AFP (ng/mL) | 0.49 ± 0.9a | 0.98 ± 0.4a | 2.3 ± 0.97c | < 0.0001 |
| Hemoglobin (g/dL) | 13.6 ± 1.8a | 14.1 ± 1.5a | 10.7 ± 1.2c | < 0.0001 |
| Platelet count × 103 (/mm3) | 236.5 ± 21.6a | 158.2 ± 52.6c | 131.2 ± 75.5c | < 0.0001 |
| leukocyte count × 103 (/mm3) | 6.3 ± 2.22a | 6.4 ± 2.41a | 5.7 ± 2.4a | 0.460 |
| Prothrombin concentration (%) | 103 ± 4.7a | 84.4 ± 8.0c | 69.9 ± 19.3e | < 0.0001 |
| Albumin (g/dL) | 4.7 ± 0.8a | 3.9 ± 0.9c | 3.28 ± 0.6e | < 0.0001 |
HCC patients yielded a broad range of AFP from normal healthy to abnormal values. Healthy AFP level up to 10 ng/mL was present in 31 patients, 168 patients had AFP more than 20 ng/mL and 103 patients showed AFP more than 400 ng/mL. By CT imaging, 112 patients showed one focal lesion in liver, with 64 of them showing focal size more than 5 cm, and 48 patients showed portal vein thrombosis, as shown in Table 2.
| Factor | n |
| AFP level | |
| Less than 20 ng/mL | 56 |
| Between 20 and 400 ng/mL | 64 |
| More than 400 ng/mL | 104 |
| Child-Pugh grade | |
| A | 128 |
| B | 80 |
| C | 16 |
| Barcelona Clinic Liver Cancer staging | |
| 0 | 0 |
| A | 8 |
| B | 160 |
| C | 40 |
| D | 16 |
| Focal lesions | |
| Single | 112 |
| Multiple | 112 |
| Focal size by CT | |
| < 3 cm | 48 |
| > 5 cm | 64 |
| Thrombosis in portal vein | |
| Present | 48 |
| Absent | 176 |
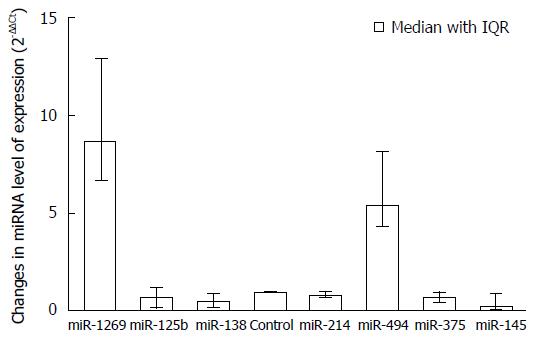
Differential serum miRNA levels were measured in HCC patients and compared to healthy controls and HCV-infected patients (Mann-Whitney U-test). The results showed that miR-1269 and miR-494 were positively regulated in HCC patients, with median fold-change of 8.71 and 5.45 respectively compared to healthy non-infected controls (P < 0.0001), while miR-125b, miR-138b, miR-214-5p, miR-375 and miR-145 were negatively regulated in HCC patients, with median fold-change of 0.51, 0.46, 0.23, 0.25 and 0.6 respectively, in comparison to healthy controls and as shown in Figure 1.
Serum miRNA profiles were studied from mild fibrosis progression to HCC. The possible changes in miRNA levels from mild fibrosis to HCC were tested by Mann-Whitney U-test. The present data show no significant difference in levels of miRNA between mild and moderate fibrosis groups, except for miR-214-5p. Moreover, the data shows that all tested miRNAs levels were significantly different between HCC and late fibrosis (P < 0.05). It was found that miR-214-5p were significantly downregulated in late fibrosis F4 compared to F3 (P < 0.05) and continued the downregulation trend during HCC progression versus late fibrosis F4 (P < 0.05). Furthermore, miR-375, miR-145 and miR-1269 levels were significantly different in HCC and late fibrosis F4 groups (P < 0.05). Conversely, miR-125 and miR-138-5p were not affected by case progression, as shown in Table 3 and Figure 2.
| Serum miRNA | AUC value | Sensitivity | Specificity | PPV | NPV | Accuracy |
| miR-214-5P | ||||||
| HCC against ctrl | 0.721 | 61.0% | 89.0% | 81.0% | 75.0% | 77.0% |
| HCC against HCV | 0.842 | 92.9% | 75.5% | 51.0% | 97.5% | 80.0% |
| miR-494 | ||||||
| HCC against ctrl | 0.813 | 77.0% | 76% | 69.0% | 82.0% | 76.0% |
| HCC against HCV | 0.631 | 77.0% | 56% | 32.0% | 90.0% | 60.0% |
| miR-138p | ||||||
| HCC against ctrl | 0.842 | 96.4% | 78.4% | 77.1% | 96.0% | 86.0% |
| HCC against HCV | 0.642 | 68.2% | 58.2% | 34.0% | 90.0% | 62.0% |
| miR-125b | ||||||
| HCC against ctrl | 0.702 | 66.7% | 75.7% | 66.7% | 75.7% | 72.0% |
| HCC against HCV | 0.769 | 92.6% | 55.4% | 35.7% | 96.6% | 63.3% |
| miR-1269 | ||||||
| HCC against ctrl | 0.862 | 96.4% | 78.4% | 77.1% | 96.7% | 86.2% |
| HCC against HCV | 0.691 | 78.6% | 59.8% | 34.9% | 91.0% | 63.8% |
| miR-145 | ||||||
| HCC against ctrl | 0.892 | 100.0% | 89.2% | 87.1% | 100.0% | 93.8% |
| HCC against HCV | 0.624 | 81.5% | 51.5% | 30.6% | 91.4% | 57.7% |
| miR-375 | ||||||
| HCC against ctrl | 0.741 | 96.4% | 59.5% | 63.4% | 95.7% | 75.4% |
| HCC against HCV | 0.811 | 96.4% | 69.3% | 46.6% | 98.6% | 75.2% |
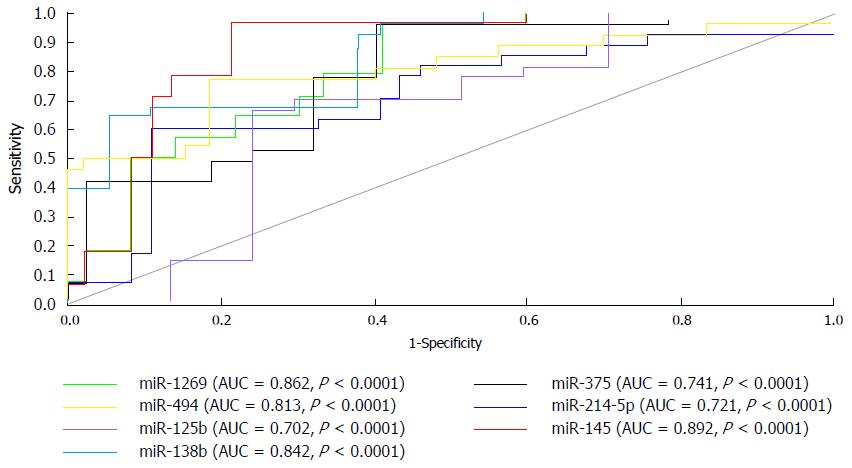
Analysis of AUC indicated that miRNAs assessed in this study might be useful to categorize HCC cases among healthy non-malignant controls. All studied circulating miRNAs showed significance difference (P < 0.0001 and 95%CI for values of AUC as 0.721, 0.813, 0.842, 0.702, 0.862, 0.892 and 0.741 for miR-214-5p, miR-494, miR-138b, miR-125b, miR-1269, miR-145 and miR-375 respectively; Figure 2).
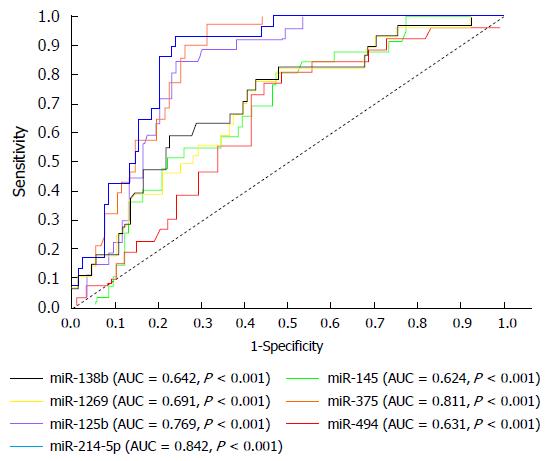
Also, the diagnostic efficacy for miRNA was studied and the obtained data showed differential expression between HCV and HCC groups, which helped to categorize each group separately. Analysis using ROC curve showed that the studied miRNAs were capable of distinguishing HCC from late fibrosis, with AUC values of 0.842, 0.631, 0.642, 0.769, 0.691, 0.624 and 0.811 for miR-214-5p, miR-494, miR-138b, miR-125b, miR-1269, miR-145 and miR-375 respectively, with 95%CI and P-value less than 0.001 (Figure 3). The accuracy, sensitivity and specificity for each studied miRNA were investigated to diagnostically discriminate HCC among HCV fibrotic patients or normal healthy controls and this result is presented in Table 3.
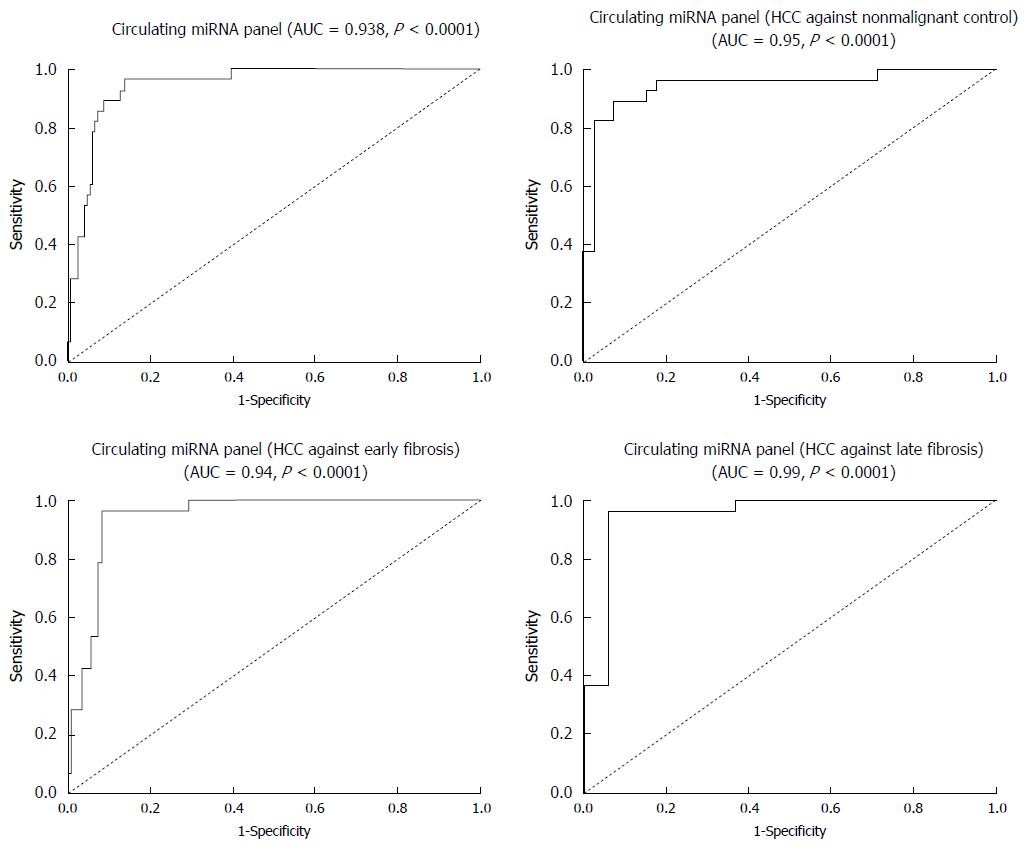
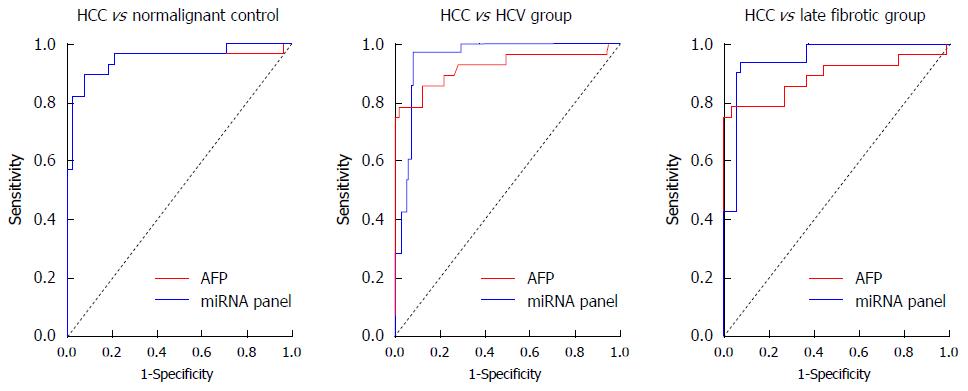
Univariate and multivariate logistic regression analyses were calculated to determine the possible association of the studied miRNAs with diagnosis of HCV-related HCC progression. Levels of expression for miR-214-5p, miR-125b, miR-1269 and miR-375 were found in univariate analysis to be a significant possible predictor of HCC diagnosis, so that they may serve as a possible miRNA panel. Logit (P) = -0.21-1.123_miR-214-5p-0.076_miR-125b + 0.58_miR-1269 + 0.046_miR-375 was used to formulate the ROC curve. The miRNA panel’s diagnostic efficacy was assessed by ROC analysis, and showed AUC of 0.938 with 95%CI specificity = 92%, sensitivity = 97.01% and accuracy = 92.3%. This panel can categorize HCC cases among healthy controls (AUC = 0.95 with 95%CI specificity = 96.9%, sensitivity = 83.2% and accuracy = 86.8%), HCV early fibrotic patients (AUC = 0.945 with 95%CI specificity = 94.3%, sensitivity = 96.7% and accuracy = 95.1%) and the late fibrotic groups (AUC = 0.99 with 95%CI, specificity = 98.3%, sensitivity = 98.7%, accuracy = 98.4%). Comparison of each of the studied miRNAs’ AUC data against the miRNA panel indicated that the panel provided superior discrimination for HCC, either from nonmalignant controls and HCV fibrotic patients. The data is listed in Table 4 and shown in Figure 4.
| Coefficient | Standard error | P value | |
| Univariate analysis | |||
| miR-214-5p | -0.570 | 0.105 | < 0.0001 |
| miR-494 | -0.005 | 0.025 | 0.825 |
| miR-125b | -0.064 | 0.013 | < 0.0001 |
| miR-1269 | 0.184 | 0.044 | < 0.0001 |
| miR-145 | 0.0021 | 0.011 | 0.844 |
| miR-375 | 0.066 | 0.011 | < 0.0001 |
| miR-138b | 0.0019 | 0.013 | 0.852 |
| Multivariate analysis | |||
| miR-214-5p | -1.116 | 0.229 | < 0.0001 |
| miR-125b | -0.076 | 0.028 | 0.004 |
| miR-1269 | 0.569 | 0.121 | < 0.0001 |
| miR-375 | 0.052 | 0.013 | 0.002 |
| Constant | -0.240 | ||
The AUC of AFP for each study group was assessed using the patients’ serum samples. AFP demonstrated a significant accurate discrimination between HCC and nonmalignant controls, HCV patients, and the late fibrotic groups with AUC values of 0.963, 0.921 and 0.888 respectively. AFP showed 100%, 96% and 90% specificity, in addition to 80% sensitivity to discriminate HCC from nonmalignant controls, the HCV group, and the late fibrosis groups respectively, with cut-off of > 20 ng/mL. However, at the AFP cut-off level of > 400 ng/mL, which is considered diagnostic for HCC, AFP showed specificity of 100%, 98% and 96%, and sensitivity reached 46% for the same conditions. Therefore, AUC was studied for the miRNA panel against AUC for AFP, and the result demonstrated no significant difference among them. Difference amid areas in the nonmalignant group was 0.019, P = 0.476 and difference amid areas in HCV groups was 0.024, P = 0.261; conversely, difference amid areas in late fibrotic groups was 0.083 (P = 0.034), as shown in Figure 5.
In the investigation of HCC among portal vein thrombosis patients (n = 48) it was found that miR-214-5p and miR-375 expression were significantly elevated compared to normal controls (median change: 25%-75%, P = 0.0001). There was no strong relation found for the effect of miRNAs on either number of focal lesions nor diameter of the tumor. In addition, miR-145 was positively correlated with HCV viral load (Pearson r = 0.574, P = 0.001).
There was significant promising association between the HCC group and all assessed miRNAs. miR-214-5p was correlated significantly with miR-138b (r = 0.649, P < 0.0002), miR-125b (r = 0.518, P = 0.005), miR-1269 (r = 0.621, P = 0.0005) and miR-145 (r = 0.449, P = 0.009). miR-494 was correlated with miR-138b (r = 0.471, P = 0.008), miR-1269 (r = 0.418, P = 0.021) and miR-145 (r = 0.538, P = 0.003). miR-138b was also correlated with miR-125b (r = 0.685, P < 0.0001), miR-1269 (r = 0.862, P < 0.0001) and miR-145 (r = 0.521, P = 0.006). miR-1269 was also correlated with miR-125b (r = 0.602, P = 0.001) and miR-145 (r = 0.453, P = 0.019). miR-145 was correlated with miR-375 (r = 0.460, P = 0.038), as shown in Table 5.
| miR-138b | miR-494 | miR-145 | miR-375 | miR-214-5p | miR-1269 | miR-125b | |
| miR-138b | r = 0.471 | r = 0.521 | NS | r = 0.649 | r = 0.862 | r = 0.685 | |
| P = 0.008 | P = 0.006 | P < 0.0002 | P < 0.0001 | P < 0.0001 | |||
| miR-494 | r = 0.471 | r = 0.538 | NS | NS | r = 0.418 | NS | |
| P = 0.008 | P = 0.003 | P = 0.021 | |||||
| miR-145 | r = 0.521 | r = 0.538 | r = 0.460 | r = 0.449 | r = 0.453 | NS | |
| P = 0.006 | P = 0.003 | P = 0.038 | P < 0.009 | P = 0.019 | |||
| miR-375 | NS | NS | r = 0.460 | r = 0.884 | NS | NS | |
| P = 0.038 | P < 0.0001 | ||||||
| miR-214-5p | r = 0.649 | NS | r = 0.449 | r = 0.884 | r = 0.621 | r = 0.518 | |
| P < 0.0002 | P < 0.009 | P < 0.0001 | P = 0.0005 | P = 0.005 | |||
| miR-1269 | r = 0.862 | r = 0.418 | r = 0.453 | NS | r = 0.621 | r = 0.602 | |
| P < 0.0001 | P = 0.021 | P = 0.019 | P = 0.0005 | P = 0.001 | |||
| miR-125b | r = 0.685 | NS | NS | NS | r = 0.518 | r = 0.602 | |
| P < 0.0001 | P = 0.005 | P = 0.001 |
One of the major challenges for HCC diagnosis in the early stage is the absence of reliable biomarkers, which strengthen urgent demand for efficient profitable early diagnostic assays. In our current investigation, it is revealed that the expression level of all the studied circulating miRNAs differed significantly between HCV patients and HCV/HCC patients when compared to healthy controls, suggesting miRNA biomarker activity against HCC progression. In addition, individual miRNA could be used to discriminate the HCC from HCV fibrotic patient from the healthy control. The multivariate analysis showed a panel of serum miRNAs (miR-214-5p, miR-375, miR-125b and miR-1269) that has accurate diagnostic efficacy against HCC progression. Also, it showed superior diagnostic accuracy over AFP in discriminating the late fibrosis group, which has a significant tendency to develop HCC. In addition, the data for the studied miRNA panel showed significantly higher diagnostic accuracy than has been formerly reported for AFP (specificity: 76%-94% and sensitivity: 39%-65%)[27]. More than 35% of HCC patients are missed by early AFP testing, besides its conflicting elevation in both chronic hepatitis and cirrhotic patients[27], which highlights the greater reliability of the studied miRNAs for early diagnosis and fortifies its therapeutic and serological tumor markers feature, especially for HCC control.
The present data confirm that the tested serum miRNAs significantly correlated with HCC-HCV groups and suggest their possible role in HCV pathogenesis. Supportive for the latter possibility, HCV proteins have been shown to regulate cell cycle behavior and apoptosis in HCC patients through alteration of cellular expression of miRNAs, especially in cells carrying the HCV infectious strand[28]. Circulating miR-375 and miR-214-5p were shown to be negatively regulated in HCC compared to either healthy controls or HCV fibrotic patients[28]. Also, miR-375 was reported as negatively regulated in HCC patients compared to HBV-infected patients[29], and in another report negative regulation was shown in the serum and tissues of HCC patients compared to nonmalignant controls[28,30]. Downregulation of miR-375, on the other hand, promotes tumor formation through upregulation of YAP1 and AEG-1, both of which induce cell growth (tissue invasion), and of IGF1R, which also induces cell growth (metastasis)[30]. miR-214-5p plays critical roles in the activation of stellate cells and, in turn, the progression of fibrotic liver[31], besides acting as a negative regulator for HCC metastasis by regulating fibroblast growth factor receptor 1 expression[32]. In the current investigation, we showed that elevated levels of circulating miR-375 were also correlated with portal vein thrombosis, especially in the HCC group, indicating the importance of miR-375 in HCC progression. miR-375 is involved in the Wnt/β-catenin pathway and its downregulation in patients with HCC is mediated by β-catenin[33]. So, both miR214-5p and miR-375 have a possible association with the presence of HCC satellite nodules, recurrence of HCC and survival rate after transplantation. All of these issues strengthen the possible prognostic importance of miR-375 and 214-5p in HCC.
Circulating miR-494 and miR-1269 were upregulated in HCC patients compared to controls. miR-494 was significantly lower in the HCV group compared to the HCC group, while miR-1269 was significantly higher in the HCC patients than in the HCV patients. Serum miR-214-5p, miR-375 and miR-145 were downregulated in HCC compared to controls or HCV groups. Similarly, serum miR-125b and miR-138-5p were downregulated in the HCC group and the HCV group compared to the control group, miR-1269 was also upregulated in HCC patient serum compared to nonmalignant healthy control serum. It has been reported that miR-1269a is a potential marker for making adjuvant chemotherapy decisions for colorectal cancer patients, and a potential therapeutic target to deter metastasis[34].
Dysregulation has been shown for some serum miRNAs in HCC patient serum and tissue, i.e., miR-125b was downregulated in HCC tissues compared to adjacent non-cancerous tissue[35]. These contradictory results reveal that the same miRNA can play dual tumor oncogenic and suppressor roles in the progression of cancer. Remarkably, miR-214 was downregulated throughout tumor progression, relating to metastasis in HCC; this downregulation stimulates tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma[36]. miR-125b also suppresses HCC invasion or metastasis via downregulation of VEGF[37]. This process in the endothelial cells clearly promotes vascularization and angiogenesis, and its downregulation was correlated with HCC deterioration[38]. Conversely, miR-1269[39] and miR-494 were elevated in HCC and negatively affected the anti-tumor immune response[40].
In the current study, it was found that the expression levels of 3 from the 7 studied serum miRNAs were matched with their tissue levels (hepatic) in HCC compared to healthy controls[35,41,42]; however, up to the time this work was begun, no studies had been performed to measure the expression levels of the studied miRNAs in HCC tissue against normal. miR-122 was downregulated in HCC cell lines and hepatic tissue[43] but upregulated in HCC serum patients[27,44], which suggests that miRNAs secreted from cells could be a vital part of circulating miRNA expression[45]. During liver injury, exosome secretion increases circulating miR-122, causing its hepatic expression to decrease[46]. Conversely, miRNAs reported as raised during liver injury have also shown serum level reduction[47], which has been reported as involving miRNAs, with HCC tissue level elevation and HCC serum level decline[48].
The significant variability between studies on the various miRNA levels in HCC reflects the antithetical mechanism regulating miRNA expression. For example, miR-375 enhances p21 expression and miR-214 controls p53 and XBP1, so that both miRNAs control tumor progression; mechanisms of liver injuries that activate p53 may be upregulated by miR-214 and miR-375[48,49]. The differences between different reports may be due to the variable procedures used for investigation, beginning with ways of collecting samples and ranging to various detection protocols and methods of data analysis[50]. It is noteworthy that the etiology of the disease is a mandatory matter, as HBV-related HCC differs from HCV-related HCC in the molecular mechanisms of miRNA dysregulation[51].
Briefly, impact of circulating miRNAs in cancer progression is now well established; often, the involved pathways are cell proliferation, differentiation, migration, invasion and angiogenesis, all of which are frequently dysregulated by cancer cell mechanisms and so their functions contrast basically according to tissue types. For example, miR-21 (oncomiR) expression increases growth rate of cancer (tissue invasion) through targeting of many tumor suppressor mRNAs, such as PTEN and PDCD4[52,53], while miR-214 targets many oncogenic mRNAs to limit cancer cell proliferation, such as CD44 and CDK1[48]. HCC progression includes several stages, but almost all HCV patients experience progression of fibrosis to cirrhosis, and HCC[54].
In the current study, a reduction in albumin and prothrombin (which represent liver synthetic function) was shown during liver disease progression, but we found no correlation with the circulating miRNAs studied. miRNA expression level is dysregulated in the fibrotic liver, which provides evidence that miRNAs are affected by the progression of liver fibrosis[51]. In the current study, miR-214-5p and miR-145 were positively regulated in fibrotic patients, as compared to healthy controls, with up to 5.2- and 3.8-fold increases respectively and with significant reduction during cirrhosis progression and downregulation in HCC patient serum. The latter can be attributed to the release of miRNA from HCV-inflamed hepatocytes into the circulation, followed by a dramatic decline in their level after late fibrosis due to complete damage of hepatocytes and the presence of cirrhotic tissue with accumulation of extracellular matrix[55].
A key player in fibrosis progression is the activation of hepatic stellate cells, which correlates with deregulation of specific miRNA through regulation of various fibrogenic signaling pathways[56]. It is likely that miR-214-5p and miR-145 is associated with inflammation-fibrosis in HCV-related HCC. Most of the previous publications have reported that miRNAs cannot be differentially expressed during different fibrosis-stages[48]. Conversely, miR-214-5p has been demonstrated to be differentially expressed during fibrosis progression, until HCC, with serum miR-145 being elevated during the progression of fibrosis in chronic HCV[57]. These findings could strengthen the probability of using miR-145 as a reliable prognostic marker, especially for fibrosis progression interpretation.
In the current study, the selected serum miR-214-5p was found to be correlated with fibrosis progression and it showed significant positive correlation with Barcelona Clinic Liver Cancer staging score and Child-Pugh grade, which is used to define HCC for various treatment protocols and which might outline the effectiveness of serum miR-214-5p in assessing severity of HCC for possible therapeutic interferences.
Earlier publications have reported evaluations of circulating miRNAs as HCC biomarkers compared to healthy nonmalignant controls and chronic HCV or fibrosis[9,12,15,19,25,44,58-60], but yielded insufficient discovery of miRNAs as effective biomarkers. Our study presented a panel of four miRNAs characterized by high specificity for HCC, supporting its clinical impact on diagnosis of HCV-related HCC. Moreover, most of the previous studies investigated HCC of mixed etiologies, not were not specific for HCV-related HCC. Another drawback to the existing literature is that there are limited studies evaluating the circulating miRNAs as reliable candidate biomarkers during HCV fibrosis progression to HCC.
Our study detected a pattern of serum miRNAs during the progression of HCV fibrosis to HCC. The evaluated miRNAs had significant variation from early fibrosis to late fibrosis to the formation of HCC, and the results led us to propose them as possible biomarkers for liver disease progression, especially for miR 214-5p and miR-145. Despite our findings, the miR-1269 levels were not fully explained, especially for the correlation from fibrosis progression to HCC, and additional studies are mandatory to validate this panel of miRNA in an independent patient cohort and to validate its outcome significantly in HCC prognosis. Moreover, the inferences of this panel as targets for HCC therapy and application in urine diagnosis will require detailed study.
In conclusion, the assessed panel of serum miRNAs (miR-214-5p, miR-375, miR-1269 and miR-125b) could be used to discriminate HCV-related HCC patients from late fibrotic patients, potentiating its practicality in uses as early prognostic biomarkers of HCC, especially for high-risk individuals. The present study identified a panel of four serum miRNAs that have an elevated accuracy for HCC and promising clinical impact on prognosis of HCV-related HCC. But, there is a need for further studies to assure efficacy of this panel in an independent patient cohort study and different population (other than Egyptian patients). Most of circulating miRNAs are fibrosis-stage independent; however, miR-214-5p showed significant downregulation during liver fibrosis progression, i.e., cirrhosis ending with HCC. The results suggest that serum miR-145 monitoring might be a possible diagnostic tool for progression of fibrosis to HCC. The studied miRNAs panel showed significant correlation with HCC patients. Finally, the association between miR-214-5p and miR-375 in portal vein thrombosis and cancer stage scores supported their prognostic value in HCC progression.
We thank all medical staff and technicians of the Hepatology Center who agreed to participate in this study. Also, I would like to express our deep gratitude to Dr. Marwa Khalil Ibrahim for her great support during this work and Dr. Mohammed Ahmad bahgat for his support finishing ethical committee approval.
Hepatitis C virus (HCV) infection is one of main causes of hepatocellular carcinoma (HCC) and the prevalence of HCV-associated HCC is rising worldwide, with consideration to public health malignancy-related concerns and as one of the prominent causes of death globally. Lack of compelling methods for diagnosis of the disease at an early stage led to its poor prognosis. Along these lines, the advancement of more dependable markers for diagnosis of HCC at early stage and better methodologies for HCC screening and early detection are desperately required. A collection of small non-coding circulating RNAs associated with HCC-related HCV, have been observed to be differentially communicated and involved in pathogenesis of the disease. The current study investigated the prospective importance of some serum micro (mi)RNAs - specifically, miR-125b, miR-138b, miR-1269, miR-214-5p, miR-494, miR375 and miR-145 - as early biomarkers for diagnosis of HCC related to viral hepatitis C.
The authors confirm data of previous epidemiological studies that suggest miRNAs as viable prognosis markers for HCV-related HCC progression. Also, this study sheds light on a new panel of miRNAs for early HCC prognosis.
miRNA panel should be added to the HCV-related HCC prognosis markers involved in HCC disease progression for the Egyptian population.
This manuscript aimed to look for a non-invasive method for early diagnosis of HCV-related HCC. The authors demonstrated that panel of four serum miRNAs (miR-214-5p, miR-375, miR-1269 and miR-125b) that had a significant correlation with HCC, and suggest that serum miR-145 monitoring might be a possible diagnosis tool for progression of fibrosis to HCC. The finding is very interesting and valuable.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Egypt
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C,C,C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Trovato FM, Wang XC, Yao YC S- Editor: Yu J L- Editor: Filipodia E- Editor: Wang CH
| 1. | Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485-1491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1196] [Cited by in RCA: 1325] [Article Influence: 82.8] [Reference Citation Analysis (0)] |
| 2. | Flores A, Marrero JA. Emerging trends in hepatocellular carcinoma: focus on diagnosis and therapeutics. Clin Med Insights Oncol. 2014;8:71-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 3. | Gomaa AI, Hashim MS, Waked I. Comparing staging systems for predicting prognosis and survival in patients with hepatocellular carcinoma in Egypt. PLoS One. 2014;9:e90929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 174] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 5. | Bartosch B, Thimme R, Blum HE, Zoulim F. Hepatitis C virus-induced hepatocarcinogenesis. J Hepatol. 2009;51:810-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 6. | el-Zayadi AR, Badran HM, Barakat EM, Attia Mel-D, Shawky S, Mohamed MK, Selim O, Saeid A. Hepatocellular carcinoma in Egypt: a single center study over a decade. World J Gastroenterol. 2005;11:5193-5198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 55] [Reference Citation Analysis (0)] |
| 7. | Ibrahim AS, Khaled HM, Mikhail NN, Baraka H, Kamel H. Cancer Incidence in Egypt: Results of the National Population-Based Cancer Registry Program. J Cancer Epidemiol. 2014;2014. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 289] [Cited by in RCA: 373] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 8. | Abdel-Hamid NM, Nazmy MH, Mahmoud AW, Fawzy MA, Youssof M. A survey on herbal management of hepatocellular carcinoma. World J Hepatol. 2011;3:175-183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 10. | Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 297] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 11. | Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology. 2012;56:1946-1957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 12. | Blanco-Calvo M, Calvo L, Figueroa A, Haz-Conde M, Antón-Aparicio L, Valladares-Ayerbes M. Circulating microRNAs: molecular microsensors in gastrointestinal cancer. Sensors (Basel). 2012;12:9349-9362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Ge Y, Chen G, Sun L, Liu F. MicroRNA-29 and fibrosis diseases. Zhongnan Daxue Xuebao Yixueban. 2011;36:908-912. |
| 14. | He Y, Huang C, Zhang SP, Sun X, Long XR, Li J. The potential of microRNAs in liver fibrosis. Cell Signal. 2012;24:2268-2272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 15. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 466] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 16. | Chen YP, Jin X, Xiang Z, Chen SH, Li YM. Circulating MicroRNAs as potential biomarkers for alcoholic steatohepatitis. Liver Int. 2013;33:1257-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Chen YJ, Zhu JM, Wu H, Fan J, Zhou J, Hu J, Yu Q, Liu TT, Yang L, Wu CL. Circulating microRNAs as a Fingerprint for Liver Cirrhosis. PLoS One. 2013;8:e66577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 347] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 19. | Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133:816-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825-832. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2199] [Cited by in RCA: 2159] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 22. | Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092-6099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 614] [Cited by in RCA: 637] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 23. | Motawi TK, Shaker OG, El-Maraghy SA, Senousy MA. Serum interferon-related microRNAs as biomarkers to predict the response to interferon therapy in chronic hepatitis C genotype 4. PLoS One. 2015;10:e0120794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67:875-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 136] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 25. | El-Garem H, Ammer A, Shehab H, Shaker O, Anwer M, El-Akel W, Omar H. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol. 2014;6:818-824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 26. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133558] [Article Influence: 5564.9] [Reference Citation Analysis (1)] |
| 27. | Qi J, Wang J, Katayama H, Sen S, Liu SM. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 28. | Zhou N, Wu J, Wang X, Sun Z, Han Q, Zhao L. Low-level expression of microRNA-375 predicts poor prognosis in hepatocellular carcinoma. Tumour Biol. 2016;37:2145-2152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Sarkar N, Chakravarty R. Hepatitis B Virus Infection, MicroRNAs and Liver Disease. Int J Mol Sci. 2015;16:17746-17762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 30. | Yan JW, Lin JS, He XX. The emerging role of miR-375 in cancer. Int J Cancer. 2014;135:1011-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 190] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 31. | Iizuka M, Ogawa T, Enomoto M, Motoyama H, Yoshizato K, Ikeda K, Kawada N. Induction of microRNA-214-5p in human and rodent liver fibrosis. Fibrogenesis Tissue Repair. 2012;5:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 32. | Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60:598-609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 33. | Fatima S, Lee NP, Luk JM. Dickkopfs and Wnt/β-catenin signalling in liver cancer. World J Clin Oncol. 2011;2:311-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 46] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Zhao L, Wang W. miR-125b suppresses the proliferation of hepatocellular carcinoma cells by targeting Sirtuin7. Int J Clin Exp Med. 2015;8:18469-18475. [PubMed] |
| 35. | Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC, Huang CH, Lee YS, Yen TC, Hsieh SY. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol. 2012;57:584-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 36. | Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene. 2015;34:5857-5868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 168] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 37. | Smits M, Wurdinger T, van het Hof B, Drexhage JA, Geerts D, Wesseling P, Noske DP, Vandertop WP, de Vries HE, Reijerkerk A. Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB J. 2012;26:2639-2647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 38. | Bandopadhyay M, Chakravarty R. An insight into interaction of cell cycle regulating miRNAs and Hepatitis B virus X protein. RNA & DISEASE. 2015;2. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 39. | Guo L, Zhang Y, Zhang L, Huang F, Li J, Wang S. MicroRNAs, TGF-β signaling, and the inflammatory microenvironment in cancer. Tumour Biol. 2016;37:115-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 40. | Yang XW, Zhang LJ, Huang XH, Chen LZ, Su Q, Zeng WT, Li W, Wang Q. miR-145 suppresses cell invasion in hepatocellular carcinoma cells: miR-145 targets ADAM17. Hepatol Res. 2014;44:551-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 41. | Huang B, Li H, Huang L, Luo C, Zhang Y. Clinical significance of microRNA 138 and cyclin D3 in hepatocellular carcinoma. J Surg Res. 2015;193:718-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 42. | Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, Wu F. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int. 2012;32:752-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 170] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 43. | Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 449] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 44. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 45. | Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, Feldstein AE. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:e113651. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 218] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 46. | Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402-4407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 944] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 47. | Sharma T, Hamilton R, Mandal CC. miR-214: a potential biomarker and therapeutic for different cancers. Future Oncol. 2015;11:349-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 48. | Roy S, Benz F, Luedde T, Roderburg C. The role of miRNAs in the regulation of inflammatory processes during hepatofibrogenesis. Hepatobiliary Surg Nutr. 2015;4:24-33. [PubMed] |
| 49. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 190] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 50. | Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 306] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 51. | Mandal CC, Ghosh-Choudhury T, Dey N, Choudhury GG, Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. 2012;33:1897-1908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 52. | Dey N, Das F, Mariappan MM, Mandal CC, Ghosh-Choudhury N, Kasinath BS, Choudhury GG. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem. 2011;286:25586-25603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 194] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 53. | Lok AS, Seeff LB, Morgan TR, di Bisceglie AM, Sterling RK, Curto TM, Everson GT, Lindsay KL, Lee WM, Bonkovsky HL. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136:138-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 452] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 54. | Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, Drebber U, Rockstroh J, Sauerbruch T. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 55. | Guo CJ, Pan Q, Cheng T, Jiang B, Chen GY, Li DG. Changes in microRNAs associated with hepatic stellate cell activation status identify signaling pathways. FEBS J. 2009;276:5163-5176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 56. | Shrivastava S, Steele R, Ray R, Ray RB. MicroRNAs: Role in Hepatitis C Virus pathogenesis. Genes Dis. 2015;2:35-45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 57. | Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45:355-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 58. | Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, Run W, Tian L, Jia X, Gao Y. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond). 2011;120:183-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 183] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 59. | Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 284] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 60. | Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: a retrospective cohort study. BMJ Open. 2012;2:e000825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 188] [Article Influence: 14.5] [Reference Citation Analysis (0)] |