Published online Feb 14, 2016. doi: 10.3748/wjg.v22.i6.2071
Peer-review started: January 16, 2015
First decision: March 26, 2015
Revised: August 4, 2015
Accepted: September 2, 2015
Article in press: September 2, 2015
Published online: February 14, 2016
Processing time: 373 Days and 2.9 Hours
AIM: To investigate whether IDH1R132C mutant in combination with loss of p53 and activated Notch signaling promotes intrahepatic cholangiocarcinoma (ICC) development.
METHODS: We applied hydrodynamic injection and sleeping beauty mediated somatic integration to induce loss of p53 (via shP53), activation of Notch [via intracellular domain of Notch1 (NICD)] and/or overexpression of IDH1R132C mutant together with the sleeping beauty transposase into the mouse liver. Specifically, we co-expressed shP53 and NICD (shP53/NICD, n = 4), shP53 and IDH1R132C (shP53/IDH1R132C, n = 3), NICD and IDH1R132C (NICD/IDH1R132C, n = 4), as well as NICD, shP53 and IDH1R132C (NICD/shP53/IDH1R132C, n = 9) in mice. Mice were monitored for liver tumor development and euthanized at various time points. Liver histology was analyzed by hematoxylin and eosin staining. Molecular features of NICD/shP53/IDH1R132C ICC tumor cells were characterized by Myc tag, Flag tag, Ki-67, p-Erk and p-AKT immunohistochemical staining. Desmoplastic reaction in tumor tissues was studied by Picro-Sirius red staining.
RESULTS: We found that co-expression of shP53/NICD, shP53/IDH1R132C or NICD/IDH1R132C did not lead to liver tumor formation. In striking contrast, co-expression of NICD/shP53/IDH1R132C resulted in ICC development in mice (P < 0.01). The tumors could be identified as early as 12 wk post hydrodynamic injection. Tumors rapidly progressed, and by 18 wk post hydrodynamic injection, multiple cystic lesions could be identified on the liver surface. NICD/shP53/IDH1R132C liver tumors shared multiple histological features of human ICCs, including hyperplasia of irregular glands. Importantly, all tumor cells were positive for the biliary epithelial cell marker cytokeratin 19. Extensive collagen fibers could be visualized in tumor tissues using Sirus red staining, duplicating the desmoplastic reaction observed in human ICC. Tumors were highly proliferative and expressed ectopically injected genes. Together these studies supported that NICD/shP53/IDH1R132C liver tumors were indeed ICCs. Finally, no p-AKT or p-ERK positive staining was observed, suggesting that NICD/shP53/IDH1R132C driven ICC development was independent of AKT/mTOR and Ras/MAPK signaling cascades.
CONCLUSION: We have generated a simple, non-germline murine ICC model with activated Notch, loss of p53 and IDH1R132C mutant. The study supported the oncogenic potential of IDH1R132C.
Core tip: We established a novel murine intrahepatic cholangiocarcinoma (ICC) model via hydrodynamic transfection of activated form of Notch1 (NICD), shP53 and IDH1R132C into the mouse liver. This study is the first to demonstrate that IDH1R132C mutant can cooperate with other oncogenes or tumor suppressor genes to promote ICC development in vivo. In addition, it provides the ICC research community with an innovative and convenient approach to generate IDH1R132C mutant ICC in mice. Finally, ICC induced by NICD/shP53/IDH1R132C provides a useful tool to study IDH mutant in ICC pathogenesis and a novel preclinical murine model for testing drugs against the deadly malignancy.
- Citation: Ding N, Che L, Li XL, Liu Y, Jiang LJ, Fan B, Tao JY, Chen X, Ji JF. Oncogenic potential of IDH1R132C mutant in cholangiocarcinoma development in mice. World J Gastroenterol 2016; 22(6): 2071-2080
- URL: https://www.wjgnet.com/1007-9327/full/v22/i6/2071.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i6.2071
Intrahepatic cholangiocarcinoma (ICC) is a deadly malignancy of the biliary epithelium arising within the liver. It is the second most common primary hepatic cancer, representing about 10% to 20% of all primary hepatic carcinomas[1,2]. While ICC remains a relatively rare malignancy worldwide, its incidence rate has been rising rapidly over the past several decades[3]. For the lack of apparent clinical symptoms, signs and deviant lab test results, ICC is generally diagnosed at the advanced stage. Treatment options for ICC are very limited. Indeed, there is no curative treatment except surgical resection at the early stage of ICC. The combination of gemcitabine and cisplatin is the first-line treatment for inoperable ICC patients[3-5]. However, standard chemotherapies only offer very limited benefit. Clearly, effective molecular targeted therapies are urgently needed for the treatment of ICC.
Molecular genetics underlying ICC pathogenesis remains poorly understood[6,7]. A rising number of genetics data points to a heterogeneous collection of underlying mutations in multiple oncogenes and tumor suppressor genes in human ICC, such as KRAS[8], BRAF[9], p53[10-12], SMAD4[11] and p16[13].
Recently, mutations in metabolic genes, which can reprogram tumor metabolism to stimulate cell growth and proliferation, have been identified in various human tumors[14]. These mutations are considered to be novel targets for cancer therapies. Isocitrate dehydrogenase (IDH) is an enzyme that participates in NADP+ or NAD+ dependent tricarboxylic acid (TCA) cycle. It localizes to the cytoplasm, mitochondrion, and peroxisome in cells, and catalyzes the oxidative decarboxylation of isocitrate to produce α-ketoglutarate (α-KG) and CO2. There are three isoforms in human: IDH1, IDH2 and IDH3. Mutations of IDH1 and IDH2 have been identified in multiple tumor types. For example, IDH1 mutants are found in brain tumors (including gliomas[15] and glioblastomas[16]), acute myeloid leukemia[17,18], thyroid carcinomas[19,20], cartilaginous tumors[21,22] and ICCs[23,24]. Previous studies have demonstrated that mutant IDH1 is oncogenic via regulating TCA cycle and increasing tumor cells’ dependence on oxidative mitochondrial metabolism[25]. IDH1 mutation alters its enzymatic activity, leading to the production of (D)-2-hydroxyglutarate (2-HG) rather than α-KG[26,27]. 2-HG is structurally similar to α-KG, and acts as a α-KG antagonist to competitively inhibit multiple α-KG-dependent dioxygenases, including lysine histone demethylases and the ten-eleven translocation (TET) family of DNA hydroxylases[28,29]. The oncogenic potential of IDH mutants in ICC was recently validated in vivo and the study demonstrated that IDH mutants function to block hepatocyte differentiation while promoting ICC pathogenesis[30]. However, the study utilized transgenic mice with IDH2R140Q or IDH2R172K mutant in combination with K-RasG12D to induce ICC formation in mice. In human ICCs, IDH2 mutant is relatively rare. On the other hand, IDH1R132C is the most common IDH mutant found in human ICC. According to COSMIC database, among all ICCs with IDH1 mutations, 47 of all the 76 identified mutations (about 61.8%) are substitution of arginine by cysteine at position 132, i.e., IDH1R132C. However, the in vivo oncogenic potential of IDH1R132C has not been investigated.
The abnormal activation of Notch signaling pathway plays critical roles in tumor development[31], including in ICC[32]. The contact of Notch ligands with their receptors induces proteolytic cleavage, and releases the Notch intracellular domain (NICD) resulting in the activation of the Notch pathway[33]. Previous studies demonstrate that the Notch pathway can control liver development by regulating biliary differentiation[34], and activated Notch 1 (NICD) synergizes with activated AKT signaling to promote ICC development[35]. As a canonical tumor suppressor gene, silencing of p53 has been implicated in ICC development[36,37]. The genetic interaction between Notch pathway and the tumor suppressor gene p53 in ICC development has not been studied.
In this study, we investigated the oncogenic potential of IDH1R132C in ICC development. We applied hydrodynamic transfection to overexpress IDH1R132C together with NICD1 or shP53 into mouse liver[38]. We found that co-expression of NICD/shP53, IDH1R132C/shP53 or NICD/IDH1R132C into the mouse liver did not lead to ICC formation in mice. In contrast, all NICD/shP53/IDH1R132C injected mice developed ICC starting at 12 wk post hydrodynamic transfection. Our results provided evidence, for the first time, that IDH1R132C mutant can promote ICC development in combination with activated Notch signaling and loss of p53. ICC induced by NICD/shP53/IDH1R132C therefore provides a useful tool to study IDH mutant in ICC pathogenesis and a novel preclinical murine model for testing drugs against ICC.
Hydrodynamic transfection induced mouse ICC used in this study was generated as previously described[38]. Mice were housed, fed, and monitored in accordance with protocols approved by the committee for animal research at the University of California, San Francisco (IACUC approval number: AN108577). Mice were monitored closely for liver tumor development. Mice with noticeable swelling abdominal mass or with a body condition score of 2 or less were euthanized by carbon dioxide inhalation followed by cervical dislocation according to the IACUC protocol.
All the constructs, including Myc tagged pT3-EF5α-NICD, shRNAmir-based silencing of p53 (pT2-shP53) and pCMV/sleeping beauty transposase (SB) used for mouse injection were previously described[35,38-40]. Flag tagged human IDH1 cDNA clone was kindly provided by Dr. Yue Xiong (University of Northern Carolina), and IDH1R132C mutant was generated using the QuickChange Site-Directed Mutagenesis kit (Stratagene, Santa Clara, CA). IDH1R132C was subsequently cloned into pT3-EF5α vector by the Gateway PCR cloning strategy (Invitrogen, Carlsbad, CA). Plasmids were purified using the Endotoxin-free Maxi-prep kit (Sigma, St. Louis, MO) before injecting into mice.
Wild type FVB/N mice were obtained from Charles River (Wilmington, MA). Hydrodynamic injections were performed as described previously[35,38,41]. Briefly, ten micrograms of the plasmids encoding pT3-EF5α-NICD (with Myc tag) and/or pT2-shP53 and/or pT3-EF5α-IDH1R132C (with Flag tag) along with sleeping beauty transposase (pCMV/SB) at a ratio of 25:1 were diluted in 2 mL saline (0.9% NaCl) for each mouse. Saline solution was filtered through a 0.22 μm filter and injected into the lateral tail vein of 6- to 8-wk-old FVB/N mice in 5-7 s.
Liver samples were fixed overnight in zinc formalin (Anatech Ltd.), embedded in paraffin, cut into 5-μm-thick sections, and placed on glass slides. The rabbit polyclonal anti-Myc (Invitrogen; dilution 1:1000), Flag tag (Cell Signaling Technology; dilution 1:200), anti-CK19 (Abcam; dilution 1:100), anti-Ki67 (Thermo Scientific; dilution 1:150), anti-p-AKT (Cell Signaling Technology; dilution 1:100), and anti-p-ERK1/2 (Cell Signaling Technology; dilution 1:100) antibodies were used. Briefly, slides were deparaffinized in xylene, rehydrated through a graded alcohol series and rinsed in PBS. Endogenous peroxidase was inactivated using 3% hydrogen peroxide in methanol. After boiled in 0.01 M citrate buffer (pH 6.0) for 10 min in a microwave oven, slides were incubated with primary antibodies overnight at 4 °C and subsequently with goat anti-rabbit biotin conjugated secondary antibody (1:500 dilution in PBS) for 30 min at room temperature. Then, signal was visualized using Vectastain ABC Elite kit (Vector Laboratories In, Burlingame, CA) and developed with 3,3’-diaminobenzidine (DAB). Sections were counterstained with hematoxylin (Sigma). Negative controls were performed with the same procedure, and PBS was incubated as a substitute for the primary antibodies.
Liver samples were fixed as described previously. Slides were deparaffinized in xylene, rehydrated through a graded alcohol series and rinsed in PBS, and incubated with Picro-Sirius red solution for 60 min. Slides were then rinsed in PBS and quickly dehydrated in xylene.
Students’t-test was used to compare tumor incident rates in mouse groups.
To study the oncogenic potential of IDH1R132C mutant, we generated pT3-EF5α-IDH1R132C plasmid which can be delivered into and stably expressed in mouse hepatocytes via sleeping beauty mediated somatic integration. It has been widely recognized that tumor development is a complex process and requires the activation of multiple signaling pathways. In our study, the IDH1 mutant, as a metabolic gene, is unlikely to be sufficient to promote any tumor formation. Deregulation of Notch pathway and p53 is known to be involved in human ICC pathogenesis. We attempted to develop mouse ICC models in which Notch, p53 and IDH1 are deregulated, either alone or in combination.
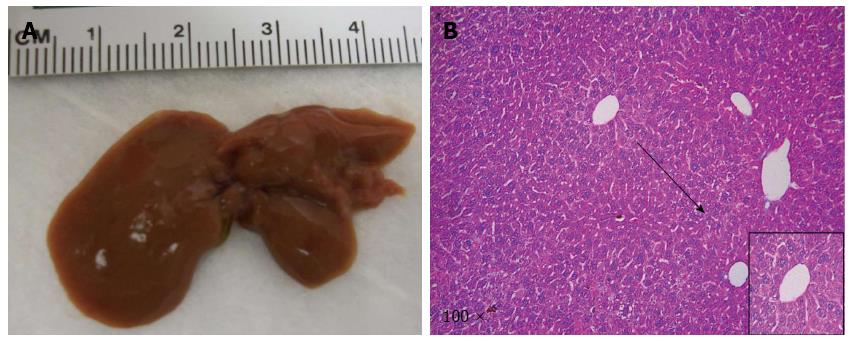
In our previous studies, we showed that NICD alone is only able to promote ICC development over long latency[35]. We therefore investigated whether co-expression of IDH1R132C accelerated NICD induced ICC development in mice. Towards this goal, we co-expressed NICD/IDH1R132C into wild type FVB/N mice (n = 4) by hydrodynamic transfection. All mice appeared to be healthy and were harvested at 22 wk post injection (Table 1). We found that the livers appeared to be normal in all mice; and none of the mice had visible nodules on the liver surface (Figure 1A). The result was corroborated by histological examination (Figure 1B). Together, the data suggest that IDH1R132C is unable to cooperate with NICD to promote liver tumor development in vivo.
| Code | Sex | Age (wk) | WPI | Tumor1 | Tumor number | Tumor size (mm) | |
| NICD/IDH1R132C | ND-F4.1 | F | 28 | 21 | N | 0 | 0 |
| ND-F4.2 | F | 28 | 22 | N | 0 | 0 | |
| ND-F4.3 | F | 28 | 22 | N | 0 | 0 | |
| ND-F4.4 | F | 28 | 22 | N | 0 | 0 | |
| NICD/shP53 | NP-F4.1 | F | 31 | 24 | N | 0 | 0 |
| NP-F4.2 | F | 31 | 24 | N | 0 | 0 | |
| NP-F4.3 | F | 31 | 24 | N | 0 | 0 | |
| NP-F4.4 | F | 31 | 24 | N | 0 | 0 | |
| shP53/IDH1R132C | PD-F3.1 | F | 27 | 20 | N | 0 | 0 |
| PD-F3.2 | F | 27 | 20 | N | 0 | 0 | |
| PD-F3.3 | F | 27 | 20 | N | 0 | 0 | |
| NICD/shP53/IDH1R132C | NDP-F4.1 | F | 16 | 9 | N | 0 | 0 |
| NDP-F4.2 | F | 19 | 12 | Y | 1 | 1.5 | |
| NDP-F4.3 | F | 19 | 12 | Y | 2 | 1, 2 | |
| NDP-F4.4 | F | 20 | 13 | Y | 2 | 1, 1 | |
| NDP-F4.5 | F | 20 | 13 | Y | 4 | 1, 1, 2, 3 | |
| NDP-F4.6 | F | 22 | 15 | Y | 3 | 1.5, 2, 2 | |
| NDP-F4.7 | F | 22 | 15 | Y | 5 | 1, 1.5, 2, 2, 3 | |
| NDP-F4.8 | F | 25 | 18 | Y | 6 | 1, 2, 2.5, 4, 4, 6 | |
| NDP-F4.9 | F | 25 | 18 | Y | 4 | 1, 1, 2, 4 |
A previous study showed that IDH mutants are associated with loss of p53 activity in human ICCs[23]. We therefore investigated whether loss of p53 expression is able to synergize with NICD and IDH1R132C to induce ICC formation in mice.
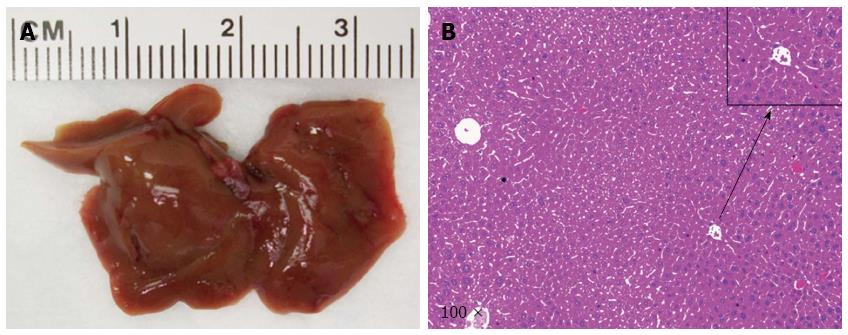
At the first step, we investigated whether loss of p53 (via shP53) accelerated NICD1 induced ICC development. We co-expressed NICD/shP53 into the mice (n = 4) by hydrodynamic transfection. All mice appeared to be healthy and were harvested at 24 wk post injection. We found that none of the mice showed any sign of liver tumor formation (Figure 2), suggesting that loss of p53 is unable to cooperate with NICD to induce ICC formation in mice.
As a second step, we examined whether loss of p53 and overexpression of IDH1R132C mutant could promote ICC development in mice. We co-expressed IDH1R132C/shP53 into the mice (n = 3) by hydrodynamic transfection. Again, all mice appeared to be healthy with no lesions on the liver (Figure 3). The result indicates that loss of p53 and overexpression of IDH1R132C cannot promote ICC development in vivo.
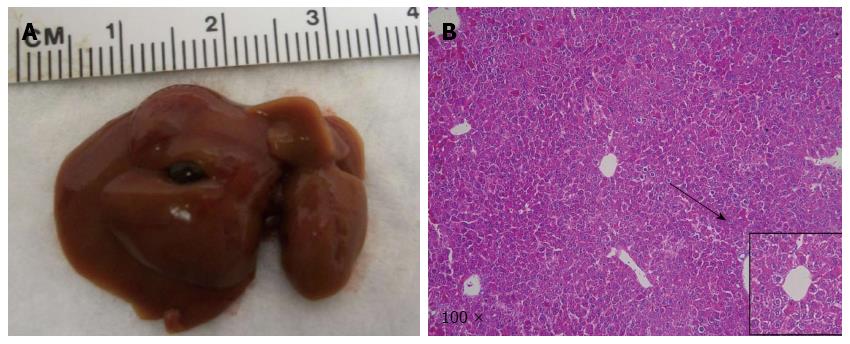
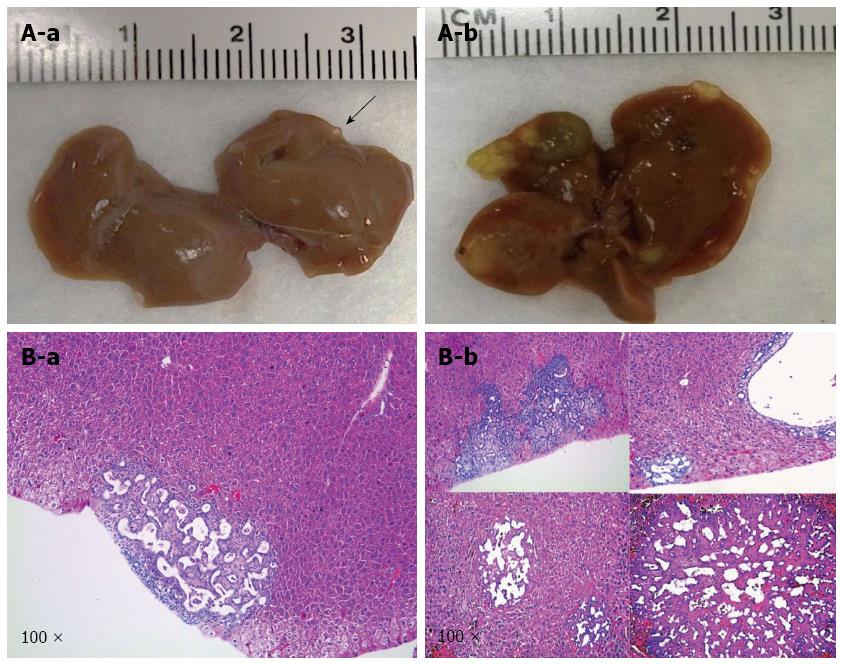
Next, we tested the hypothesis that IDH1R132C mutant cooperates with NICD and shP53 to promote ICC development in mice. In this study, a mixture of NICD/shP53/IDHR132C plasmids was hydrodynamically transfected to mice (n = 9). Mice were harvested at five time points [9 wk (n = 1), 12 wk (n = 2), 13 wk (n = 2), 15 wk (n = 2) and 18 wk (n = 2)] in order to better understand the tumor initiation and progress processes.
Macroscopically, livers of these mice appeared normal at 9 wk after injection. However, small and cyst-like lesions were present on the liver surface of both mice at 12 wk post-injection (Table 1 and Figure 4A-a). By 18 wk post injection, multiple cystic lesions could be identified on the liver surface (Table 1 and Figure 4A-b).
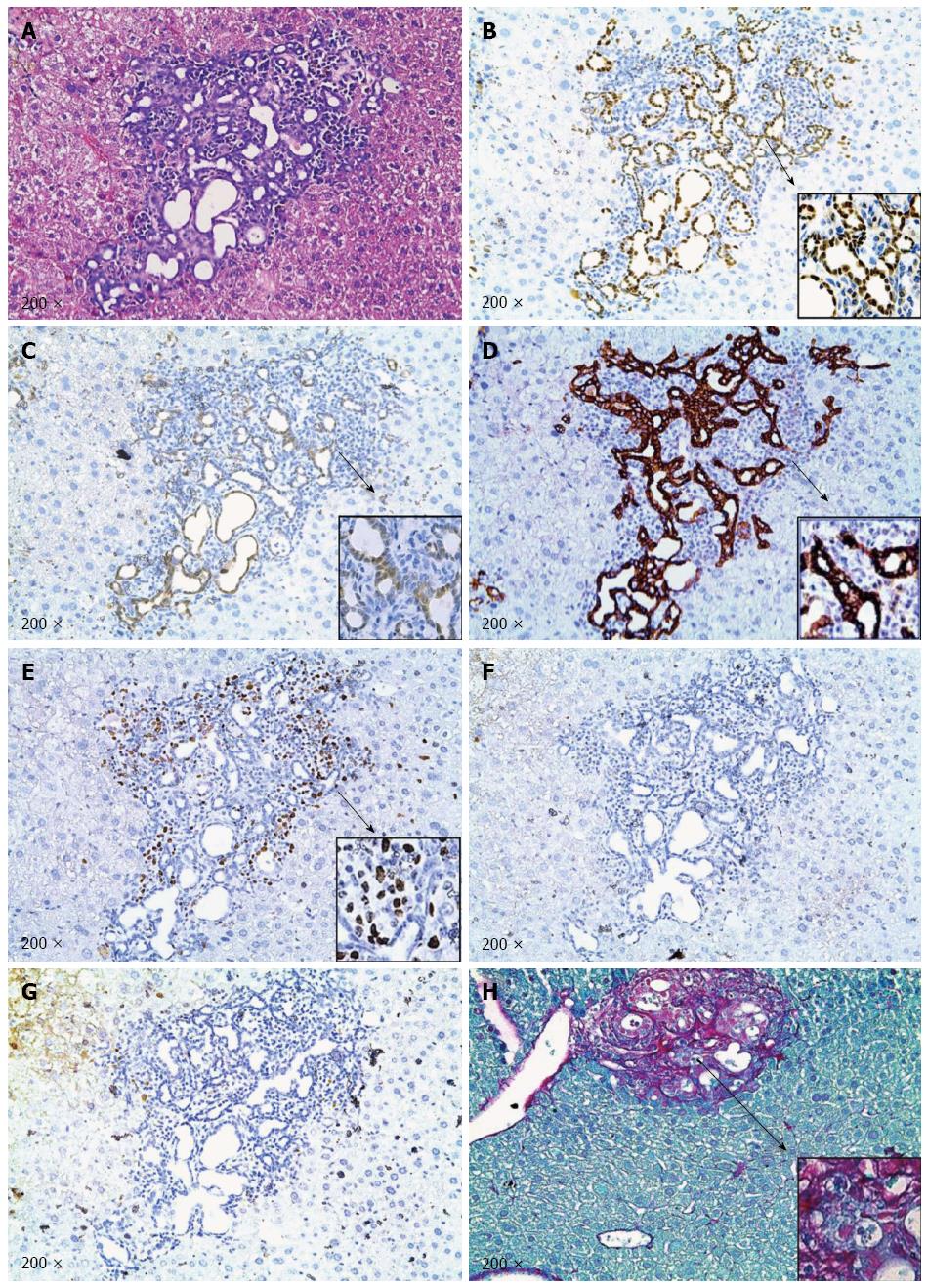
At the microscopic level, small tumors of ductular phenotype could be identified on the mouse liver at 12 wk post injection (Figure 4B-a). The tumors grew markedly by 18 wk post injection and exhibited either a ductular or cystic phenotype (Figure 4B-b). The tumors were indeed induced by the ectopically expressed genes, as tumor cells strongly and uniformly expressed Myc-tagged NICD and Flag-tagged IDHR132C (Figure 5B and C). The liver tumors shared multiple histological features of human ICCs, such as hyperplasia of irregular glands (Figure 5A). Importantly, immunohistochemial staining for the biliary marker cytokeratin 19 (CK19)[42,43] demonstrated that tumor cells were all CK19 positive (Figure 5D). Many cells in NICD/shP53/IDHR132C induced ICC were positive for the proliferation marker Ki67 (Figure 5E), which is in accordance with what has been shown in human high-grade ICC[44]. Furthermore, extensive collagen fibers could be visualized in tumor tissues using Sirus red staining (Figure 5H), duplicating the desmoplastic reaction observed in human ICC. Altogether, these results confirm that the tumors exhibited exclusively biliary differentiation (Figure 5), supporting the classification as murine ICC.
Activation of AKT/mTOR and Ras/MAPK signaling has been implicated in ICC development[35,40,41], we therefore investigated whether these pathways are activated in NICD/shP53/IDH1R132C ICC tumor cells. We found that neither p-AKT nor p-ERK1/2 was expressed in the ICC tumor cells (Figure 5F and G), suggesting that ICC development in this model was independent of activated AKT/mTOR and Ras/MAPK cascades.
In summary, our results indicate that NICD/shP53/IDH1R132C can drive ICC development in vivo (P < 0.01 when compared with other mouse cohorts). The results support the oncogenic potential of IDH1R132C mutant in ICC pathogenesis. ICC induced by NICD/shP53/IDH1R132C therefore provides a useful tool to further characterize the functional contribution of IDH1R132C mutant in ICC development. These mice also can be utilized as a novel and useful preclinical murine model for testing drugs against ICC.
Our present study was designed to evaluate the effect of mutant IDH1 in the development of liver tumors. Mutant IDH1 induces accumulation of 2-HG, which increases DNA methylation and decreases expression of tumor suppress genes, resulting in cellular proliferation in the tissue[23]. Thus, mutant IDH1 is generally considered a candidate oncogene in previous liver cancer studies. It is interesting to note that in human ICC samples, IDH1 mutant is associated with better prognosis[23], suggesting that most likely, mutant IDH1 per se is not oncogenic. Rather, it functions to modify the tumor progression initiated by other onocogenes or loss of tumor suppressor genes. In our current studies, we show that hydrodynamic transfection of NICD/shP53, shP53/IDH1R132C or NICD/IDH1R132C cannot promote liver tumor formation in mice. In striking contrast, overexpression of all the three factors, NICD, shP53 and IDH1R132C, is sufficient for ICC development in mice. The results support that mutant IDH1 is capable of stimulating ICC development in the combination of other genetic events, and in this case, activation of Notch pathway and loss of p53 tumor suppressor.
Recently, transgenic mice overexpressing IDH2 mutants (IDH2R140Q or IDH2R172K) were generated[30]. It was found that overexpression of IDH2 mutants did not lead to tumor development or any abnormal liver phenotype in the absence of liver injury. Importantly, the study demonstrated that co-expression of IDH2R192K and KRasG12D oncogene in the mouse liver was able to promote ICC development in mice. The tumor development required long latency with liver tumors detected between 33 and 58 wk of age. Mechanistically, the study suggested that it is likely that IDH2R192K and KRasG12D cooperated to initiate the activation and expansion of hepatic progenitor cells, eventually leading to ICC formation. However, IDH2 mutants are relatively rare in human ICCs, and it is not known whether IDH1 mutant can cooperate with other oncogenes to stimulate ICC development. Our study, therefore, is the first in vivo study to demonstrate that IDH1 mutant can indeed cooperate with other oncogenic stimuli, such as activated Notch and loss of p53, to promote ICC development in mice. Intriguingly, we did not observe any sign of progenitor or oval cell-like cell expansion in non-tumor liver tissues adjacent to the ICC lesions in NICD/shP53/IDH1R132C injected mice. It remains unknown whether NICD/shP53/IDH1R132C promotes ICC pathogenesis via progenitor cell expansion. As IDH1 is a metabolic gene, it would be also of great interest to further characterize the metabolic events in NICD/shP53/IDH1R132C ICC tumor cells. For example, it is important to analyze whether 2-HG accumulates in NICD/shP53/IDH1R132C tumor cells, and whether tumor cells show increased glycolysis.
Traditionally, genetically engineered mouse models, including knockout or transgenic mice, are required to demonstrate the oncogenic or tumor suppressor potential of the target genes and to illustrate how these genes contribute to tumor initiation and progression. Recently, however, hydrodynamic transfection, which combines hydrodynamic transfection and sleeping beauty mediated somatic integration, was developed and this technology has been applied to study HCC and ICC[38]. The most important features of hydrodynamic transfection reside in its flexibility and cost effectiveness. For example, using the traditional transgenic model, Saha and colleagues need to first generate LSL-IDH2R172K transgenic mice. These mice need to be maintained, and crossed to Alb-Cre and LSL-KRasG12D in order to generate the triple transgenic mice, i.e., Alb-Cre;LSL-KRasG12D;LSL-IDH2R172K to study whether IDH2R172K synergized with KRasG12D to induce ICC development in mice. Clearly, these studies are labor intensive, expansive and require a large number of mice. For other investigators to duplicate the study, one has to import the mice into his or her own institute. In contrast, hydrodynamic transfection is highly flexible, and one only needs wild type mice and plasmids required for injection in order to determine the genetic and biochemical crosstalk among multiple oncogenic pathways and their potential to promote liver tumor development in vivo. Our study therefore provides a convenient and cost-effective approach to generate IDH mutant related ICC murine models.
In our current study, we co-expressed IDH1R132C with NICD and shP53 in the mouse liver. It would be highly interesting to expand the study in order to further elucidate the functional contribution of IDH1 mutant in ICC pathogenesis. Other signaling pathways which are important for ICC tumorigenesis include activation of Yap[45], hypermethylation of Pten[46], and mutant Smad4[47], etc. Using hydrodynamic transfection, one can combine IDH1R132C with these genetic events to determine whether the combination can lead to ICC formation. In addition, it would be important to co-express other IDH1 mutants, such as IDH1R132L (about 13.2% of IDH1 mutations in human ICCs) and IDH1R132G (about 18.4% of IDH1 mutations in human ICCs) with NICD1 and shP53, and determine whether these IDH1 mutants have similar functions in vivo.
The authors would like to thank Dr. Yue Xiong of University of Northern Carolina for providing IDH1 cDNA plasmid.
Intrahepatic cholangiocarcinoma (ICC) is a deadly disease lacking effective treatment options. IDH1 mutation is identified in about 10% of all human ICC cases, and IDH1R132C represents the most frequent IDH1 mutation in ICC. However, the oncogenic potential of IDH1R132C in ICC pathogenesis remains unknown, especially in vivo.
In this study, the authors applied hydrodynamic transfection to study the oncogenic potential of IDH1R132C mutant in driving ICC development in vivo. Hydrodynamic transfection is an innovative cost effective and reliable method for generating preclinical murine models of liver cancer, including hepatocellular carcinoma and cholangiocarcinoma. Furthermore, IDH1 mutants are identified as critical genetic events in ICC pathogenesis, and it is important to clarify the oncogenic potential of IDH1 mutants using in vivo approaches.
In this manuscript, the authors described the establishment of a novel murine ICC model via hydrodynamic transfection of activated form of Notch1 (NICD), silencing of p53 (shP53) and IDH1R132C (NICD/shP53/IDH1R132C) into the mouse liver. This study is the first to demonstrate that IDH1R132C mutant can cooperate with other oncogenes or tumor suppressor genes to promote ICC development in vivo, supporting the oncogenic potential of IDH1R132C mutant during ICC pathogenesis.
ICC induced by NICD/shP53/IDH1R132C provides a useful tool to further characterize the functional contribution of IDH1R132C mutant in ICC development. These mice also can be utilized as a novel and useful preclinical murine model for testing drugs against ICC.
Hydrodynamic transfection: it is a novel approach for stable gene expression in mouse hepatocytes by hydrodynamic injection in combination with sleeping beauty mediated somatic integration. Specifically, to achieve the goal of long-term gene expression in hepatocytes, two plasmids are needed: one encoding the SB transposase, and the other encoding the gene of interest under a mammalian promoter and flanked by inverted repeats. The two plasmids are then mixed together, diluted into saline, and injected into lateral vein of mouse tail via hydrodynamic injection. Hydrodynamic transfection is now widely used to generate novel murine models of liver cancer.
Good article and interesting topic. This study provides a useful tool to further characterize the functional contribution of IDH1R132C mutant in ICC development.
P- Reviewer: Chen XP, Garancini M S- Editor: Yu J L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Brandi G, Farioli A, Astolfi A, Biasco G, Tavolari S. Genetic heterogeneity in cholangiocarcinoma: a major challenge for targeted therapies. Oncotarget. 2015;6:14744-14753. [PubMed] |
| 2. | Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1072] [Cited by in RCA: 1376] [Article Influence: 125.1] [Reference Citation Analysis (1)] |
| 3. | Mansour JC, Aloia TA, Crane CH, Heimbach JK, Nagino M, Vauthey JN. Hilar cholangiocarcinoma: expert consensus statement. HPB (Oxford). 2015;17:691-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 279] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 4. | Woo SM, Lee WJ, Kim JH, Kim DH, Han SS, Park SJ, Kim TH, Lee JH, Koh YH, Hong EK. Gemcitabine plus cisplatin versus capecitabine plus cisplatin as first-line chemotherapy for advanced biliary tract cancer: a retrospective cohort study. Chemotherapy. 2013;59:232-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Valle JW. BINGO: targeted therapy for advanced biliary-tract cancer. Lancet Oncol. 2014;15:778-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 6. | Kongpetch S, Jusakul A, Ong CK, Lim WK, Rozen SG, Tan P, Teh BT. Pathogenesis of cholangiocarcinoma: From genetics to signalling pathways. Best Pract Res Clin Gastroenterol. 2015;29:233-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Rizvi S, Borad MJ, Patel T, Gores GJ. Cholangiocarcinoma: molecular pathways and therapeutic opportunities. Semin Liver Dis. 2014;34:456-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 100] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 8. | Deshpande V, Nduaguba A, Zimmerman SM, Kehoe SM, Macconaill LE, Lauwers GY, Ferrone C, Bardeesy N, Zhu AX, Hezel AF. Mutational profiling reveals PIK3CA mutations in gallbladder carcinoma. BMC Cancer. 2011;11:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706-712. [PubMed] |
| 10. | Tannapfel A, Weinans L, Geissler F, Schütz A, Katalinic A, Köckerling F, Hauss J, Wittekind C. Mutations of p53 tumor suppressor gene, apoptosis, and proliferation in intrahepatic cholangiocellular carcinoma of the liver. Dig Dis Sci. 2000;45:317-324. [PubMed] |
| 11. | Hezel AF, Deshpande V, Zhu AX. Genetics of biliary tract cancers and emerging targeted therapies. J Clin Oncol. 2010;28:3531-3540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 171] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 12. | Khan SA, Thomas HC, Toledano MB, Cox IJ, Taylor-Robinson SD. p53 Mutations in human cholangiocarcinoma: a review. Liver Int. 2005;25:704-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Tannapfel A, Benicke M, Katalinic A, Uhlmann D, Köckerling F, Hauss J, Wittekind C. Frequency of p16(INK4A) alterations and K-ras mutations in intrahepatic cholangiocarcinoma of the liver. Gut. 2000;47:721-727. [PubMed] |
| 14. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47023] [Article Influence: 3358.8] [Reference Citation Analysis (5)] |
| 15. | Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 311] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 16. | Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807-1812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4786] [Cited by in RCA: 4462] [Article Influence: 262.5] [Reference Citation Analysis (0)] |
| 17. | Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1728] [Cited by in RCA: 1781] [Article Influence: 111.3] [Reference Citation Analysis (0)] |
| 18. | Wagner K, Damm F, Göhring G, Görlich K, Heuser M, Schäfer I, Ottmann O, Lübbert M, Heit W, Kanz L. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J Clin Oncol. 2010;28:2356-2364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 19. | Hemerly JP, Bastos AU, Cerutti JM. Identification of several novel non-p.R132 IDH1 variants in thyroid carcinomas. Eur J Endocrinol. 2010;163:747-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 20. | Murugan AK, Bojdani E, Xing M. Identification and functional characterization of isocitrate dehydrogenase 1 (IDH1) mutations in thyroid cancer. Biochem Biophys Res Commun. 2010;393:555-559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, Pollock R, O’Donnell P, Grigoriadis A, Diss T. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 703] [Cited by in RCA: 762] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 22. | Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 286] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 23. | Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, Andersen JB, Jiang W, Savich GL, Tan TX. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091-3100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 315] [Cited by in RCA: 318] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 24. | Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 515] [Cited by in RCA: 596] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 25. | Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, Slocum KL, Pu M, Lin F, Vickers C. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74:3317-3331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 219] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 26. | Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1720] [Cited by in RCA: 1623] [Article Influence: 108.2] [Reference Citation Analysis (0)] |
| 27. | Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739-744. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2913] [Cited by in RCA: 3008] [Article Influence: 188.0] [Reference Citation Analysis (0)] |
| 28. | Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2370] [Cited by in RCA: 2248] [Article Influence: 160.6] [Reference Citation Analysis (0)] |
| 29. | Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1562] [Article Influence: 120.2] [Reference Citation Analysis (0)] |
| 30. | Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, Gurumurthy S, Akbay EA, Sia D, Cornella H. Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 372] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 31. | Koch U, Radtke F. Notch signaling in solid tumors. Curr Top Dev Biol. 2010;92:411-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 91] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 32. | Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 202] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 33. | Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770-776. [PubMed] |
| 34. | Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 347] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 35. | Fan B, Malato Y, Calvisi DF, Naqvi S, Razumilava N, Ribback S, Gores GJ, Dombrowski F, Evert M, Chen X. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122:2911-2915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 401] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 36. | Furubo S, Harada K, Shimonishi T, Katayanagi K, Tsui W, Nakanuma Y. Protein expression and genetic alterations of p53 and ras in intrahepatic cholangiocarcinoma. Histopathology. 1999;35:230-240. [PubMed] |
| 37. | Hsu M, Sasaki M, Igarashi S, Sato Y, Nakanuma Y. KRAS and GNAS mutations and p53 overexpression in biliary intraepithelial neoplasia and intrahepatic cholangiocarcinomas. Cancer. 2013;119:1669-1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 38. | Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol. 2014;184:912-923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 39. | Wangensteen KJ, Wilber A, Keng VW, He Z, Matise I, Wangensteen L, Carson CM, Chen Y, Steer CJ, McIvor RS. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology. 2008;47:1714-1724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 40. | Evert M, Dombrowski F, Fan B, Ribback S, Chen X, Calvisi DF. On the role of notch1 and adult hepatocytes in murine intrahepatic cholangiocarcinoma development. Hepatology. 2013;58:1857-1859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc Natl Acad Sci USA. 2005;102:17059-17064. [PubMed] |
| 42. | Jain R, Fischer S, Serra S, Chetty R. The use of Cytokeratin 19 (CK19) immunohistochemistry in lesions of the pancreas, gastrointestinal tract, and liver. Appl Immunohistochem Mol Morphol. 2010;18:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 101] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 43. | Malato Y, Naqvi S, Schürmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121:4850-4860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 343] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 44. | Settakorn J, Kaewpila N, Burns GF, Leong AS. FAT, E-cadherin, beta catenin, HER 2/neu, Ki67 immuno-expression, and histological grade in intrahepatic cholangiocarcinoma. J Clin Pathol. 2005;58:1249-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 59] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 45. | Tao J, Calvisi DF, Ranganathan S, Cigliano A, Zhou L, Singh S, Jiang L, Fan B, Terracciano L, Armeanu-Ebinger S. Activation of β-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 46. | Sriraksa R, Zeller C, El-Bahrawy MA, Dai W, Daduang J, Jearanaikoon P, Chau-In S, Brown R, Limpaiboon T. CpG-island methylation study of liver fluke-related cholangiocarcinoma. Br J Cancer. 2011;104:1313-1318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, Myint SS, Rajasegaran V, Heng HL, Gan A, Zang ZJ, Wu Y, Wu J, Lee MH, Huang D, Ong P, Chan-on W, Cao Y, Qian CN, Lim KH, Ooi A, Dykema K, Furge K, Kukongviriyapan V, Sripa B, Wongkham C, Yongvanit P, Futreal PA, Bhudhisawasdi V, Rozen S, Tan P, Teh BT. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 397] [Article Influence: 30.5] [Reference Citation Analysis (0)] |