Published online Mar 7, 2016. doi: 10.3748/wjg.v22.i9.2771
Peer-review started: April 24, 2015
First decision: July 10, 2015
Revised: August 31, 2015
Accepted: December 1, 2015
Article in press: December 1, 2015
Published online: March 7, 2016
Processing time: 314 Days and 1.9 Hours
AIM: To investigate the targeted inhibition of proliferation and migration of SW620 human colon cancer cells by upregulating miRNA-145 (miR-145).
METHODS: Forty-five samples of colon cancer tissues and 45 normal control samples were obtained from the biological database of the First Affiliated Hospital of Liaoning Medical University. We performed quantitative analysis of miR-145 and N-ras expression in tissues; reverse transcriptase polymerase chain reaction analysis of miR-145 expression in SW620 colon cancer cells and normal colonic epithelial cells; construction of miR-145 lentiviral vector and determination of miR-145 expression in SW620 cells transduced with miR-145 vector; analysis of the effect of miR-145 overexpression on SW620 cell proliferation; analysis of the effect of miR-145 overexpression on SW620 cell migration using a wound healing assay; and analysis of the effect of miR-145 on N-ras expression using Western blotting.
RESULTS: miR-145 expression was significantly downregulated in colon cancer tissues, with its expression in normal colonic tissues being 4-5-fold higher (two sample t test, P < 0.05), whereas N-ras expression showed the opposite trend. miR-145 expression in SW620 cells was downregulated, which was significantly lower compared to that in colonic epithelial cells (two sample t test, P < 0.05). miR-145 vector and control were successfully packaged; expression of miR-145 in SW620 cells transduced with miR-145 was 8.2-fold of that in control cells (two sample t test, P < 0.05). The proliferation of miR-145-transduced SW620 cells was significantly decreased compared to control cells (two sample t test, P < 0.05). At 48 h in the wound healing experiment, the migration indexes and controls were (97.27% ± 9.25%) and (70.22% ± 6.53%), respectively (two sample t test, P < 0.05). N-ras expression in miR-145-tranduced SW620 cells was significantly lower than others (one-way analysis of variance, P < 0.05).
CONCLUSION: miR-145 is important in inhibiting colon cancer cell proliferation and migration. This is a good foundation for development of colon cancer therapy by targeting tumor suppressor miR-145.
Core tip: miRNA-145 (miR-145) may play an important role in inhibiting colon cancer proliferation and invasive migration. This finding lays a good foundation for further investigation of targeting miR-145 as a tumor suppressor in colon cancer treatment, and provides novel evidence for the anti-cancer effects and therapeutic potential of miR-145.
- Citation: Li C, Xu N, Li YQ, Wang Y, Zhu ZT. Inhibition of SW620 human colon cancer cells by upregulating miRNA-145. World J Gastroenterol 2016; 22(9): 2771-2778
- URL: https://www.wjgnet.com/1007-9327/full/v22/i9/2771.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i9.2771
Colorectal cancer is currently one of the most common malignancies worldwide[1,2]. Invasive migration at early stages is a critical factor affecting the prognosis and survival of colorectal cancer patients[3,4]. Despite improved diagnosis and medical technologies, treatment effects and clinical prognosis are still not ideal. With the development of genetic therapy, targeted therapy has become the main specific anti-cancer treatment. Blocking invasive migration by modifying the microenvironment at the molecular level is the frontier of current research in colorectal cancer therapeutics[5-7].
Recently, the discovery of a class of small non-coding RNAs known as miRNAs that regulates gene expression at the transcription level shed new light on cancer research[8,9]. miRNAs are endogenous, evolutionarily conserved, single chain non-coding RNAs consisting of 19-22 nucleotides, which inhibit mRNA transcription or induce mRNA degradation by base-paring with the 3’ untranslated region of target mRNA, and thus play an important role in cell proliferation, differentiation and apoptosis, gene regulation, and tumor formation by suppressing the expression of target genes[10,11]. Although current understanding of the biological function of miRNAs is still limited, we know for certain that miRNAs play a critical role in many aspects including modulating development, growth, differentiation, apoptosis, and tumorigenesis. In some malignant tumors, miRNAs function like oncogenes or tumor suppressors by regulating different gene targets. The function of specific miRNA during tumor formation and development is becoming a hot topic in cancer research. Some experts have predicted that targeting miRNA in biological cancer therapeutics will be more effective than targeting coding genes[12].
Earlier studies reported that miRNA-145 (miR-145) expression is downregulated in many cancers including breast cancer, melanoma, ovarian cancer, and liver cancer, suggesting that miR-145 may function as a tumor suppressor[13-16]. However, so far, there has been no systemic research on the role of miR-145 in colorectal cancer. Therefore, we investigated the expression of miR-145 and its target gene N-ras in colon cancer and normal control tissues, and postulated that miR-145 may play a biological role in colon cancer development and metastasis. Next, we constructed recombinant lentiviral vector expressing miR-145 and examined the inhibitory function of miR-145 after its transduction into SW620 colon cancer cells as an experimental basis for continued research on targeting miR-145 in colon cancer treatment. Lastly, we analyzed the expression of miR-145 target N-ras to clarify the regulatory role of miR-145 in colon cancer.
Forty-five samples of colon cancer tissues and 45 normal control samples were obtained from the biological database of the First Affiliated Hospital of Liaoning Medical University. These samples were obtained from colon cancer patients attending the hospital between January and December 2013. Patients had no treatment before surgery. All patients gave written informed consent to the study, which was reviewed and approved by the First Affiliated Hospital of Liaoning Medical University Institutional Review Board. pSilencerTM 4.1 lentiviral vector system and TRIzol reagent were purchased from Invitrogen (Carlsbad, CA, United States). HEK293T cell line, SW620 colon cancer cell line, and normal human colonic epithelial cell line were purchased from the American Type Culture Collection (Manassas, VA, United States). Dulbecco’s modified Eagle’s medium, fetal bovine serum, and trypsin were from Gibco. Ultra-pure plasmid extraction kit was from Promega (Madison, WI, United States). Restriction endonucleases, DNA polymerase, plasmid miniprep kit, one-step reverse transcriptase polymerase chain reaction (RT-PCR) kit, and gel extraction kit were from TaKaRa (Tokyo, Japan). Monoclonal rabbit anti-human primary antibodies, secondary antibodies, and internal controls were from Santa Cruz Biotechnology (Santa Cruz, CA, United States). miR-145 and internal control U6 primer were designed and synthesized by TaKaRa (Table 1). Other common reagents were provided by the Central Laboratory of the First Affiliated Hospital of Liaoning Medical University.
| Primers | Sequences (5’-3’) |
| miR-145-F | ACACTCCAGCTGGGGTCCAGTTTTCCCAGGA |
| miR-145-R | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGGATTC |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| URP | TGGTGTCGTGGAGTCG |
One milliliter of TRIzol reagent was added to ≤ 100 mg of frozen samples. After grinding in liquid nitrogen, total RNA was extracted using the phenol-chloroform protocol. Quality of RNA was analyzed on 1% agarose gels. RNA samples were sent to TaKaRa for sequencing and quantitative analysis of miR-145 and N-ras expression in colon cancer and control tissues.
SW620 and normal colonic epithelial cells were grown to the third passage of the exponential growth phase, and total RNA was extracted using the phenol-chloroform protocol. Semi-quantitative one-step RT-PCR analysis of miR-145 expression was carried out according to the instruction provided by the manufacturer (TaKaRa). PCR products were analyzed on 1% agarose gels and OD values were read on a gel imaging system. Experiments were repeated three times to compare miR-145 expression levels in these two cell lines.
The complete sequence of miR-145 was PCR-amplified using up- and downstream primers and purified by gel extraction. pSilencerTM 4.1 vector was linearized and purified according to the instruction provided with the lentiviral vector kit, and then ligated with miR-145 sequence. DH5α competent cells were then transformed with the ligation reaction. Positive clones were selected, and the sequences were amplified and linearized. miR-145 lentiviral vector was then packaged in HEK293T cells using Lipofectamine 2000, and viral titer was determined. The control vector containing a scrambled unrelated sequence was constructed and packaged in the same way.
SW620 cells that were 90% confluent were passaged at 1:3 at 18-24 h before transduction so that cells were 70%-80% confluent and at the exponential growth stage at the time of transduction. SW620 cells were transduced with packaged miR-145 lentiviral vector or control vector with 50% multiplicity of infection and cultured for 48-72 h. When the cytopathic effect occurred, total RNA was extracted from each of the two groups. PCR products were analyzed on 1% agarose gels and OD values were read on a gel imaging system. Experiments were repeated three times to compare miR-145 expression levels in two groups of cells to confirm successful construction of the expression system.
SW620 colon cancer cells were grown to the third passage of the exponential growth phase, and seeded into 96-well plates. Cells were transduced with either of the two lentiviral vectors mentioned above. Twenty microliters of 5 mg/mL MTT solution was added to each well. After incubation for 4 h at 37 °C, medium was removed and 150 μL DMSO was added. The absorption was then read on a plate reader at 490 nm, and proliferation curves were generated.
SW620 cells at the third passage were plated into two sets of dishes. When cells were confluent, a homogeneous scratch wound was created on the monolayer cells using a 1-mL pipette tip to form a cell-free area. miR-145 lentiviral solution was then added to the experimental group. Medium containing no lentivirus was used for the blank control group. Cells were imaged at 0 or 48 h after incubation. The width of the open area was measured at three different positions (top, middle and bottom) and the average width was calculated. The speed of cell migration was described using migration index (MI).
Using bioinformatics, we observed that miR-145 induced colon cancer cell apoptosis via N-ras. In other words, N-ras gene might be a target of miR-145. We therefore further tested this at protein level. Total proteins were extracted from miR-145-tranduced SW620 cells and quantified. After separation on 5% stacking gel and 8% separating gel at 60 V for 30 min and 100 V for 1.5 h, proteins were transferred to a membrane and Ponceau stained. After eluting three times, blots were incubated with primary antibody (1:1000, monoclonal rabbit-anti-human) at 4 °C overnight. After incubation for 1 h at room temperature with secondary antibody (1:1500, polyclonal goat-anti-rabbit) and BCIP/NBT staining in the dark for 3 h, blots were analyzed on a gel imaging system for target bands and internal reference bands. Experiments were repeated three times and OD values were calculated.
SPSS for Windows version 19.0 was used for statistical analysis. Data were expressed as mean ± SD. Differences among groups were compared using one-way ANOVA. P < 0.05 was considered statistically significant.
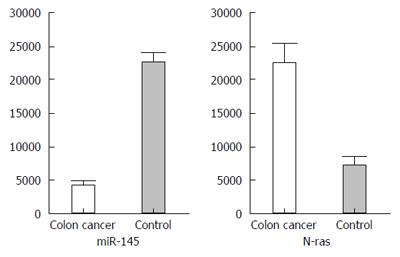
RNA sequencing and quantitative analysis indicated that miR-145 expression was markedly decreased in colon cancer tissues as compared to normal colonic tissues. miR-145 level in control tissues was 4-5-fold higher than that in colon cancer tissues (P < 0.05). In contrast, N-ras expression level was significantly higher in colon cancer then in control tissues (Figure 1).
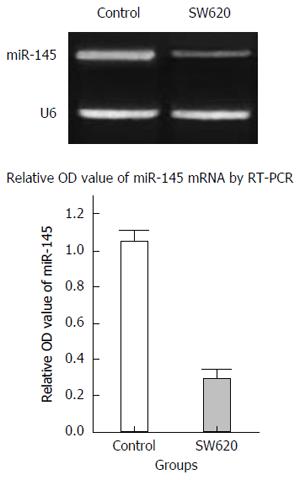
RT-PCR analysis revealed that miR145 expression in normal colonic epithelial cells was relatively high, whereas its expression in SW620 colon cancer cells was markedly downregulated. There was an approximately fivefold difference, which was statistically significant (P < 0.05) (Figure 2).
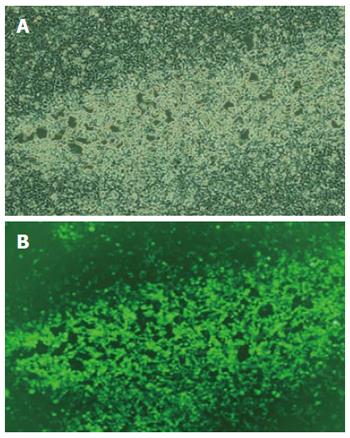
Construction of lentiviral vector and negative control sequence was successful, and sequencing results indicated no mutations. Both sequences were consistent with the expectation, indicating that insertion of miR-145 sequence was successful, and could be used for further experiments. Sequencing was done by TaKaRa. At 72 h post transfection of HEK293T cells with the lentiviral vector system, we observed green fluorescence under fluorescence microscopy. Eighty to ninety percent of cells expressed green fluorescence. We observed a cytopathic effect of shrinking, swelling and rounding of cells, and some cells were detached and floating (Figure 3A and B). Viral titers were 2.08 × 109 TU/mL in the experimental group and 1.92 × 109 TU/mL in the control group. Both met the requirement of the study and could be used for further experiments.
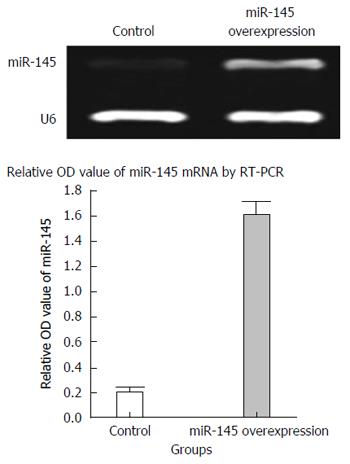
RT-PCR data showed that miR-145 expression in SW620 cells transduced with control lentiviral vector was low, whereas its expression in miR-145-transduced SW620 cells was 8.2-fold higher. The difference was statistically significant (P < 0.05) (Figure 4), indicating the success of generating the miR-145 lentiviral transfection system for future experiments.
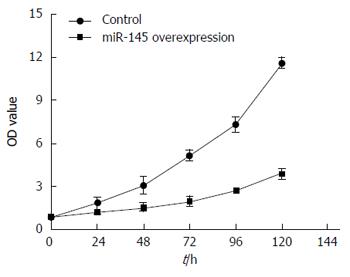
Two groups of SW620 cells were cultured separately for 24, 48, 72, 96, or 120 h followed by MTT analysis. Plates were read on a plate reader and proliferation curves were generated based on OD values. Results indicated that SW620 cells of the control group proliferated rapidly, whereas cells of the experimental group had significantly slower growth (P < 0.05), suggesting that miR-145 lentiviral vector can markedly inhibit the proliferation of SW620 colon cancer cells (Figure 5).
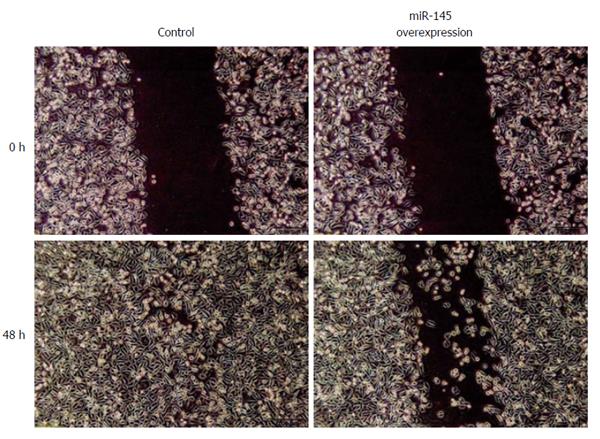
Our data showed that at 48 h after scratching the monolayer of cells, the wound in the control group was basically healed. The miR-145 group showed a small number of cells migrating to the open area, and the wound was still apparent. The MI in the control and experimental groups was 97.27% ± 9.25% and 70.22% ± 6.53%, respectively. The differences between the two groups were statistically significant (P < 0.05), suggesting that upregulating expression of miR-145 can markedly inhibit migration of SW620 cells (Figure 6).
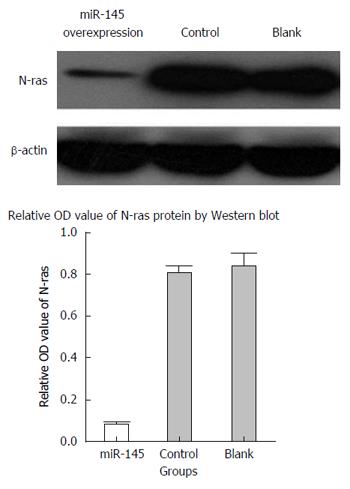
Western blot analysis revealed that N-ras expression level in miR-145-transduced SW620 cells was significantly reduced as compared to the empty vector and blank control groups (P < 0.05), whereas the empty vector and blank control groups had similar N-ras expression levels (P < 0.05), suggesting that miR-145 downregulated N-ras protein expression by effectively suppressing N-ras mRNA translation (Figure 7).
Currently, the genes and molecular mechanisms involved in colon cancer development and progression have become a hot research topic[17]. Studies have shown that many miRNAs are abnormally expressed in colon cancer tissues and cell lines. Among them, miR-143 is downregulated, whereas miR-21 and miR-223 are upregulated in colon cancer. They each function by regulating different target genes and are tightly related to each other during colon cancer development and progression[18-20]. It has been demonstrated that miRNAs downregulated in cancer often function as tumor suppressors, while miRNAs upregulated in cancer often function as oncogenes[21]. Various miRNAs and their target genes play important roles in colon cancer, but their specific mechanisms remain to be investigated.
miR-145, like miR-143, is downregulated in various cancers, but the degree of downregulation varies in different cancers[22]. Research has shown that miR-145 is downregulated by approximately 2.5-fold in breast cancer, 10-fold in nasal cancer and 20-fold in bladder cancer[23-26]. We have observed that miR-145 expression in colon cancer is 4-5-fold lower than in normal colonic tissues, and the degree of downregulation is positively correlated with colon cancer progression. In the present study, we demonstrated miR-145 downregulation in SW620 colon cancer cells and found that expression of miR-145 in normal human colonic epithelial cells was five times higher than that in SW620 cells. The above evidence supports that miR-145 may play an important role in the development and progression of various cancers.
miR-145 expression is decreased in SW620 cells, thus, we chose to examine its function further by upregulating its expression. In this study, we used lentiviral vector to express miR-145. Lentiviral vector is a novel vector type developed in recent years, which has high transfection efficiency and low cellular immune response and exerts its silencing effect via gene integration[27,28]. After successful construction of lentiviral vectors, HEK293T cells were transfected for viral packaging. Both transfection efficiency and viral titers met experimental requirements, so, the vectors could be used for further experiments.
In terms of inhibiting SW620 cell proliferation, we found that cells in the blank control group proliferated exponentially at 24, 48, 72, 96 and 120 h, while the proliferation rate of cells in the experimental group (miR-145-tranduced SW620 cells) was significantly lower compared to that in the blank control group, indicating that miR-145 can markedly suppress SW620 cell proliferation. This suggests that miR-145 plays a critical role in inhibiting colon cancer growth.
Cancer cells with metastatic potential have migratory ability, and cells with high metastatic potential often have more active migratory ability[29,30]. We used a wound healing assay to examine colon cancer cell migration. Our data showed that overexpression of miR-145 using recombinant lentiviral vector decreased the MI of SW620 cells by > 20% and markedly suppressed wound healing ability, indicating that miR-145 can suppress colon cancer cell migration and distant invasion. This is an important finding regarding cancer cell migration, which agrees with our expectation.
Ras oncogene is involved in human cancer development and progression. Although Ras mutation in colon cancer mainly involves K-ras, and N-ras mutation accounts for only 5%, the role of N-ras in colon cancer cannot be replaced by K-ras. K-ras expression promotes colon cancer cell proliferation, but it also makes these cancer cells prone to apoptosis. In contrast, high expression of N-ras does not promote cancer cell proliferation, but markedly inhibits cancer cell apoptosis. This function of N-ras cannot be replaced by K-ras[31,32]. By determining N-ras expression level in colon cancer tissues, we found that N-ras expression in cancer tissues is increased as compared to normal tissues, and this trend is the opposite from miR-145. This is consistent with our finding using bioinformatics that N-ras is a target gene of miR-145. miR-145 may induce apoptosis of SW620 colon cancer cells by regulating N-ras. We postulate that downregulation of miR-145 during colon cancer development may lead to decreased suppression of N-ras, and therefore, increased N-ras expression in cancer cells. N-ras signaling then leads to resistance to proapoptotic signals and promotes colon cancer development.
In conclusion, miR-145 may play an important role in inhibiting colon cancer proliferation and invasive migration. This finding lays a good foundation for further investigation of targeting miR-145 as a tumor suppressor in colon cancer treatment, and provides new evidence for the anti-cancer effects and therapeutic potential of miR-145.
Invasive migration at early stages is a critical factor affecting the prognosis and survival of colorectal cancer patients. With the development of genetic therapy, targeted therapy has become the main specific anti-cancer treatment. Blocking invasive migration by modifying the microenvironment at the molecular level is the frontier of current research in colorectal cancer therapeutics. Some experts have predicted that targeting miRNA in biological cancer therapeutics will be more effective than targeting coding genes.
miRNA-145 (miR-145) expression is downregulated in many cancers including breast cancer, melanoma, ovarian cancer, and liver cancer, suggesting that miR-145 may function as a tumor suppressor. However, so far, there has been no systematic research on the role of miR-145 in colorectal cancer.
miR-145 may induce apoptosis of SW620 colon cancer cells by regulating N-ras. The authors postulate that downregulation of miR-145 during colon cancer development may lead to decreased suppression of N-ras and therefore increased N-ras expression in cancer cells. N-ras signaling then leads to resistance to proapoptotic signals and promotes colon cancer development.
miR-145 may play an important role in inhibiting colon cancer proliferation and invasive migration. This finding lays a good foundation for further investigation on targeting miR-145 as a tumor suppressor in colon cancer treatment, and provides novel evidence for the anti-cancer effects and therapeutic potential of miR-145.
miRNAs are endogenous, evolutionarily conserved, single chain non-coding RNAs consisting of 19-22 nucleotides, which inhibit mRNA transcription or induce mRNA degradation by base-paring with the 3’ untranslated region of target mRNA, and thus play an important role in cell proliferation, differentiation and apoptosis, gene regulation, and tumor formation by suppressing the expression of target genes.
Authors presented a study in which they evaluated the role of microRNA-145 on colon cancer. They showed evidences of the tumor suppressor activity of microRNA-145 in colon cancer using several approaches.
P- Reviewer: Chiurillo MA S- Editor: Yu J L- Editor: Kerr C E- Editor: Wang CH
| 1. | Swiderska M, Choromańska B, Dąbrowska E, Konarzewska-Duchnowska E, Choromańska K, Szczurko G, Myśliwiec P, Dadan J, Ladny JR, Zwierz K. The diagnostics of colorectal cancer. Contemp Oncol (Pozn). 2014;18:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 2. | Winkels RM, Heine-Bröring RC, van Zutphen M, van Harten-Gerritsen S, Kok DE, van Duijnhoven FJ, Kampman E. The COLON study: Colorectal cancer: Longitudinal, Observational study on Nutritional and lifestyle factors that may influence colorectal tumour recurrence, survival and quality of life. BMC Cancer. 2014;14:374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 3. | Janakiram NB, Rao CV. The role of inflammation in colon cancer. Adv Exp Med Biol. 2014;816:25-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 4. | Tsikitis VL, Larson DW, Huebner M, Lohse CM, Thompson PA. Predictors of recurrence free survival for patients with stage II and III colon cancer. BMC Cancer. 2014;14:336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 5. | Bhatt VR. Targeted therapy in metastatic colon cancer. Future Oncol. 2014;10:707-711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Mahon SM. Microsatellite testing in colon cancer. Oncol Nurs Forum. 2014;41:331-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 7. | Mathonnet M, Perraud A, Christou N, Akil H, Melin C, Battu S, Jauberteau MO, Denizot Y. Hallmarks in colorectal cancer: angiogenesis and cancer stem-like cells. World J Gastroenterol. 2014;20:4189-4196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 68] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 8. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9285] [Article Influence: 464.3] [Reference Citation Analysis (0)] |
| 9. | Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2843] [Cited by in RCA: 3084] [Article Influence: 181.4] [Reference Citation Analysis (0)] |
| 10. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7370] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 11. | Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 740] [Cited by in RCA: 745] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 12. | Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, Abdelnaby AA, Balch GC, Mansour JC, Yopp AC. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 13. | Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699-8707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1168] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 14. | Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, Nam SJ, Chun KH. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011;155:427-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 15. | Duan X, Hu J, Wang Y, Gao J, Peng D, Xia L. MicroRNA-145: a promising biomarker for hepatocellular carcinoma (HCC). Gene. 2014;541:67-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Li Y, Huang Q, Shi X, Jin X, Shen L, Xu X, Wei W. MicroRNA 145 may play an important role in uveal melanoma cell growth by potentially targeting insulin receptor substrate-1. Chin Med J (Engl). 2014;127:1410-1416. [PubMed] |
| 17. | Djaldetti M, Bessler H. Modulators affecting the immune dialogue between human immune and colon cancer cells. World J Gastrointest Oncol. 2014;6:129-138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Gregersen LH, Jacobsen A, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 19. | Li T, Leong MH, Harms B, Kennedy G, Chen L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J Gastroenterol. 2013;19:5615-5621. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 53] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 20. | Zhang J, Luo X, Li H, Yue X, Deng L, Cui Y, Lu Y. MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol Rep. 2014;32:115-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 21. | Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 601] [Cited by in RCA: 623] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 22. | Iio A, Takagi T, Miki K, Naoe T, Nakayama A, Akao Y. DDX6 post-transcriptionally down-regulates miR-143/145 expression through host gene NCR143/145 in cancer cells. Biochim Biophys Acta. 2013;1829:1102-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416-6424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 388] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 24. | Chen HC, Chen GH, Chen YH, Liao WL, Liu CY, Chang KP, Chang YS, Chen SJ. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100:1002-1011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 222] [Cited by in RCA: 236] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 25. | Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345-352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 316] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 26. | Sachdeva M, Mo YY. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70:378-387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 307] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 27. | Liu L, Zhang N, Liu J, Min J, Ma N, Liu N, Liu Y, Zhang H. Lentivirus-mediated siRNA interference targeting SGO-1 inhibits human NSCLC cell growth. Tumour Biol. 2012;33:515-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Prados J, Melguizo C, Roldan H, Alvarez PJ, Ortiz R, Arias JL, Aranega A. RNA interference in the treatment of colon cancer. BioDrugs. 2013;27:317-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 29. | Bailey KM, Wojtkowiak JW, Cornnell HH, Ribeiro MC, Balagurunathan Y, Hashim AI, Gillies RJ. Mechanisms of buffer therapy resistance. Neoplasia. 2014;16:354-364.e1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 30. | Tang X, Kuhlenschmidt TB, Li Q, Ali S, Lezmi S, Chen H, Pires-Alves M, Laegreid WW, Saif TA, Kuhlenschmidt MS. A mechanically-induced colon cancer cell population shows increased metastatic potential. Mol Cancer. 2014;13:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Yin Y, Yan ZP, Lu NN, Xu Q, He J, Qian X, Yu J, Guan X, Jiang BH, Liu LZ. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim Biophys Acta. 2013;1829:239-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Panza A, Votino C, Gentile A, Valvano MR, Colangelo T, Pancione M, Micale L, Merla G, Andriulli A, Sabatino L. Peroxisome proliferator-activated receptor γ-mediated induction of microRNA-145 opposes tumor phenotype in colorectal cancer. Biochim Biophys Acta. 2014;1843:1225-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |