Published online Mar 14, 2017. doi: 10.3748/wjg.v23.i10.1787
Peer-review started: November 16, 2016
First decision: December 19, 2016
Revised: January 17, 2017
Accepted: February 16, 2017
Article in press: February 17, 2017
Published online: March 14, 2017
Processing time: 117 Days and 22.9 Hours
To identify the miRNA-mRNA regulatory network in hepatitis B virus X (HBx)-expressing hepatic cells.
A stable HBx-expressing human liver cell line L02 was established. The mRNA and miRNA expression profiles of L02/HBx and L02/pcDNA liver cells were identified by RNA-sequencing analysis. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis was performed to investigate the function of candidate biomarkers, and the relationship between miRNA and mRNA was studied by network analysis.
Compared with L02/pcDNA cells, 742 unregulated genes and 501 downregulated genes were determined as differentially expressed in L02/HBx cells. Gene ontology analysis suggested that the differentially expressed genes were relevant to different biological processes. Concurrently, 22 differential miRNAs were also determined in L02/HBx cells. Furthermore, integrated analysis of miRNA and mRNA expression profiles identified a core miRNA-mRNA regulatory network that is correlated with the carcinogenic role of HBx.
Collectively, the miRNA-mRNA network-based analysis could be useful to elucidate the potential role of HBx in liver cell malignant transformation and shed light on the underlying molecular mechanism and novel therapy targets for hepatocellular carcinoma.
Core tip: A number of miRNAs have been identified to be substantially involved in hepatocellular carcinoma cell proliferation, migration and invasion. Thus, an integrated analysis of the expression and function of miRNA and mRNA makes it possible to successfully identify the predicted miRNA-target network pattern and functional candidates of miRNA-mRNA pairs associated with HBx-related hepatocarcinogenesis.
- Citation: Chen RC, Wang J, Kuang XY, Peng F, Fu YM, Huang Y, Li N, Fan XG. Integrated analysis of microRNA and mRNA expression profiles in HBx-expressing hepatic cells. World J Gastroenterol 2017; 23(10): 1787-1795
- URL: https://www.wjgnet.com/1007-9327/full/v23/i10/1787.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i10.1787
Hepatocellular carcinoma (HCC) is one of the most common malignancies around the world, accounting for more than 745000 deaths per year[1,2]. As a typical inflammation-driven tumor, approximately 53% of HCC cases happen in the setting of chronic virus infection (mostly the hepatitis B virus (HBV) and hepatitis C virus), especially in Asia[3]. The HBV genome has 4 open reading frames (ORFs), namely X, S, C and P genes. The X gene, which encodes hepatitis B virus X (HBx) protein, correlates most with HCC occurrence and development[4,5]. Mounting evidence indicates that the integration of HBx into the host genome in hepatocytes could lead to gene transcription, cell proliferation, cell signaling transduction, protein degradation and apoptosis[6-9]. Moreover, several studies in vivo have confirmed the high morbidity of HCC in HBx-expressing transgenic mice[10-13]. However, the exact mechanism of HBx-induced hepatocarcinogenesis remains relatively poorly defined.
The miRNAs are a group of endogenously expressed RNAs with small molecular length, which play vital roles in various biological and pathological processes[14]. Mounting evidence has suggested the importance of miRNAs in the modulation of gene expression, cellular proliferation, cellular mobility, cellular differentiation, apoptosis and tumorigenesis[15]. A number of miRNAs have been identified to be substantially involved in HCC cell proliferation, migration and invasion, among which miR-122, miR-125, miR-199 family members and so on are closely related with HBV-associated HCC, especially[16]. As has been widely interpreted, the expression of miRNAs and their corresponding target genes are often inversely modulated in different backgrounds[17]. Meanwhile, increasing evidence has highlighted the success of a combined approach to investigate the miRNA-mediated mRNA regulation in various diseases[18,19]. Thus, an integrated analysis of the expression and functional interaction involving miRNA and mRNA makes it possible to successfully identify the predicted miRNA-target network pattern and functional candidates of miRNA-mRNA pairs associated with HBx-related hepatocarcinogenesis.
In this study, we conducted a comprehensive analysis for the first time to identify the functional miRNA-mRNA interactive network in HBx-transfected liver cells. By integrating the transcriptome and miRNAome, our study shed light on the potential molecular mechanism of HBx-related liver cell malignant transformation.
The human liver cell line L02 (purchased from the China Center for Type Culture Collection, China) was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin and 10% fetal bovine serum (Gibco, Thermo Fisher, Waltham, MA, United States) in a cell incubator with 5% CO2 at 37 °C. L02 cells were then transfected with empty plasmids pcDNA3.0 (as a control) and pcDNA/HBx (the experiment group) by LipofectamineTM 2000 (Invitrogen, Carlsbad, CA, United States) and cell clones were selected with Geneticin® (G418) according to the manufacturer’s instructions (Gibco). The efficiency of transfection with empty vector (termed L02/pcDNA) or pcDNA/HBx (termed L02/HBx) was validated by Western blot.
L02/pcDNA and L02/HBx cells were cultured and grown to 70%-90% confluency. Total cell RNA was extracted by using Trizol reagents (Invitrogen) following the manufacturer’s instructions. The RNA quality was validated by agarose gel electrophoresis. The mRNA and miRNA expression profile was identified through RNA-sequencing (RNA-seq) analysis of the total RNA sample (Novel Bioinformatics, China).
As a fast splice junction mapper for RNA-seq reads, TopHat was used for RNA-seq alignment in our study. Based on the ultra-high throughput short read aligner Bowtie, TopHat aligns RNA-Seq reads to reference genomes and further analyzes the mapping reads to determine the possible splice junctions between exons. In contrast, the unmapped reads are separated into small parts, which enable them to align to the reference genome and define splice junctions[20].
Limma algorithm was used to filter the differentially expressed mRNAs and miRNAs, according to the significant analysis and false discovery rate (FDR) analysis[21]. All data analysis meets the following two criteria: (1) fold-change > 2 or < 0.5; and (2) FDR < 0.05[22].
TargetScan and miRnada were utilized as analysis tools for miRNA target prediction based on the differentially expressed mRNAs and miRNAs[20]. The complex relationship between mRNAs and miRNAs was elucidated to build the miRNA-mRNA network according to differential expression values as well as the interactions of miRNA and their target genes listed in the Sanger MicroRNA database. In the miRNA-mRNA interaction network, the shape of square represents miRNAs and the circle represents target genes. The key genes and miRNAs usually possess the biggest degrees in the entire network.
We then performed gene ontology (GO) analysis to help elucidate the concrete biological functions of specific genes with significant differences in the representative profiles of the miRNA target genes[23]. GO annotations were downloaded from Gene Ontology (http://www.geneontology.org/), UniProt (http://www.uniprot.org/), and NCBI (http://www.ncbi.nlm.nih.gov/). Fisher’s exact test was employed to determine the significant GO categories and P values were corrected by FDR.
Pathway analysis was employed to identify the critical signal pathways of the differential expressed genes based on the resources from Kyoto Encyclopedia of Genes and Genomes (KEGG) database[24]. Fisher’s exact test was used to pick out the significant pathways according to P value and FDR.
The cDNA was synthesized using primers with the PrimeScript Real-Time Reagent Kit (TaKaRa, Shiga, Japan) in accordance with the manufacturer’s instructions. Quantitative real-time PCR (Q-PCR) reactions were conducted with an Applied Biosystem-7500 Real-Time PCR System, using Taq PCR Master Mix (Zhongtai, China). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The Ct values of different mRNAs and miRNAs were normalized to GAPDH and analyzed via the Applied Biosystems 7500 Fast Software. The primers of miRNAs were purchased from Yingrun Biotechnologies Inc. (Changsha, China). The primer sequences of selected genes are shown in Table 1.
| Real-time PCR primer | Primer sequence (5'-3') |
| Alpha-2-macroglobulin (A2M) | F: CGGAGAATGACGTACTCCACT |
| R: TGGGTTGGTCCTTTCACTTGG | |
| KIAA1522 (KIAA1522) | F: CCAGGACAACGTCTTCTTTCC |
| R: CAGCCACCCTTGTTCAGTTTC | |
| Procollagen C-endopeptidase enhancer (PCOLCE) | F: GTGCGGAGGGGATGTGAAG |
| R: CGAAGACTCGGAATGAGAGGG | |
| Solute carrier family 22, member 14 (SLC22A14) | F: TGGAGATGCTGTTACGCAGAT |
| R: CTGGAATGTGCCAAACTCCC | |
| Asparagine synthetase (glutamine-hydrolyzing) (ASNS) | F: GGAAGACAGCCCCGATTTACT |
| R: AGCACGAACTGTTGTAATGTCA | |
| Claudine 1 (CLDN1) | F: CCTCCTGGGAGTGATAGCAAT |
| R: GGCAACTAAAATAGCCAGACCT | |
| Origin recognition complex, subunit 4 (ORC4) | F: AGGTGACCGAACTAGCAGTG |
| R: CTGCCGGTGAGAAAATCTTGA | |
| Forkhead box C1 (FOXC1) | F: GGCGAGCAGAGCTACTACC |
| R: TGCGAGTACACGCTCATGG | |
| Spermatid perinuclear RNA binding protein (STRBP) | F: GTGTGGTGTAATGAGGATTGGC |
| R: GTGGGCTCTTTTGTATTCCGAA |
Two-way ANOVA test was performed for statistical analysis, using SPSS16.0 software. P < 0.05 was considered to be statistically significant.
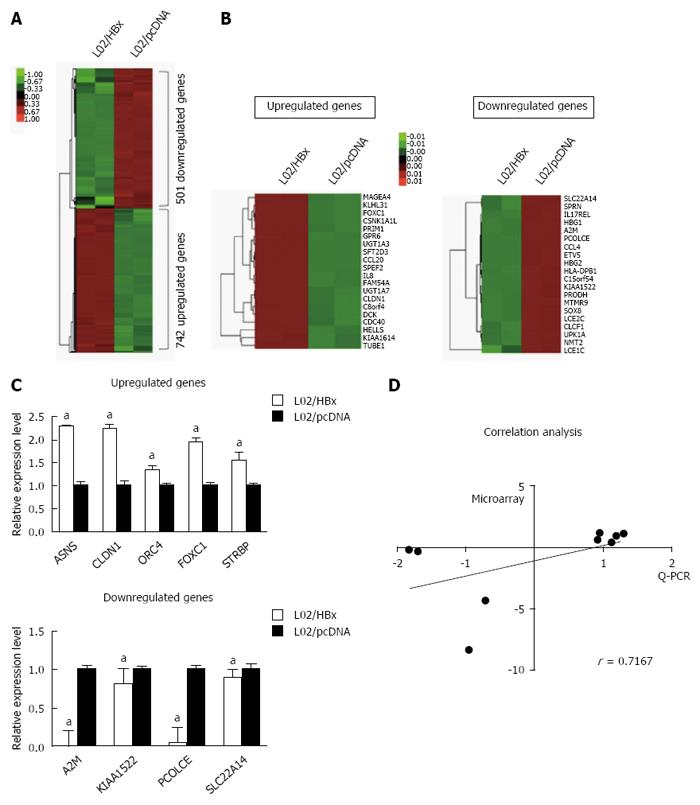
To elucidate the molecular mechanism underlying HBx-mediated hepatocarcinogenesis, we first conducted the RNA-seq analysis to compare the mRNA expression profile of L02/HBx cells with that of L02/pcDNA L02 cells. Of 1243 differentially expressed genes, the expression of 742 genes were upregulated and 501 genes expression were downregulated (fold-change ≥ 2, P < 0.05) in L02/HBx cells (Figure 1A). The 20 top-ranking differentially expressed genes were subjected to a cluster analysis (Figure 1B), which determined several groups of genes with identical expression patterns. According to comparison with the control L02/pcDNA cells, the expression of GPR6, KLHL31, FOXC1, UGT1A7 and MAGEA4 was most upregulated and of SLC22A14, HBG2, PCOLCE, A2M and KIAA1522 was most significantly downregulated in L02/HBx cells. These differentially expressed genes were diversely involved in DNA replication, cell cycle, cell proliferation, cell adhesion, cell-cell signaling, metabolic process, tumor transformation and several other signaling pathways.
Next, we performed Q-PCR to confirm the relative mRNA expression of the 9 selected genes in which we were interested for further study. As shown in Figure 1C, the relative mRNA expression levels of 5 upregulated and 4 downregulated genes measured by Q-PCR were totally consistent with the RNA-seq results. Taking the sensitivity of two methods into consideration, the gene expression changes in RNA-seq were not exactly the same with that in Q-PCR analysis; however, there was a highly similar tendency between the two groups (Figure 1D; r = 0.7167, P = 0.0369).
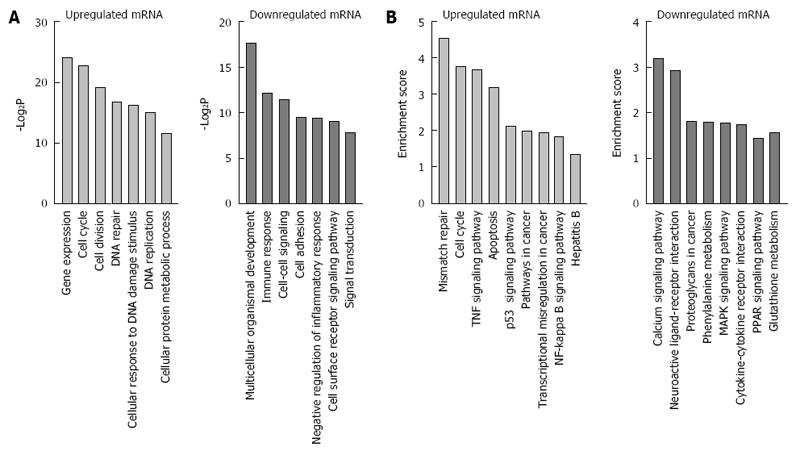
In order to elucidate the correlation between differentially expressed genes and HBx-related hepatocarcigenesis, GO and signaling pathway analyses were applied respectively. GO analyses demonstrated that the 1243 annotated genes displayed diverse biological functions. The 742 upregulated genes were majorly correlated with the response to gene expression, DNA replication, DNA repair, cell cycle, cell division, and cellular response to DNA damage stimulus, while 501 downregulated genes were mainly related to immune response, cell-cell signaling, cell adhesion, signal transduction and cell surface receptor signaling pathway (Figure 2A). Of note, further KEGG signal pathway analysis revealed that upregulated genes were briefly related to hepatitis B, mismatch repair, cell cycle, NF-κB signaling pathway and p53 signaling pathway, while downregulated genes were mostly involved in glutathione metabolism, PPAR signaling pathway, cytokine-cytokine receptor interaction, proteoglycans in cancer and MAPK signaling pathway (Figure 2B).
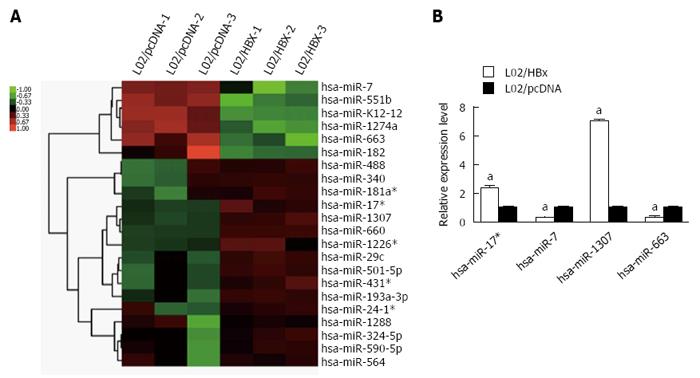
Compared with L02/pcDNA cells, 6 miRNAs were decreased and 16 miRNAs were upregulated in L02/HBx cells (Figure 3A). Specially, hsa-miR-7, hsa-miR-551b, hshv-miR-K12-12, hsa-miR-1274a, hsa-miR-663 and hsa-miR-182 were found to be downregulated; meanwhile, hsa-miR-488, hsa-miR-340, hsa-miR-181a*, hsa-miR-17*, hsa-miR-1307, hsa-miR-660, hsa-miR-1226*, hsa-miR-29c, hsa-miR-501-5p, hsa-miR-431, hsa-miR-193a-3p, hsa-miR-24-1*, hsa-miR-1288, hsa-miR-324-5p, hsa-miR-590-5p and hsa-miR-564 were upregulated in L02/HBx cells. Then, the expression of 4 differentially expressed miRNAs was further validated through Q-PCR analysis, namely hsa-miR-17*, hsa-miR-7, hsa-miR-1307 and hsa-miR-663. As shown in Figure 3B, there was a consistent tendency of expression changes between miRNA-seq and Q-PCR data.
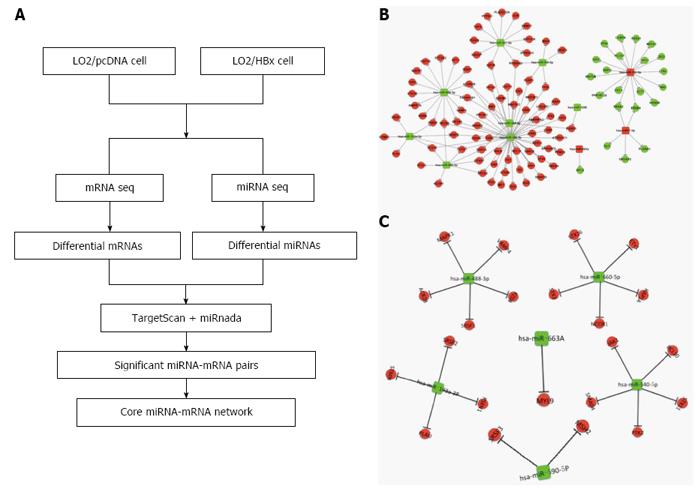
It is generally believed that the network-based identification of hub miRNA patterns possesses higher accuracy compared with non-network-based methods. Thus, we employed the strategy illustrated in Figure 4A to identify the miRNA-mRNA regulatory network in L02 cells transfected with HBx. Through analysis using miRnada and TargetScan software, potential miRNA targets were successfully predicted. In combination with the expression pattern of target genes (Figure 1A), we generated several significant miRNA-mRNA pairs, which exhibited opposite expression patterns in L02/pcDNA and L02/HBx cells.
Hubs refer to the central parts of a network, and they play a substantial role in biological system progression. Hub miRNAs are generally defined as the top 10%-15% of the nodes by degree. Eleven differentially expressed miRNAs were identified as hub miRNAs in our study (Figure 4B). We found that miR340-5p is the one that regulates the most genes, as many as 51 predicted target genes. Furthermore, we also found that several miRNAs modulate a few common target genes through a combinational manner (Figure 4B). In order to identify the core miRNA-mRNA regulatory network, which is most functionally related to HCC development, the functional enrichment of these predicted target genes was analyzed. Genes closely related with cancer-associated pathways, such as DNA replication, gene expression, cell cycle, cell division, metabolic processes, signal transduction and so on, were especially picked out to build up the essential miRNA-mRNA regulatory network. Significantly, the miRNAs and their target genes displayed opposite expression patterns in L02/pcDNA and L02/HBx cells. As an example, the increased miRNAs in L02/HBx cells, such as hsa-miR-182-5p and hsa-miR-7-5p, may repress the expression of negative modulators, for instance, MEF2C and GLI3, to promote the cell transformation (Figure 4C). In contrast, the decreased miRNAs, including hsa-miR-501-5p, hsa-miR-340-5p, has-miR324-5p and has-miR-488-3p, may release the suppression of their target genes, for example, JAK1, MAPK1, STX3 and BCL10, whose expression was upregulated in L02/HBx cells. The activation of these potential miRNA target genes might help to establish HBx-induced HCC progression.
Summarizing all the results, we have demonstrated that these core miRNAs may function as crucial regulators by modulating DNA replication, gene expression, cell cycle, cell division, metabolic processes and signal transduction, thus playing vital roles in tumorigenesis.
HCC is one of the deadliest malignancies, with dismal prognosis around the world. In southeast Africa and China, chronic HBV infection is the most important cause of HCC. At present, the molecular mechanism of HBV-related HCC development is not fully understood, and the HBx gene is frequently regarded as a vital regulator in HBV-related hepatocarcinogenesis[25,26]. Due to being integrated into the host genome, the HBx gene can act as a transactivator of numerous host cellular genes related with growth control, cell cycle and malignant transformation[10]. Therefore, a better understanding of the specific role of HBx in hepatocarcinogenesis is vital to the advancement of HCC treatment. Although a number of studies have been investigating the molecular mechanism underlying HBx-associated HCC development, the conclusions from different groups have varied from among each other[8,27-29]. Thus, a more reliable strategy was employed in our study to help improve the understanding of gene regulations on HBX-related progression on the basis of the combined analysis of the miRNA-mRNA regulatory network.
Of all the differentially expressed genes, a substantial proportion were associated with gene expression, DNA repair, cell cycle, cell division, metabolic processes, cell-cell signaling, cell adhesion and signal transduction (Figure 1C and D). These differentially expressed mRNA might vigorously participate in the hepatic malignant cell transformation through regulating multiple cellular processes. We further obtained the associated signal pathways modulated by the differential expressed genes with KEGG analysis. Among the 52 pathways mediated by the upregulated genes, 5 pathways have been confirmed to be HBx-related, including the hepatitis B pathway, cell cycle, apoptosis, and NF-kappa B and p53 signaling pathways. There is only one HBx-related signaling pathway among the 21 pathways mediated by downregulated genes, namely MAPK signal pathway[7]. Through regulating these 6 pathways and possibly other potential pathways as well, HBx might play a crucial role in hepatocarcinogenesis via modulating chromosome integrity[30], cell proliferation[29], cell cycle[8], apoptosis and oncogene expression[27,28]. Combining the function and pathway analysis for differentially expressed genes in L02/HBx cells, we may arrive at a conclusion that liver cells might develop various cellular responses and gain malignant potentials given the transfection of HBx.
With regard to the altered miRNAs, we successfully identified 6 downregulated and 16 upregulated miRNAs in L02/HBx cells by the application of custom microarray. Of these altered miRNAs, several, such as hsa-miR-338-3p and hsa-miR-29c, have been previously reported to be involved in HBx-associated HCC development[31]. Despite the fact that several research groups have independently established the HBx-regulated mRNA or miRNA expression pattern by identifying different mRNAs as well as miRNAs in the presence of HBx[31-34], few studies have focused on the miRNA-mRNA modulation and interaction.
Actually, hepatocarcinogenesis is a complex process in response to external and internal stimuli, and miRNA-target gene modulation is a crucial means of cellular response. Therefore, identification of miRNA-target with opposite expression patterns facilitated our selection of the potential miRNA as well as their corresponding target genes in HBx-related cell transformation. Bioinformatics analysis demonstrated that 11 hub miRNAs participated in constructing the miRNA-mRNA regulatory network (Figure 4B). Next, we will carry out more specific research on the function of those hub miRNAs in order to gain a better understanding of their role in HCC. Moreover, in this study, we also acquired an understanding of the essential miRNA-mRNA interactive network on the basis of functional enrichment (Figure 4C). The corresponding target genes for the 11 hub miRNAs, including JAK1, MAPK1, STX3 and BCL10, are crucial regulators in cell cycle, gene expression, signal transduction and apoptosis[35-37]. Thus, identification of the target genes modulated by the dysregulated miRNAs will greatly promote our understanding of HBx-associated hepatocarcinogenesis and facilitate design of new targeted therapeutic agents.
Collectively, we not only identified the mRNAs and miRNAs that may become the potential predictors and diagnostically valuable biomarkers of HBV-related HCC but we also established a core functional miRNA-mRNA regulatory network in the context of HBx-induced cell transformation. This study shows that HBx transfection of liver cells is correlated with gene expression, cell cycle, signal transduction and apoptosis disturbance. The established network might reveal the regulation from miRNAs to target genes in the development of HBx-induced malignant transformation in HCC cells. Thus, it will be helpful to further investigate the novel function of HBx in hepatocarcinogenesis with the selection of miRNA-mRNA pairs.
We thank Novel Bioinformatics Co. Ltd. for providing technical assistance in deep sequencing and bioinformatics analysis.
Chronic hepatitis B virus (HBV) infection is one of the leading causes of hepatocellular carcinoma (HCC), especially in Asia. Accumulated evidence has suggested that hepatitis B virus X (HBx) protein plays a potentially oncogenic role in hepatocarcinogenesis; however, the exact mechanism remains exclusive.
Mounting evidence indicates that the integration of HBx into the host genome in hepatocytes could lead to gene transcription, cell proliferation, cell signaling transduction, protein degradation and apoptosis.
In this study, the authors conducted a comprehensive analysis for the first time to identify the functional miRNA-mRNA interactive network in HBx-transfected liver cells. This study shows that HBx transfection of liver cells is correlated with gene expression, cell cycle, signal transduction and apoptosis disturbance.
By integrating the transcriptome and miRNAome, this study shed light on the potential molecular mechanism of HBx-related liver cell malignant transformation.
Kyoto Encyclopedia of Genes and Genomes is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high throughput experimental technologies.
The authors’ approach to clarify the association of miRNA and mRNA in HBx-carcinogenesis using software seems interesting.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Ohkoshi S, Rojas E S- Editor: Yu J L- Editor: Filipodia E- Editor: Wang CH
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23762] [Cited by in RCA: 25540] [Article Influence: 1824.3] [Reference Citation Analysis (7)] |
| 2. | Ma L, Wang X, Jia T, Wei W, Chua MS, So S. Tankyrase inhibitors attenuate WNT/β-catenin signaling and inhibit growth of hepatocellular carcinoma cells. Oncotarget. 2015;6:25390-25401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 3. | Okuda K. Hepatocellular carcinoma. J Hepatol. 2000;32:225-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 398] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 4. | Yang ST, Yen CJ, Lai CH, Lin YJ, Chang KC, Lee JC, Liu YW, Chang-Liao PY, Hsu LS, Chang WC. SUMOylated CPAP is required for IKK-mediated NF-κB activation and enhances HBx-induced NF-κB signaling in HCC. J Hepatol. 2013;58:1157-1164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Shi L, Peng F, Tao Y, Fan X, Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 6. | Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651-660. [PubMed] |
| 7. | Matsuda Y, Ichida T. Impact of hepatitis B virus X protein on the DNA damage response during hepatocarcinogenesis. Med Mol Morphol. 2009;42:138-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 8. | Cheng P, Li Y, Yang L, Wen Y, Shi W, Mao Y, Chen P, Lv H, Tang Q, Wei Y. Hepatitis B virus X protein (HBx) induces G2/M arrest and apoptosis through sustained activation of cyclin B1-CDK1 kinase. Oncol Rep. 2009;22:1101-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res. 2008;14:5061-5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 10. | Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, Han YM, Lee CS, Park JS, Lee CH. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 214] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 11. | Kim JH, Alam MM, Park DB, Cho M, Lee SH, Jeon YJ, Yu DY, Kim TD, Kim HY, Cho CG. The Effect of Metformin Treatment on CRBP-I Level and Cancer Development in the Liver of HBx Transgenic Mice. Korean J Physiol Pharmacol. 2013;17:455-461. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Wang C, Yang W, Yan HX, Luo T, Zhang J, Tang L, Wu FQ, Zhang HL, Yu LX, Zheng LY. Hepatitis B virus X (HBx) induces tumorigenicity of hepatic progenitor cells in 3,5-diethoxycarbonyl-1,4-dihydrocollidine-treated HBx transgenic mice. Hepatology. 2012;55:108-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 125] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 13. | Koff RS. Hepatocellular carcinoma in transgenic mice: a consequence of continued expression of the HBx gene? Gastroenterology. 1992;102:1081-1082. [PubMed] |
| 14. | Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1353] [Cited by in RCA: 1268] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 15. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 16. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2191] [Cited by in RCA: 2184] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 17. | Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 665] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 18. | Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z, Shao T, Jiang T, Ren H, Kang C. Comprehensive analysis of the functional microRNA-mRNA regulatory network identifies miRNA signatures associated with glioma malignant progression. Nucleic Acids Res. 2013;41:e203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 19. | Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. 2008;18:823-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 148] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 20. | Shen Y, Pan Y, Xu L, Chen L, Liu L, Chen H, Chen Z, Meng Z. Identifying microRNA-mRNA regulatory network in gemcitabine-resistant cells derived from human pancreatic cancer cells. Tumour Biol. 2015;36:4525-4534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Wettenhall JM, Simpson KM, Satterley K, Smyth GK. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22:897-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 172] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 22. | Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279-284. [PubMed] |
| 23. | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29963] [Cited by in RCA: 28928] [Article Influence: 1157.1] [Reference Citation Analysis (1)] |
| 24. | Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537-1545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 806] [Cited by in RCA: 938] [Article Influence: 52.1] [Reference Citation Analysis (0)] |
| 25. | Feitelson MA, Duan LX. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141-1157. [PubMed] |
| 26. | Su Q, Schröder CH, Hofmann WJ, Otto G, Pichlmayr R, Bannasch P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology. 1998;27:1109-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 176] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 27. | Shukla R, Yue J, Siouda M, Gheit T, Hantz O, Merle P, Zoulim F, Krutovskikh V, Tommasino M, Sylla BS. Proinflammatory cytokine TNF-α increases the stability of hepatitis B virus X protein through NF-κB signaling. Carcinogenesis. 2011;32:978-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 28. | Park SG, Min JY, Chung C, Hsieh A, Jung G. Tumor suppressor protein p53 induces degradation of the oncogenic protein HBx. Cancer Lett. 2009;282:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Bouchard MJ, Wang LH, Schneider RJ. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science. 2001;294:2376-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 326] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 30. | Haruki K, Shiba H, Fujiwara Y, Furukawa K, Iwase R, Uwagawa T, Misawa T, Ohashi T, Yanaga K. Inhibition of nuclear factor-κB enhances the antitumor effect of tumor necrosis factor-α gene therapy for hepatocellular carcinoma in mice. Surgery. 2013;154:468-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 31. | Lamontagne J, Steel LF, Bouchard MJ. Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J Gastroenterol. 2015;21:7375-7399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 49] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 32. | Xu G, Gao Z, He W, Ma Y, Feng X, Cai T, Lu F, Liu L, Li W. microRNA expression in hepatitis B virus infected primary treeshrew hepatocytes and the independence of intracellular miR-122 level for de novo HBV infection in culture. Virology. 2014;448:247-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 33. | Gao P, Wong CC, Tung EK, Lee JM, Wong CM, Ng IO. Deregulation of microRNA expression occurs early and accumulates in early stages of HBV-associated multistep hepatocarcinogenesis. J Hepatol. 2011;54:1177-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 34. | Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin Med J (Engl). 2009;122:10-14. [PubMed] |
| 35. | Wu S, Xue J, Yang Y, Zhu H, Chen F, Wang J, Lou G, Liu Y, Shi Y, Yu Y. Isoliquiritigenin Inhibits Interferon-γ-Inducible Genes Expression in Hepatocytes through Down-Regulating Activation of JAK1/STAT1, IRF3/MyD88, ERK/MAPK, JNK/MAPK and PI3K/Akt Signaling Pathways. Cell Physiol Biochem. 2015;37:501-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Yiwei T, Hua H, Hui G, Mao M, Xiang L. HOTAIR Interacting with MAPK1 Regulates Ovarian Cancer skov3 Cell Proliferation, Migration, and Invasion. Med Sci Monit. 2015;21:1856-1863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 37. | Chiarini A, Liu D, Armato U, Dal Prà I. Bcl10 crucially nucleates the pro-apoptotic complexes comprising PDK1, PKCζ and caspase-3 at the nuclear envelope of etoposide-treated human cervical carcinoma C4-I cells. Int J Mol Med. 2015;36:845-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |