Published online Apr 14, 2017. doi: 10.3748/wjg.v23.i14.2585
Peer-review started: January 1, 2017
First decision: February 9, 2017
Revised: February 19, 2017
Accepted: March 2, 2017
Article in press: March 2, 2017
Published online: April 14, 2017
Processing time: 104 Days and 20.7 Hours
To evaluate PIK3CA gene mutational status in Northwest Chinese esophageal squamous cell carcinoma (ESCC) patients, and examine the associations of PIK3CA gene mutations with clinicopathological characteristics and clinical outcome.
A total of 210 patients with ESCC who underwent curative resection were enrolled in this study. Pyrosequencing was applied to investigate mutations in exons 9 and 20 of PIK3CA gene in 210 Northwest Chinese ESCCs. The associations of PIK3CA gene mutations with clinicopathological characteristics and clinical outcome were examined.
PIK3CA gene mutations in exon 9 were detected in 48 cases (22.9%) of a non-biased database of 210 curatively resected Northwest Chinese ESCCs. PIK3CA gene mutations were not associated with sex, tobacco use, alcohol use, tumor location, stage, or local recurrence. When compared with wild-type PIK3CA gene cases, patients with PIK3CA gene mutations in exons 9 experienced significantly better disease-free survival and overall survival rates.
The results of this study suggest that PIK3CA gene mutations could act as a prognostic biomarker in Northwest Chinese ESCC patients.
Core tip:PIK3CA gene mutations have been associated with various prognoses in patients with different cancers. However, no large-scale study has examined the prognostic impact of PIK3CA gene mutations in Northwest Chinese esophageal squamous cell carcinoma (ESCC). In this study, we quantified PIK3CA gene mutations via pyrosequencing technology using a non-biased database of 210 curatively resected ESCCs. It was found that PIK3CA gene mutations in Northwest Chinese ESCC are associated with favorable prognoses. It has been suggested that PIK3CA gene mutational status can have a potential role as a prognostic biomarker for ESCC.
- Citation: Liu SY, Chen W, Chughtai EA, Qiao Z, Jiang JT, Li SM, Zhang W, Zhang J. PIK3CA gene mutations in Northwest Chinese esophageal squamous cell carcinoma. World J Gastroenterol 2017; 23(14): 2585-2591
- URL: https://www.wjgnet.com/1007-9327/full/v23/i14/2585.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i14.2585
Esophageal squamous cell carcinoma (ESCC) is a major histologic type of esophageal cancer that is one of the most aggressive malignant tumors worldwide, especially in East Asian countries, and accounts for most esophageal malignancies in China and Japan[1,2]. As one of the most commonly diagnosed cancers among men in China, the estimated number of new cases of esophageal cancer was 291238 in 2011, while the numbers of deaths was 218957 in the same year[3]; by 2015, these numbers had increased to 477900 and 375000, respectively[4]. Both the incidence and mortality rates were higher in rural areas than in urban areas. Despite the continuing development of cancer multimodality therapies, including surgery, radiotherapy, and chemotherapy, the prognosis of ESCC patients remains poor, even for those who undergo complete resection of their carcinomas[5].
Phosphatidylinositol 3-kinases (PI3Ks) are expressed as heterodimers of p110 catalytic subunits and p85 regulatory subunits that interact with phosphatidylinositol-3-phosphate at the membrane and catalyze the phosphorylation of protein kinase B (PKB, also known as AKT), which activates the downstream signaling pathway[6]. Activation of the PI3K/AKT signaling pathway plays an important role in the development of a variety of human carcinomas[7]. The catalytic subunits of PI3K are encoded by three genes (α, β, γ), with p110α subunit (PIK3CA) amplification being reported in a number of different tumor types. The mutant PIK3CA gene stimulates the AKT pathway and promotes cell growth and invasion in various types of human cancer[8,9] (Samuels, 2004 #620; Samuels, 2005 #638), including lung, breast, gastric, and colon[10-17].
PIK3CA gene mutations have also been detected in Japanese and Korean ESCCs[18,19]. Although independently associated with a poor prognosis in Chinese breast cancer patients[13], it was found to be associated with improved outcome in breast cancer patients in the United States[20]; this seeming contradiction requires an intensive study of this gene in future research. In addition, PIK3CA gene mutations and their prognostic role in Chinese ESCC patients have been rarely reported. We therefore quantified PIK3CA gene mutations in 210 samples of curatively resected ESCCs using pyrosequencing, and examined the prognostic significance of PIK3CA gene mutations in Northwest Chinese ESCC patients.
A total of 210 patients with ESCC who underwent curative resection at the Second Affiliated Hospital of Xi’an Jiaotong University between 2009 and 2015 were enrolled in this study. Patients were observed at 1 to 3 mo intervals until either death or December 30, 2015. Tumor staging was carried out according to the 7th American Joint Committee Cancer Staging Manual[21]. Disease-free survival was defined as the length of time after surgical treatment of the cancer during which the patient survived with no sign of cancer recurrence. Cancer-specific survival was defined as the time between the date of operation and the date of death, which was confirmed to be attributable to ESCC. Overall survival was defined as the time between the date of the operation and the date of death. Written consent was obtained from each subject and the study procedures were approved by the ethical committees of the Second Affiliated Hospital of Xi’an Jiaotong University.
Genomic DNA was extracted from 210 paraffin-embedded tissue specimens of surgically resected esophageal cancers using the QIAamp DNA Mini kit (Qiagen, Hilgen, Germany) according to the manufacturer’s instructions.
Polymerase chain reaction (PCR) amplifications targeting the PIK3CA gene (exon 9 and 20) were performed. Two sets of primers (Table 1) were used for the detection of any mutation points in exons 9 and 20 of the PIK3CA gene. PCR was carried out in a total volume of 20 μL. The mixture included 1x HotStarTaq buffer, 2.0 mmol/L Mg2+, 0.2 mmol/L dNTP, 0.2 μmol/L of each primer, 1U HotStarTaq polymerase (Qiagen, Hilgen, Germany), and 1 μL template DNA. The cycling program for exon 9 was initial denaturation at 95 °C for 15 min, followed by 11 cycles at 94 °C for 20 s, 62 °C-0.5 °C per cycle for 40 s, and 72 °C for 1 min. The cycling program for exon 20 was initial denaturation at 95 °C for 15 min, followed by 27 cycles at 94 °C for 20 s, 56 °C for 30 s, and 72 °C for 1 min. The PCR products were electrophoresed on agarose gels to confirm successful amplification of the 81 (exon 9) and 74 bp (exon 20) products.
| Exon | Primers | |
| Exon 9 | Forward | 5’CAAAGCAATTTCTACACGAGATCC 3’ |
| Reverse | 5’GTAAAAACATGCTGAGATCAGCCACAT 3’ | |
| Exon 20 | Forward | 5’TGGAATGCCAGAACTACAATCTTT 3’ |
| Reverse | 5’GGTCTTTGCCTGCTGAGAGTT 3’ | |
PIK3CA pyrosequencing was carried out using the Pyro-Mark Q24 System (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Primers of PIK3CA gene (exon 9 and exon 20) for pyrosequencing are shown in Table 2.
| Exon | Primers | |
| Exon 9 RS1 | Nucleotide dispensation order | 5’ CCATAGAAAATCTTTCTCCT 3’ |
| 5’ ATCGACTACACTGACTGACTGACTGACTGACTGACTG 3’ | ||
| Exon 9 RS2 | Nucleotide dispensation order | 5’ TTCTCCTTGCTTCAGTGATTT 3’ |
| 5’ ATACACATGTCAGTCAGACTAGCTAGCTAGCTAG 3’ | ||
| Exon 9 RS3 | Nucleotide dispensation order | 5’ TAGAAAATCTTTCTCCTGCT 3’ |
| 5’ ATAGCACTGACTGACTGACTACTGACTGACTGACTG 3’ | ||
| Exon 20 RS | Nucleotide dispensation order | 5’TGGAATGCCAGAACTACAATCTTT 3’ |
| 5’GGTCTTTGCCTGCTGAGAGTT 3’ | ||
For the statistical analysis, we used GraphPad Prism 5 software (GraphPad Software, La Jolla, CA). The association between PIK3CA gene mutations and clinicopathological variables were performed using the χ2-test or Fisher’s exact probability test. All P values were two-tailed, with a P-value less than 0.01 being considered significant. Estimation of overall survival was calculated using the Kaplan-Meier method, with statistical differences analyzed via the log-rank test.
For 210 patients who had undergone curative resection of stage I to III ESCC, we examined PIK3CA gene mutations (exon 9 and exon 20) by pyrosequencing technology. In this study, PIK3CA gene mutations were only observed in exon 9 in 48 (22.9%) of 210 Northwest Chinese ESCC samples. The most common mutation of PIK3CA exon 9 was the c.1634A>C (p.E545A) mutation, which was present in 35 tumors, followed by c.1633G>A (p.E545K) in 13 tumors.
We examined whether the influence of PIK3CA gene mutations on cancer-specific survival was modified by any of the evaluated clinical, pathologic, or epidemiologic variables of the ESCCs. As a result, we found that PIK3CA gene mutations were not significantly associated with any of the evaluated characteristics of ESCCs, namely sex (male vs female), tobacco use (yes vs no), alcohol use (yes vs no), tumor location (upper, middle vs lower thoracic), preoperative treatment (yes vs no), lymph node metastasis (yes vs no), or local recurrence (yes vs no) (all P > 0.01; Table 3).
| Clinical, epidemiologic, or pathologic feature | Total, n | PIK3CA | P value | |
| Mutant | Wild-type | |||
| All cases | 210 | 48 | 162 | |
| Sex | 0.4756 | |||
| Male | 137 (65.3) | 34 (70.8) | 123 (75.9) | |
| Female | 73 (34.7) | 14 (29.2) | 39 (24.1) | |
| Tobacco use | 0.2684 | |||
| Yes | 149 (71.0) | 31 (64.6) | 118 (72.9) | |
| No | 61 (29.0) | 17 (35.4) | 44 (28.1) | |
| Alcohol use | 0.3778 | |||
| Yes | 175 (83.3) | 38 (79.2) | 137 (84.6) | |
| No | 35 (16.7) | 10 (20.8) | 25 (15.4) | |
| Preoperative treatment | 0.8467 | |||
| Yes | 28 (13.3) | 6 (12.5) | 22 (13.6) | |
| No | 182 (86.7) | 42 (87.5) | 140 (86.4) | |
| Tumor location | 0.9651 | |||
| Upper thoracic | 20 (9.5) | 5 (10.4) | 15 (9.2) | |
| Middle thoracic | 109 (51.9) | 25 (52.1) | 84 (51.9) | |
| Lower thoracic | 81 (38.6) | 18 (37.5) | 63 (38.9) | |
| Stage | 0.1641 | |||
| IA | 16 (7.6) | 3 (6.3) | 13 (8.0) | |
| IB | 20 (9.5) | 5 (10.4) | 15 (9.3) | |
| IIA | 28 (13.3) | 11 (22.9) | 17 (10.5) | |
| IIB | 44 (21.0) | 10 (20.8) | 34 (21.0) | |
| IIIA | 49 (23.3) | 12 (25.0) | 37 (22.8) | |
| IIIB | 23 (11.0) | 1 (2.1) | 22 (13.6) | |
| IIIC | 30 (14.3) | 6 (12.5) | 24 (14.8) | |
| Lymph node metastasis | 0.2663 | |||
| Yes | 121 (57.6) | 31 (64.6) | 90 (55.6) | |
| No | 89 (42.4) | 17 (35.4) | 72 (44.4) | |
| Local recurrence | 0.7368 | |||
| Yes | 43 (20.5) | 9 (18.8) | 34 (21.0) | |
| No | 167 (79.5) | 39 (81.2) | 128 (79.0) | |
| Prognosis | 0.0885 | |||
| Dead | 88 (41.9) | 15 (31.3) | 73 (45.1) | |
| Survived | 122 (58.1) | 33 (68.7) | 89 (54.9) | |
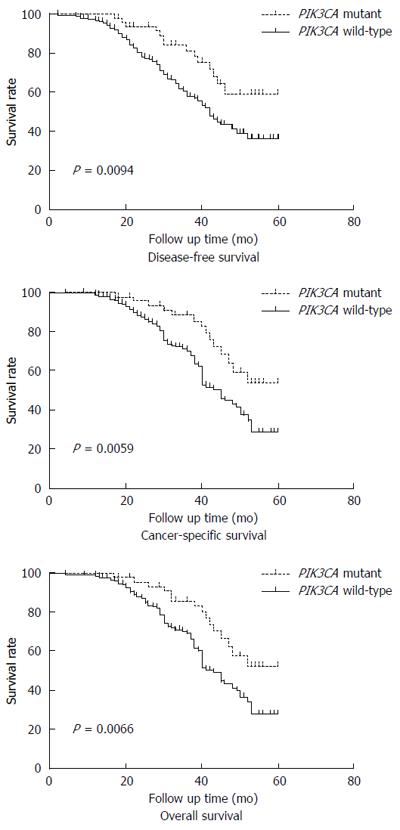
We assessed the influence of PIK3CA gene mutations on clinical outcome in Northwest Chinese patients with curatively resected ESCC. During the follow-up of the 210 patients, there were a total of 46 deaths confirmed to be attributable to esophageal cancer. The median follow-up time for censored patients was 36.5 mo. In the Kaplan-Meier analysis, patients with PIK3CA gene mutations experienced significantly longer disease-free survival (log rank P = 0.0094), cancer-specific survival (log rank P = 0.0059), and overall survival (log rank P = 0.0066) rates than those with the wild-type PIK3CA gene (Figure 1).
Numerous genetic and functional studies have clearly established a fundamental role for the PI3K signaling pathway in the development of neoplasia. As an oncogene in various human cancers, PIK3CA is one of the most genetically mutated genes in human cancers (including colorectal, brain, and gastric cancers)[22], having been displayed as mutated in various tumors, thereby making it a possible therapeutic marker. PIK3CA gene mutations and the subsequent activation of the PI3K/AKT pathway are considered to play a crucial role in cancer cell signaling pathways downstream of growth factors, cytokines, and other cellular stimuli in human neoplasm[6,23]. We therefore conducted this study to examine the prognostic impact of PIK3CA gene mutations among 210 Northwest Chinese patients with curatively resected ESCC.
In this study, we identified PIK3CA gene mutations in 48 out of 210 (22.9%) Northwest Chinese patients with curatively resected ESCC, which is a rate similar to that previously observed in ESCC (21%)[24], colorectal cancer (32%)[9], and breast cancer (25%-40%)[25,26], but slightly higher than that for gastric cancers (4.3%)[27] and brain tumors (5%)[28]. Additionally, we also found that c.1634A>C (p.E545A) was the dominant mutation type, which was consistent with a previous study in China[29]. The PIK3CA gene mutation frequency of ESCC in this study is slightly high when compared with those of previous studies; we believe this may be due to a difference in the patient cohorts, sample sizes, or methods used to assess PIK3CA gene mutation. When identifying PIK3CA gene mutations, other researchers typically use direct sequencing rather than the pyrosequencing used in the current study, which is a reliable high-throughput method that could be used as an alternative method for genotyping mutation studies[30]. There is also a non-electrophoretic nucleotide extension sequencing technology that can be used for mutation detection in tumors. Additionally, pyrosequencing has been shown to be more sensitive than regular sequencing in detecting EGFR and KRAS mutations in lung cancer patients[31,32]. PIK3CA gene mutational status was not identified as being associated with any clinicopathological characteristics of Northwest Chinese ESCC patients in our study, which is consistent with two other studies in Korea and China[19,33].
Identifying prognostic factors or biomarkers plays a crucial role in cancer research and clinical treatment[34-36]. Previous studies examining the relationship between PIK3CA gene mutations and prognosis in human cancers have yielded variable results and showed that PIK3CA gene mutational status is not associated with ESCC patient survival, although it does denote a better prognosis in breast cancer and ovarian cancer[37,38]. This discrepancy might be due to differences in tumor histologic type. We conducted this study to explore the prognostic impact of PIK3CA gene mutations among 210 Northwest Chinese patients with curatively resected ESCC. It was revealed that PIK3CA gene mutations were associated with a favorable prognosis among patients with curatively resected ESCC, suggesting PIK3CA gene mutational status may be a prognostic biomarker for Northwest Chinese ESCC patients that can be used to identify the clinical outcome of patients with curatively resected ESCC, which is consistent with its roles in Japanese ESCC patients[24]. Nonetheless, our findings regarding the correlation between PIK3CA mutations and favorable prognosis in esophageal cancer requires further confirmation by future independent studies using much larger non-biased cohorts of ESCCs.
In summary, this study suggests that PIK3CA gene mutations are associated with a favorable clinical outcome in operational resected ESCC, which supports the PIK3CA gene’s role as a prognostic biomarker for ESCC. Our data correlates with that of previous studies suggesting that the acquisition of PIK3CA gene mutations may be an important molecular event in the etiology of a wide range of tumor types and highlights the potential broad applicability that the PIK3CA gene may have in the clinical outcome of human cancers. Future studies are needed to confirm this association and clarify the exact molecular mechanisms by which PIK3CA gene mutations affects human cancer behavior.
Esophageal squamous cell carcinoma (ESCC) is the predominant histological subtype of esophageal cancer in East Asian countries, where it accounts for more than 90% of total esophageal cancer cases. Despite the development of multimodality therapies, the prognosis of ESCC patients remains poor, even for those who undergo complete resection of their carcinomas. The 5-year survival rates of ESCC are between 11.1% and 56.5%, depending on the clinical stage at the time of diagnosis. With the development of high-throughput genome sequencing and screening technologies, an increasing number of cancer-associated genes have been identified to serve as potential therapeutic targets or prognostic indicators. High frequencies of somatic mutations conferring oncogenic potential have been found in the PIK3CA gene, which is associated with poor prognosis in patients with colorectal or lung cancer. In contrast, a relationship between PIK3CA gene mutations and favorable prognoses has been shown in breast cancer. However, no large-scale study has examined the prognostic impact of PIK3CA gene mutations in Northwest Chinese ESCC patients.
The frequency of PIK3CA gene mutation in ESCC varied from 0% to 21%, which could likely introduce some bias in the statistical analyses of their clinical significance. More than 80% of PIK3CA gene mutations detected were localized in exons 9 and 20 (helical and kinase domain), with three “hot-spot” mutations: E542K, E545K, and H1047R. A recent report correlated with previous studies suggesting that the acquisition of PIK3CA mutations may be an important molecular event in the etiology of ESCC, and that mutations are associated with their clinical outcome.
This is, by far, one of the largest studies on the prognostic role of PIK3CA gene mutations in Northwest Chinese ESCC to date, and it shows that PIK3CA gene mutations in ESCC are associated with a favorable prognoses. It has been suggested that PIK3CA gene mutational status can have a potential role as a prognostic biomarker for ESCC patients.
PIK3CA gene mutations are associated with a favorable clinical outcome in operational resected Northwest Chinese ESCC patients, thereby suggesting that the acquisition of PIK3CA gene mutations may be an important molecular event in the etiology of a wide range of tumor types and highlighting the potential broad applicability that PIK3CA gene may have in the clinical outcome of human cancers.
The PIK3CA gene is located on the 3q26.3 chromosome and encodes the catalytic p110 alpha subunit of phosphoinositide 3-kinase (PI3K). The PI3K signaling pathway is deregulated in many types of cancer, with only the PIK3CA gene being reported as mutated and amplified.
The authors examined the associations of PIK3CA gene mutations with clinicopathological characteristics and clinical outcome in esophageal squamous cell carcinoma patients in Northwest China. The authors exploited the most recent literature concerning the subject. The study suggests that PIK3CA gene mutations are associated with a favorable clinical outcome in esophageal squamous cell cancer and that in the future the evaluation of PIK3CA gene mutations may be potentially applied as a prognostic marker. The manuscript is worth sharing with other researchers. It is concise, clear, comprehensive, and convincing.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Ciesielski M, Ribas G S- Editor: Yu J L- Editor: Rutherford A E- Editor: Wang CH
| 1. | Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241-2252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2115] [Cited by in RCA: 2219] [Article Influence: 100.9] [Reference Citation Analysis (0)] |
| 2. | Sasaki Y, Tamura M, Koyama R, Nakagaki T, Adachi Y, Tokino T. Genomic characterization of esophageal squamous cell carcinoma: Insights from next-generation sequencing. World J Gastroenterol. 2016;22:2284-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 43] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 3. | Zeng H, Zheng R, Zhang S, Zuo T, Xia C, Zou X, Chen W. Esophageal cancer statistics in China, 2011: Estimates based on 177 cancer registries. Thorac Cancer. 2016;7:232-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 212] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 4. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13214] [Article Influence: 1468.2] [Reference Citation Analysis (3)] |
| 5. | Gertler R, Stein HJ, Langer R, Nettelmann M, Schuster T, Hoefler H, Siewert JR, Feith M. Long-term outcome of 2920 patients with cancers of the esophagus and esophagogastric junction: evaluation of the New Union Internationale Contre le Cancer/American Joint Cancer Committee staging system. Ann Surg. 2011;253:689-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 6. | Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261-1274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4880] [Cited by in RCA: 4821] [Article Influence: 267.8] [Reference Citation Analysis (0)] |
| 7. | Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2247] [Cited by in RCA: 2560] [Article Influence: 134.7] [Reference Citation Analysis (0)] |
| 8. | Samuels Y, Diaz LA, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 669] [Cited by in RCA: 744] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 9. | Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2599] [Cited by in RCA: 2761] [Article Influence: 131.5] [Reference Citation Analysis (0)] |
| 10. | Nam SK, Yun S, Koh J, Kwak Y, Seo AN, Park KU, Kim DW, Kang SB, Kim WH, Lee HS. BRAF, PIK3CA, and HER2 Oncogenic Alterations According to KRAS Mutation Status in Advanced Colorectal Cancers with Distant Metastasis. PLoS One. 2016;11:e0151865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 11. | Jelovac D, Beaver JA, Balukrishna S, Wong HY, Toro PV, Cimino-Mathews A, Argani P, Stearns V, Jacobs L, VanDenBerg D. A PIK3CA mutation detected in plasma from a patient with synchronous primary breast and lung cancers. Hum Pathol. 2014;45:880-883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 12. | Whitehall VL, Rickman C, Bond CE, Ramsnes I, Greco SA, Umapathy A, McKeone D, Faleiro RJ, Buttenshaw RL, Worthley DL. Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J Cancer. 2012;131:813-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Lai YL, Mau BL, Cheng WH, Chen HM, Chiu HH, Tzen CY. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15:1064-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 84] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 14. | Abubaker J, Bavi P, Al-Harbi S, Ibrahim M, Siraj AK, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F. Clinicopathological analysis of colorectal cancers with PIK3CA mutations in Middle Eastern population. Oncogene. 2008;27:3539-3545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 15. | Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, Soh J, Suzuki M, Wistuba II, Fong KM. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913-6921. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 346] [Cited by in RCA: 347] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 16. | Miyaki M, Iijima T, Yamaguchi T, Takahashi K, Matsumoto H, Yasutome M, Funata N, Mori T. Mutations of the PIK3CA gene in hereditary colorectal cancers. Int J Cancer. 2007;121:1627-1630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Fang WL, Huang KH, Lan YT, Lin CH, Chang SC, Chen MH, Chao Y, Lin WC, Lo SS, Li AF. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7:6201-6220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 18. | Mori R, Ishiguro H, Kimura M, Mitsui A, Sasaki H, Tomoda K, Mori Y, Ogawa R, Katada T, Kawano O. PIK3CA mutation status in Japanese esophageal squamous cell carcinoma. J Surg Res. 2008;145:320-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Maeng CH, Lee J, van Hummelen P, Park SH, Palescandolo E, Jang J, Park HY, Kang SY, MacConaill L, Kim KM. High-throughput genotyping in metastatic esophageal squamous cell carcinoma identifies phosphoinositide-3-kinase and BRAF mutations. PLoS One. 2012;7:e41655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, Hedvat CV, Traina TA, Solit D, Gerald W. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15:5049-5059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 322] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 21. | Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC Cancer Staging Manual: esophagus and esophagogastric junction. Ann Surg Oncol. 2010;17:1721-1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 571] [Cited by in RCA: 639] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 22. | Murugan AK, Munirajan AK, Tsuchida N. Genetic deregulation of the PIK3CA oncogene in oral cancer. Cancer Lett. 2013;338:193-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle. 2004;3:1221-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 372] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 24. | Shigaki H, Baba Y, Watanabe M, Murata A, Ishimoto T, Iwatsuki M, Iwagami S, Nosho K, Baba H. PIK3CA mutation is associated with a favorable prognosis among patients with curatively resected esophageal squamous cell carcinoma. Clin Cancer Res. 2013;19:2451-2459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 25. | Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678-7681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 719] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 26. | Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554-2559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 662] [Cited by in RCA: 698] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 27. | Li VS, Wong CW, Chan TL, Chan AS, Zhao W, Chu KM, So S, Chen X, Yuen ST, Leung SY. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer. 2005;5:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 140] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048-5050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 248] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 29. | Wang L, Shan L, Zhang S, Ying J, Xue L, Yuan Y, Xie Y, Lu N. PIK3CA gene mutations and overexpression: implications for prognostic biomarker and therapeutic target in Chinese esophageal squamous cell carcinoma. PLoS One. 2014;9:e103021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Zhou Z, Poe AC, Limor J, Grady KK, Goldman I, McCollum AM, Escalante AA, Barnwell JW, Udhayakumar V. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J Clin Microbiol. 2006;44:3900-3910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 49] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Lee SE, Lee SY, Park HK, Oh SY, Kim HJ, Lee KY, Kim WS. Detection of EGFR and KRAS Mutation by Pyrosequencing Analysis in Cytologic Samples of Non-Small Cell Lung Cancer. J Korean Med Sci. 2016;31:1224-1230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 32. | Xie G, Xie F, Wu P, Yuan X, Ma Y, Xu Y, Li L, Xu L, Yang M, Shen L. The mutation rates of EGFR in non-small cell lung cancer and KRAS in colorectal cancer of Chinese patients as detected by pyrosequencing using a novel dispensation order. J Exp Clin Cancer Res. 2015;34:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Wang WF, Xie Y, Zhou ZH, Qin ZH, Wu JC, He JK. PIK3CA hypomethylation plays a key role in activation of the PI3K/AKT pathway in esophageal cancer in Chinese patients. Acta Pharmacol Sin. 2013;34:1560-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Sutcliffe P, Hummel S, Simpson E, Young T, Rees A, Wilkinson A, Hamdy F, Clarke N, Staffurth J. Use of classical and novel biomarkers as prognostic risk factors for localised prostate cancer: a systematic review. Health Technol Assess. 2009;13:iii, xi-xiii 1-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 35. | Trapé J, Montesinos J, Catot S, Buxó J, Franquesa J, Sala M, Domenech M, Sant F, Badal JM, Arnau A. A prognostic score based on clinical factors and biomarkers for advanced non-small cell lung cancer. Int J Biol Markers. 2012;27:e257-e262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 36. | Muc-Wierzgoń M, Nowakowska-Zajdel E, Dzięgielewska-Gęsiak S, Kokot T, Klakla K, Fatyga E, Grochowska-Niedworok E, Waniczek D, Wierzgoń J. Specific metabolic biomarkers as risk and prognostic factors in colorectal cancer. World J Gastroenterol. 2014;20:9759-9774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (2)] |
| 37. | Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Del Grammastro M, Ferro A, Dalla Palma P, Galligioni E, Marchetti A. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13:6064-6069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 160] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 38. | Rahman M, Nakayama K, Rahman MT, Nakayama N, Ishikawa M, Katagiri A, Iida K, Nakayama S, Otsuki Y, Shih IeM, Miyazaki K. Clinicopathologic and biological analysis of PIK3CA mutation in ovarian clear cell carcinoma. Hum Pathol. 2012;43:2197-2206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |