Published online May 7, 2017. doi: 10.3748/wjg.v23.i17.3022
Peer-review started: October 28, 2016
First decision: March 3, 2017
Revised: March 19, 2017
Accepted: April 12, 2017
Article in press: April 12, 2017
Published online: May 7, 2017
Processing time: 193 Days and 13.6 Hours
Colonic polyps may arise from BRAF inhibitor treatment of melanoma, possibly due to paradoxical activation of the mitogen-activated protein (MAP)-kinase pathway. In an alternative evidence based scenario, tubular colonic adenomas with APC gene mutations have also been identified in the context of BRAF inhibitor treatment, in the absence of mutations of MAPK genes. A minority of colorectal cancers develop by an alternative “serrated polyp pathway”. This article postulates a novel hypothesis, that the established phenotypic and molecular characteristics of serrated colonic polyps/CRC offer an intriguing insight into the pathobiology of BRAF inhibitor induced colonic polyps. Serrated polyps are characterized by a CpG island methylation phenotype, MLH1 silencing and cellular senescence. They also have BRAF mutations. The contention is that BRAF inhibitor induced polyps mimic the afore-described histology and molecular features of serrated polyps with the exception that instead of the presence of BRAF mutations they induce C-RAF homodimers and B-RAF: C-RAF heterodimers.
Core tip: In this article, we focus on BRAF inhibitors, and their relationship to colonic polyps. As is already known, colonic polyps may arise from BRAF inhibitor treatment of melanoma, possibly due to paradoxical activation of the mitogen-activated protein-kinase pathway. In this article, we postulate a novel hypothesis, that the established phenotypic and molecular characteristics of serrated colonic polyps offer an intriguing insight into the pathobiology of BRAF inhibitor induced colonic polyps.
- Citation: Kelleher FC, Callaghan G, Gallagher C, O’Sullivan H. BRAF inhibitor treatment of melanoma causing colonic polyps: An alternative hypothesis. World J Gastroenterol 2017; 23(17): 3022-3029
- URL: https://www.wjgnet.com/1007-9327/full/v23/i17/3022.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i17.3022
Classically, 70%-75% of colorectal cancers (CRCs) arise through the adenoma-carcinoma sequence[1]. Inactivation of APC is the initial molecular change with subsequent alterations including KRAS mutation, Chr. 18q loss (DCC and DPC4) and subsequent inactivation of TP53[2,3]. An alternative route to colorectal cancer development is the “serrated polyp pathway”. This is characterized by BRAF mutations, a CpG island methylation phenotype (CIMP), and cellular senescence. This distinct tumor subgroup accounts for 7.5% of all CRC and 17.5% of proximal CRC[4-6].
Of all cases of metastatic CRC, 10% have BRAF mutations, with 95% of BRAF missense mutations causing amino acid substitutions at V600 in the expressed protein[7,8]. The hypothesis is that BRAF inhibitor treatment of melanoma will not cause “serrated polyp pathway” lesions per say, but by molecular mimicry, creates lesions with the clinical and molecular features of serrated lesions. The only difference is that these polyps will not have BRAF mutations but C-RAF homodimers and C-RAF: B-RAF heterodimers.
At the time of writing this manuscript a publication emerged evaluating multiple gastrointestinal polyps in patients treated with BRAF inhibitors[9]. Fourteen patients treated with BRAF inhibitors had endoscopic assessment for polyps. All patients that were treated with BRAF inhibitors for greater than 2 years, and who were in excess of 40 years of age, had colonic tubular adenomas. Hyperplastic polyps were also identified and the temporal evolution of polyps was suggestive of a causal association with BRAF inhibition. Next generation sequencing of the polyps did not identify mutations within MAPK pathway genes but did identify APC mutations in all tubular adenomas. This was most commonly a truncating mutation in the β-catenin binding domain (R1450X). In an Apc Min +/- mouse model there was an increased number of polyps compared to controls (20.8 vs 12.8, P = 0.016) respectively.
Altered stochastic relations in RAF dimers and epigenetic changes form an important part of our contention that some BRAF inhibitor induced polyps arise via the serrated poly pathway. Epigenetics affects gene expression without altering the DNA nucleotide sequence. Next generation sequencing would fail to detect such changes. In addition, nuclear β-catenin was perhaps unexpectedly not detected in the human colonic polyps. The contention is that these findings of a possible role for the classical adeno- to carcinoma sequence does not exclude the possibility that corruption, or molecular mimicry of the serrated polyp pathway may account for some BRAF inhibitor induced polyps.
The “serrated polyp pathway” is vicariously informative of a subgroup of colorectal cancers in which MAP-kinase activation is important, as exemplified by their molecular signature with characteristic BRAF mutants. This informs of a molecular circumstance in which RAF is dysregulated in colonic polyps. Paradoxical BRAF activation and upregulation of MAP-kinase signaling, is due to RAF inhibitors trans activating RAF dimers with increased ERK signaling in cells, which are BRAF wild-type[10]. There is preclinical evidence in other tumor types that RAF inhibitors increase MAP kinase pathway activity by inducing C-RAF heterodimers or B-RAF homodimers[10-12].
Clinically this molecular phenomenon is only brought to phenotypic manifestation in specific circumstances. Most phenomenon described so far have been in the context of pre-existing “primed NRAS” mutations in keratinocytes causing cutaneous squamous cell carcinomas, or the interesting case of progression of a RAS-mutant leukemia during treatment with a RAF inhibitor[13]. A case has also been described of a new RAS-mutant pancreatic adenocarcinoma in a patient receiving combined BRAF and MEK inhibitor treatment for metastatic melanoma[14]. The first described premalignant colonic adenomas and gastric polyps associated with inhibition of BRAF were reported in 2012[15]. Four of eight patients with V600E mutant metastatic melanoma on BRAF inhibitor treatment greater than 2 years had a colonoscopy. Three of the four patients were found to have multiple colonic adenomas and two had hyperplastic polyps. One of these patients had a negative colonoscopy five months prior to initiation of Vemurafenib, but at colonoscopy four colonic adenomas and one hyperplastic polyp were identified.
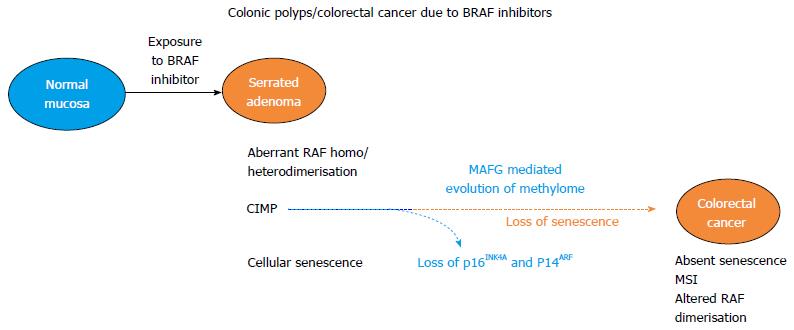
The postulate in this article is that colonic polyps may arise due to BRAF inhibitors because of altered gene expression due to epigenetic dysregulation of the methylome. Implicated genes need to be further delineated. In a simple comparator example the tumor suppressor, and RAS domain family member, RASSF1A undergoes hypermethylation in thyroid cancer with an activating mutation of BRAF gene[16]. RASSF1A is also methylated in CIMP CRC and its methylation is correlated with CRC liver metastasis[17]. Also unlike the static pre-primed RAS mutant models the CpG island methylated gene profile is one of temporal evolution, mediated by BRAF and changes with progression along the serrated polyp pathway. An adenoma to carcinoma progression model with tumors arising in the context of BRAF inhibitor induction is illustrated in Figure 1.
The World Health Organization classifies serrated polyps into 3 subtypes: hyperplastic polyps, sessile serrated adenomas, and traditional serrated adenomas. The terms serrated polyps and serrated adenomas are synonymous. Serrated CRC simply is a histologic descriptor of a sawtooth-like infolding of the intestinal lesion epithelium. In one series they accounted for 30% of colorectal cancers, and they arise from serrated polyps via the “serrated polyp pathway”[18]. Serrated tumors can also be sub-categorized using molecular descriptors. These are: (1) KRAS mutant, CIMP-low, MSS/MSI-low (microsatellite stable/microsatellite instability-low); (2) BRAF mutant, CIMP-high, MSI-high; and (3) BRAF mutant, CIMP-low, MSS/MSI-low.
Silencing of MLH1 by CpG island promoter methylation causes microsatellite instability and a hyper mutable phenotype, as it is a mismatch repair gene. Serrated polyps have a co-association with gastric metaplasia. Using the aberrant CpG island methylation phenotype as a categorical arbiter, colorectal cancers can be sub-divided into CIMP-high (CIMP-1), CIMP-low (CIMP-2), and CIMP negative (CIMP-3)[19,20]. The BRAF mutation, MLH1 methylation and CpG island methylation phenotype describes an aggressive subgroup of colorectal cancer.
A study led by Fred Hutchinson Cancer Research Center, WA, assessed the BRAF mutant, MLH1 silenced, CIMP, molecular signature in a series of polyps. Participants underwent an initial index colonoscopy for any indication. There were 580 conventional adenomas and 419 serrated lesions identified. CIMP methylation was determined by assessing methylation status of the following genes: IGF2, NEUROG1, CACNA1G, RUNX3, and SOCS1[21]. The prevalence of the mutations leading to BRAF V600E, MLH1 methylation and CIMP in the adenomas was < 1%. In contrast, 55% of serrated lesions had mutations leading to BRAF V600E, 5% had MLH1 methylation and 26% were CIMP-high. The highest prevalence of these markers occurred within the sessile-serrated polyps. Sessile serrated polyps were BRAF mutant in 68%, MLH1 methylated 11%, and CIMP-high 49%. In a series of sporadic classical serrated adenomas the promoter of SLC5A8 was methylated in 82.5%[22]. The extent of widespread aberrant CpG island methylation increased with histological progression of serrated adenomas. Methylation of genes encoding p14, p16, MGMT, FHIT, and TIMP3 were found to be important tumorigenic steps in the serrated neoplastic pathway. A longitudinal study, which evaluated patients with sessile serrated polyps, demonstrated that 12.5% developed colorectal cancer within 5 years[23]. Metastatic BRAF mutant colorectal cancer have a particularly poor prognosis with a median overall survival of 20 mo compared to 47 mo for those which are BRAF wildtype[24].
A simple analogy remains however with previously primed NRAS mutant colonic epithelium but the frequency of this mutation is low. An inverse relationship also exists for KRAS and BRAF mutations in serrated adenomas[25]. Though the theme of histopathology and molecular mimicry of serrated colonic lesions suffuses this hypothesis article, BRAF inhibitors may also be relevant to progression of KRAS mutant adenomas to carcinoma by removal of the senescence barrier to developing cancer. Expression of oncogenic K-rasG12D in mice induces serrated hyperplasia with overexpression of p16ink4a and induction of senescence[26]. When Ink4a/Arf is deleted in K-rasG12D expressing mice senescence is prevented with consequent invasion and metastasis as well as molecular and morphologic changes consistent with KRAS mutated serrated tumors. The reader is reminded that CDKN2A the gene that encodes p14 and p16 is a participant gene of the CpG island methylated phenotype.
This paper postulates that colonic polyps arising from BRAF inhibitor treatment are due to paradoxical MAP-kinase upregulation. However, the exact molecular mechanism of how this causes colonic polyps has been elusive. A conceptual advance may be inferred from findings by investigators at Howard Hughes Medical Institute and the University of Massachusetts, MA[27]. Aberrant CpG island methylation of MLH1, was selected as a prototypical epigenetic gene dysregulation event, with silencing of MLH1 in CIMP-1 colorectal cancer. This gene is a member of the CIMP gene spectrum, which characterizes a subset of CRC. Using an RNAi screen the transcriptional repressor MAFG (v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G) was established as a decisive requirement for MLH1 silencing and establishing the CpG island methylation phenotype of BRAF (V600E) colorectal cancer. In BRAF mutant colorectal cancer cell lines MAFG bound to the promoter of MLH1 as well as other CIMP genes with recruitment of a co-repressor complex including its’ heterodimeric partner BACH1, the DNA methyltransferase DNMT3B and the chromatin remodelling factor CHD8. This caused hypermethylation and transcriptional silencing. In a BRAF mutant cell line, treatment with a BRAF inhibitor decreased MAFG protein. Not all genes of CIMP co-associate with mutant BRAF in individual serrated colonic lesions. An inference could be drawn that in BRAF mutant sporadic colonic lesions with co-association of CIMP, BRAF inhibitors could de-repress genes within the CIMP phenotype in a binary way.
There is the alternative scenario of patients developing iatrogenic colonic polyps from exposure to BRAF inhibitors indicated for treating melanoma. In BRAF wild type colonic epithelium, BRAF inhibitors could, through C-RAF homodimers or C-RAF: B-RAF heterodimers, upregulate BRAF-MEK-ERK activity. Upregulation of this pathway leads to ERK1 phosphorylation of S124 of MAFG with increased MAFG stability and protein levels. There is consequential MAFG binding to DNA with transcriptional silencing of genes possibly replicating the CIMP gene signature. MAFG levels are also increased by prevention of polyubiquitation and proteosomal destruction.
Representative genes of CIMP gene promoters in CRC include DAPK1, PRDM2, AOX1, CACNA1G, CHFR, EFEMP1, HAND1, IRF8, LOX and p16INK4A. PAT-ChIP analysis of MAFG binding to these 10 representative CIMP genes’ promoters, and adjacent normal tissue, in BRAF mutant colorectal cancers demonstrated that MAFG was substantially enriched compared to matched normal tissue. In another study DNA methylation of 16 CpG islands in 904 colorectal cancers was quantitated[28]. The 5 markers (SOCS1, IGF2, NEUROG1, RUNX3 and CACNA1G), CDKN2A, MINT31, CRABP1, MLH1, p14 and WRN usually clustered on unsupervised hierarchical clustering analysis. These co clustered with microsatellite instability and mutant BRAF. Multivariate logistic regression analysis found CIMP-high to be independently associated with proximal tumor location, older age, MSI- high, poor differentiation, BRAF mutation, and inversely with CTNNB1 and LINE-1 hypo-methylation. p53-negativity, signet ring cells and mucinous histology only co-associated with CIMP on univariate analysis. BRAF inhibitors paradoxically trans activate RAF dimers and this is the mechanism for paradoxical MAP-kinase up regulation in BRAF wild type cells. This is mediated by drug binding to the ATP-binding site of one kinase of the RAF dimer C-RAF: C-RAF or CRAF: BRAF. Inhibition of one promoter leads to transactivation of the drug-free promoter[10].
Epigenetic alterations are heritable changes in gene expression in the absence of changes in the DNA sequence. They usually are secondary to methylation of DNA in gene promoter regions or modifications in histone acetylation. Aberrant DNA methylation can consist of either site-specific hypermethylation of DNA or global hypomethylation of DNA. One example of hypomethylation in CRC is the gene LINE-1. LINE-1 hypomethylation in CRC is associated with inferior survival, with a hazard ratio of 2.45 in MSI-high cancers[29]. Widespread hypermethylation of CpG islands is described as the CpG island methylation phenotype. CDKN2A is a member gene of the CpG island methylation phenotype of the serrated pathway and its transcription is silenced in a subset of serrated polyps. In an interesting comparator, CDKN2A is also silenced in a minority of melanomas with loss of p16INK4A causing de-repression of cyclin D - CDK4/6. Selected panel genes of the CIMP profile in CRC are detailed in Table 1 below, however these only represent a small proportion of the probable 100 s-1000 s of methylated CIMP genes. This CIMP-gene panel in Table 1 was supplemented by selected other genes in one study in 2013[30]. Additional genes of potential relevance identified then included RASSF1A, APC, PTEN and TWIST1. The methylome of 100s to 1000s of genes remains to be further interrogated to establish the genes of greatest importance in BRAF inhibitor induced colonic polyps.
| MINT1 | BRAF | P14 | RASSF2 |
| MINT2 | TIMP3 | SOCS1 | HAND1 |
| MINT12 | RIZ1 | LOX | CACNA10 |
| MINT17 | HIC1 | WRN | MGMT |
| MINT25 | IGF2 | ADAMTS1 | FBN2 |
| MINT27 | IGFB3 | EDIL3 | THBD |
| MINT31 | CHFR | ELM01 | UCHL1 |
| P16 | NEUROG1 | DUSP26 | STOX2 |
| MLH1 | CRABP1 | RUNX3 |
BRAF inhibitor treatment decreases MAFG protein in BRAF mutant CRC cells, whereas BRAF inhibitor treatment has been demonstrated to paradoxically increase MAP-kinase activity in BRAF wild type keratinocytes with pre-primed RAS mutations. Therefore the effects are context dependent but ultimately will alter the spectrum of repressed and expressed genes, in some circumstances epigenetically.
Some genes are deserving of some further detail however acknowledging the limitations of current knowledge. The INK4a/ARF locus on Chromosome 9p21 encodes both p16INK4A and p14ARF. Despite these tumor suppressor genes sharing exons, their encoded proteins do not have amino acid homology. This is because of differences in their reading frames. Progression of sessile serrated adenomas to CRC is restrained by p16INK4A mediated senescence as well as by p53, a downstream effector of p14ARF. p16INK4A and p14ARF are frequently inactivated in CRC by aberrant promoter methylation of their encoding gene CDKN2A[31]. In a tumor progression model paradoxical MAP-kinase mediated loss of oncogene induced senescence was found to be attributable to functional loss of p16INK4A. Removal of the senescence barrier permits progression to colon cancer.
Genes specifically methylated by mutant BRAF in CRC have been identified32. These include forkhead box (FOX) transcription factors that associate with the PI3 kinase pathway, smoothened (part of the Hedgehog signaling pathway) and MLH1, as is illustrated in Table 2 below. The repressed expression of FOXD3 in colorectal tumors has been attributed also to promoter hypermethylation[32]. The average CRC methylome comprises hundreds to thousands of genes but identification of the oncogenic drivers is difficult. p16INK4a is up and down regulated in a context dependent manner, with implications for loss of senescence, and would appear a likely culprit indirect repressor of FOXD3. In mammalian cells p16INK4A inhibits activity of cyclin D-CDK4/6 complexes. When active these complexes phosphorylate and regulate the mammalian Forkhead Box transcription factor (FOXM1) which itself inhibits senescence. FOXM1 promotes the growth and metastasis of colon cancer cells in orthotopic mouse models[33].
| BRAF mutations specific promoter | Associated genes |
| PI3 kinase pathway | FOXB1; FOXB2, FOXD3, CCND1, GSK3A |
| Insulin/IGF pathway | FOXB1; FOXB2, FOXD3; GSK3A |
| Hedgehog signaling pathway | SMO; GSK3A; CREBBP |
| Wnt signaling | NKD2; GNG4; CCND1; GSK3A; CREBBP |
| Transcription-regulation by bZIP transcription factor | CREBBP, MTERF; TAF7 |
In a study of CRC cell lines for all CpG rich regions, 389 had co-occurrence of mutant BRAF and CIMP, 369 mutant BRAF alone and 360 CIMP alone[32]. When filtered using H3K27me binding within embryonic stem cells it was found that 96 had co-occurrence of mutant BRAF and CIMP, 90 had mutant BRAF alone and 112 CIMP alone. Pathways enriched for BRAF mutation associated promoter methylation once ES cell K3K27me binding promoter regions were excluded, and are detailed in Table 2. Considering just one selected gene, Cyclin D1 is commonly over activated in CRC. Preclinical studies found that Aspirin likely inhibits Cyclin D1/CDK4 in CRC cells through the p38 MAP-kinase pathway. This inhibition causes NF-κB mediated induction of nucleolar translocation of RelA (p65) -a component of NF-κB-, as well as apoptosis[34]. BRAF mutations correlate with Cyclin D1 overexpression in metastatic colorectal cancer[35].
Hayflick and Moorhead first observed cellular senescence in 1961 in experiments where serial in vitro cultivation of human fibroblasts caused then to enter an irreversible state of arrested growth[36]. The eponymously named Hayflick factors that characterize senescence, record a cells or tissues proliferative history. These include telomere shortening, de-repression of the INK4a/ARF locus and accumulation of DNA damage. Senescence and p16INK4A/ p14ARF are of particular interest as senescence is a characteristic feature of serrated polyps. As previously detailed, silencing of CDKN2A the gene encoding these tumor suppressors is part of the CpG island methylation phenotype. In a study of BRAF mutated colonic serrated lesions, p16Ink4a was upregulated in premalignant lesions only to be later lost in invasive serrated carcinomas[37]. Progression of the malignant phenotype in serrated lesions was accompanied by increased methylation of the CDKN2A gene promoter. Simply stated, progression from adenoma to BRAF mutant CRC partly involves epigenetic loss of senescence. As a physiologic comparator, in development epigenetic regulators of the Polycomb family are partially responsible for low expression levels of p16INK4A and ARF[38,39].
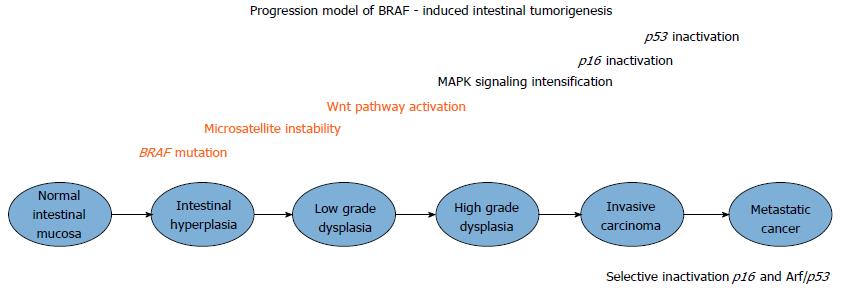
An experimental mouse model of BrafV600E induced intestinal carcinogenesis has been created and described, using a conditional Cre activated Braf knochin allele[40]. The inferred findings arising from this murine model are illustrated in Figure 2. Murine BrafV637E in exon 18 is orthologous to human BRAFV600E exon 15 mutations. One hundred percent of Villin-Cre; BrafLSL-V673/+ mice developed crypt hyperplasia (restricted to mid-upper intestinal crypts) without apoptosis. In a phenomenon determined by increasing age, hyperplasia progressed to dysplasia, and typical serrated adenomas but not sessile serrated adenomas. This may be because these mice models have a predilection for the development of small intestinal rather than colonic polyps, and sessile serrated adenomas usually only occur in the colon. Dysplasia progressed to carcinoma in 16% of BrafV637E knockin mice. Intercross experiments to generate progeny were also undertaken using BrafLSL-V637E/+;p53LSL-R172H/+ and Vil-Cre; BrafLSL-V637E/+ mice. These were performed because of the long latency for cancer formation and to assess the functional consequences of p53 inactivation. The arising progeny were Villin-Cre; BrafV637E/+; p53LSL-R172H/+. No comparator difference was found in the proportion of such progenitor mice developing serrated adenomas but invasive cancer were more frequent with the average number of cancers 5.2 times more frequent in the BrafLSL-V637E/+;p53LSL-R172H/+ group (P = 0.007; Mann-Whitney rank sum test). In the interval from 10-20 mo, 56% of compound mutant mice developed cancer. The inference is that p53 does not have an impact on early stages of BRAFV637E induced tumorigenesis, but is important in late stages including invasiveness. In the molecular progression from dysplasia to adenoma with subsequent carcinoma, selective pressure for p53 inactivation developed at more advanced stages of tumor evolution with p16 inactivation promoting advanced phases of BrafV637E induced intestinal tumorigenesis. P16Ink4a compound homozygous mutant mice had a 6.4-fold greater increase in carcinoma compared to mice with p16Ink4a expression. The serrated cancer progression model discovered by Rad and colleagues is illustrated in Figure 2.
The described genetic progression model of BrafV600E-intestinal tumorigenesis in mice demonstrates selective pressure for inactivation of the p16/Rb and Arf/p53 pathways late in the progression path. A paradoxical increase in MAP-kinase activity would through C-RAF: B-RAF heterodimers and C-RAF homodimerisation cause MAFG mediated silencing of p16INK4A expression. This would have the effect of loss of senescence in later stages of progression from colonic polyps to CRC.
A comprehensive compilation of a series of patients on BRAF inhibitor treatment for melanoma in whom colonic polyps arise is mandated. This will permit histological characterization as to whether these polyps are serrated or adenomatous. Bi-sulfite sequencing analysis of genes that are proposed to be epigenetically silenced by CpG island promoter methylation should inform on the relative merits of the epigentic component of the iatrogenic disease model. DNA and RNA of the selected genes of interest are suggested. Lastly RNAi screening to assess the level of MAFG in these polyps, itself a silencer of p16INK4A should provide evidence supporting the senescent tenet of the hypothesis.
The prospect of efficacious medical treatment of colorectal cancers arising from BRAF inhibitor induced polyps is appealing. Endoscopic removal of colonic polyps is the optimal initial intervention. However the emergent molecular biology in this theory, which needs future evidential substantiation, suggests some molecular treatment approaches. One potential treatment modality in development is ERK inhibitors, which can decrease ERK1 phosphorylation of MAFG. The exciting recent findings of Amaravadi and colleagues suggest a classical adeno-carcinoma cause for gastrointestinal polyps arising from BRAF inhibitor treatment. The postulated theory of corrupted molecular mimicry of the serrated polyp pathway detailed above is supplementary rather than contradictory and deserves experimental investigation.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Ireland
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Sammour T S- Editor: Qi Y L- Editor: A E- Editor: Zhang FF
| 1. | Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159-170. [PubMed] |
| 2. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [PubMed] |
| 3. | Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1163] [Cited by in RCA: 1269] [Article Influence: 90.6] [Reference Citation Analysis (1)] |
| 4. | Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 955] [Cited by in RCA: 1004] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 5. | Mäkinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 208] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 6. | Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol. 2009;4:343-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 7. | Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. New England J Med. 2009;361:98-99. [RCA] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 405] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 8. | Ikenoue T, Hikiba Y, Kanai F, Tanaka Y, Imamura J, Imamura T, Ohta M, Ijichi H, Tateishi K, Kawakami T. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003;63:8132-8137. [PubMed] |
| 9. | Amaravadi RK, Hamilton KE, Ma X, Piao S, Portillo AD, Nathanson KL, Carlino MS, Long GV, Puzanov I, Xu X. Multiple Gastrointestinal Polyps in Patients Treated with BRAF Inhibitors. Clin Cancer Res. 2015;21:5215-5221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1536] [Cited by in RCA: 1429] [Article Influence: 95.3] [Reference Citation Analysis (0)] |
| 11. | Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1208] [Cited by in RCA: 1260] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 12. | Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1241] [Cited by in RCA: 1182] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 13. | Callahan MK, Rampal R, Harding JJ, Klimek VM, Chung YR, Merghoub T, Wolchok JD, Solit DB, Rosen N, Abdel-Wahab O. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N Engl J Med. 2012;367:2316-2321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 190] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 14. | Carlino MS, Kwan V, Miller DK, Saunders CA, Yip D, Nagrial AM, Tomlinson J, Grimmond SM, Scolyer RA, Kefford RF. New RAS-mutant pancreatic adenocarcinoma with combined BRAF and MEK inhibition for metastatic melanoma. J Clin Oncol. 2015;33:e52-e56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Chapman P, Sepulvede AR. Development of colonic adenomas and gastric poyps in BRAF mutant melanoma patients treated with Vemurafenib. Society for Melanoma Research Congress, Los Angeles, CA, 2012. . |
| 16. | Xing M, Cohen Y, Mambo E, Tallini G, Udelsman R, Ladenson PW, Sidransky D. Early occurrence of RASSF1A hypermethylation and its mutual exclusion with BRAF mutation in thyroid tumorigenesis. Cancer Res. 2004;64:1664-1668. [PubMed] |
| 17. | Tommasi S, Pinto R, Petriella D, Pilato B, Lacalamita R, Santini D, Zito F, Colucci G, Paradiso A, Silvestris N. Oncosuppressor methylation: a possible key role in colon metastatic progression. J Cell Physiol. 2011;226:1934-1939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, Goldblum JR, Guillem JG, Kahi CJ, Kalady MF. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315-1329; quiz 1314, 1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 828] [Article Influence: 63.7] [Reference Citation Analysis (0)] |
| 19. | Kaneda A, Yagi K. Two groups of DNA methylation markers to classify colorectal cancer into three epigenotypes. Cancer Sci. 2011;102:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 20. | Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 187] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 21. | Burnett-Hartman AN, Newcomb PA, Potter JD, Passarelli MN, Phipps AI, Wurscher MA, Grady WM, Zhu LC, Upton MP, Makar KW. Genomic aberrations occurring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer Res. 2013;73:2863-2872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 22. | Dong SM, Lee EJ, Jeon ES, Park CK, Kim KM. Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol. 2005;18:170-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 96] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Lu FI, van Niekerk de W, Owen D, Tha SP, Turbin DA, Webber DL. Longitudinal outcome study of sessile serrated adenomas of the colorectum: an increased risk for subsequent right-sided colorectal carcinoma. Am J Surg Pathol. 2010;34:927-934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 136] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 24. | Yaeger R, Cercek A, Chou JF, Sylvester BE, Kemeny NE, Hechtman JF, Ladanyi M, Rosen N, Weiser MR, Capanu M. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer. 2014;120:2316-2324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 167] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 25. | Chan TL, Zhao W, Leung SY, Yuen ST. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63:4878-4881. [PubMed] |
| 26. | Bennecke M, Kriegl L, Bajbouj M, Retzlaff K, Robine S, Jung A, Arkan MC, Kirchner T, Greten FR. Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell. 2010;18:135-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 27. | Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG Island Methylator phenotype. Mol Cell. 2014;55:904-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 177] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 28. | Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One. 2008;3:e3698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 262] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 29. | Inamura K, Yamauchi M, Nishihara R, Lochhead P, Qian ZR, Kuchiba A, Kim SA, Mima K, Sukawa Y, Jung S. Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis. J Natl Cancer Inst. 2014;106:pii. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 30. | Ashktorab H, Rahi H, Wansley D, Varma S, Shokrani B, Lee E, Daremipouran M, Laiyemo A, Goel A, Carethers JM. Toward a comprehensive and systematic methylome signature in colorectal cancers. Epigenetics. 2013;8:807-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 31. | Martin V, Jørgensen HF, Chaubert AS, Berger J, Barr H, Shaw P, Bird A, Chaubert P. MBD2-mediated transcriptional repression of the p14ARF tumor suppressor gene in human colon cancer cells. Pathobiology. 2008;75:281-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 32. | van Roon EH, Boot A, Dihal AA, Ernst RF, van Wezel T, Morreau H, Boer JM. BRAF mutation-specific promoter methylation of FOX genes in colorectal cancer. Clin Epigenetics. 2013;5:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Li D, Wei P, Peng Z, Huang C, Tang H, Jia Z, Cui J, Le X, Huang S, Xie K. The critical role of dysregulated FOXM1-PLAUR signaling in human colon cancer progression and metastasis. Clin Cancer Res. 2013;19:62-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 452] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 34. | Thoms HC, Dunlop MG, Stark LA. p38-mediated inactivation of cyclin D1/cyclin-dependent kinase 4 stimulates nucleolar translocation of RelA and apoptosis in colorectal cancer cells. Cancer Res. 2007;67:1660-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 35. | Saridaki Z, Papadatos-Pastos D, Tzardi M, Mavroudis D, Bairaktari E, Arvanity H, Stathopoulos E, Georgoulias V, Souglakos J. BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients’ outcome. Br J Cancer. 2010;102:1762-1768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 36. | Hayflick l, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585-621. [PubMed] |
| 37. | Kriegl L, Neumann J, Vieth M, Greten FR, Reu S, Jung A, Kirchner T. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol. 2011;24:1015-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 38. | Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Mönch K, Minucci S, Porse BT, Marine JC. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 700] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 39. | Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1208] [Cited by in RCA: 1242] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 40. | Rad R, Cadiñanos J, Rad L, Varela I, Strong A, Kriegl L, Constantino-Casas F, Eser S, Hieber M, Seidler B. A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell. 2013;24:15-29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 171] [Article Influence: 14.3] [Reference Citation Analysis (0)] |