Published online Dec 21, 2016. doi: 10.3748/wjg.v22.i47.10341
Peer-review started: June 27, 2016
First decision: August 29, 2016
Revised: October 1, 2016
Accepted: November 16, 2016
Article in press: November 16, 2016
Published online: December 21, 2016
Processing time: 176 Days and 19.8 Hours
Toll like receptors plays a significant anti-viral role in different infections. The aim of this study was to look into the role of toll like receptor 4 (TLR4) in hepatitis B virus (HBV) infection.
Real time PCR was used to analyze the transcription of TLR4 signaling molecules, cell cycle regulators and HBV DNA viral load after triggering the HepG2.2.15 cells with TLR4 specific ligand. Nuclear factor (NF)-κB translocation on TLR4 activation was analyzed using microscopic techniques. Protein and cell cycle analysis was done using Western Blot and FACS respectively.
The present study shows that TLR4 activation represses HBV infection. As a result of HBV suppression, there are several changes in host factors which include partial release in G1/S cell cycle arrest and changes in host epigenetic marks. Finally, it was observed that anti-viral action of TLR4 takes place through the NF-κB pathway.
The study shows that TLR4 activation in HBV infection brings about changes in hepatocyte microenvironment and can be used for developing a promising therapeutic target in future.
Core tip: The study delves into the role of toll like receptor 4 (TLR4) in hepatitis B virus (HBV) infection. Inciting TLR4 brings about drastic changes in viral and host responses in hepatocyte niche. Stimulating TLR4 represses viral replication and alters important host gene expression. Virus induced G1/S cell cycle arrest is partially released on viral elimination which is corroborated with the expression of chief cell cycle regulators. Further, we have observed activation of epigenetic signatures H3K9Ac/H3K18Ac, on viral suppression post TLR4 activation. Thus, this work is important in perspective of the role of TLR4 during HBV infection in hepatocyte microenvironment.
- Citation: Das D, Sarkar N, Sengupta I, Pal A, Saha D, Bandopadhyay M, Das C, Narayan J, Singh SP, Chakravarty R. Anti-viral role of toll like receptor 4 in hepatitis B virus infection: An in vitro study. World J Gastroenterol 2016; 22(47): 10341-10352
- URL: https://www.wjgnet.com/1007-9327/full/v22/i47/10341.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i47.10341
Hepatitis B viral infection is a major global health problem. Hepatitis B virus (HBV) causes liver diseases in humans with variable degree of severity, which mostly depends on the strength of the host immune response against the virus. Though a large number of anti-HBV drugs and effective vaccines are available, Hepatitis B infection continues to pose a severe threat worldwide[1].
Toll like receptors (TLRs) are pattern recognition receptors, which are responsible for limiting the spread of pathogen by initiating an innate immune response, and promoting effective adaptive immune response[2]. Since, TLRs are weakly activated in HBV infection; several studies have shown that experimental activation of the TLRs using its specific ligands can be effective in HBV suppression, both in vitro and in vivo[3-6]. TLR4 is a cell surface receptor and initiates crucial innate immune response pathways[7]. Bacterial endotoxin liposaccharide are well-recognized TLR4 ligands[8]. Previous studies have shown that TLR4 plays crucial role in initiating an innate immune response against HBV and the virus strategically target this receptor to evade immune response[9]. An earlier report showed that saturated fatty acid serving as a potential ligand for TLR4 activated TLR4 pathway and accelerate the mechanism of inhibiting HBV viremia[10]. TLR4 and the signaling molecules participate in the pathogenesis of HBV related advanced liver diseases like liver cirrhosis and HCC[11].
The present study focuses on the role of TLR4 in HBV infection. Triggering TLR4 with its specific ligand activates a cascade of immune pathways and production of relevant inflammatory cytokines. This in turn represses the HBV infection and results in the release of G1/S arrest allowing the progression of host cell cycle stages. Epigenetic transcription activation marks like H3K9Ac and H3K18Ac that remain suppressed by the infection, is significantly restored on viral evasion. It was also investigated that the anti-viral activity of TLR4 is probably executed through the nuclear factor-κB (NF-κB) pathway. Taken together this study indicates that activation of TLR4 exhibits viral clearance by modulating several host factors. Thus, immuno-targeting of TLR4 could be a novel mechanism in treatment of hepatitis B infection.
A total of 8 liver biopsy samples from patients with chronic HBV infection were collected from Kalinga Gastroenterology Foundation (Cuttack, Orissa, India). Signed informed consent was obtained from patients and the study was approved by the institutional ethical committee. The diagnosis of patients with CHB was conformed according to the AASLD guidelines 2009.
Control human liver total RNA was purchased commercially from Ambion with a concentration of 1 mg/mL(AM7960). The RNA was extracted from the liver tissue of a patient having no history of HBV, HCV or HIV infection.
Hepatoblastoma cell lines HepG2 and HepG2.2.15 was maintained and plated in Dulbecco’s Modified Eagle Medium (DMEM) and Roswell Park Memorial Institute (RPMI 1640) media in 10% Fetal Bovine Serum in a 5% CO2 humidifier incubator at 37 °C.
TLR4 signaling pathway was stimulated with its synthetic ligand LPS-B5 Ultrapure (Invivogen San Diego, CA, United States). Previous studies have used this ligand to trigger TLR4 pathway to combat infections[12]. Cell culture supernatant was collected from HepG2.2.15 cells after treatment with 1, 2 and 4 μg/mL of LPS-B5 Ultrapure for 72 h.
Total DNA was extracted using Qiagen Blood mini-kit (Hilden, Germany). HBV viral load was quantified by real-time TaqMan PCR assay from the culture supernatant using NIBSC standards as described earlier[13]. HBeAg and HBsAg levels were analyzed by using commercial ELISA kits (Diasorin, S.P.A., Saluggia, Italy).
For pathway screening, HepG2.2.15 cells were stimulated with 4 μg/mL of LPS-B5 Ultrapuresingly or in conjunction with various inhibitors purchased from Sigma-Aldrich (St. Louis, MO, United States). 10 μmol/L NF-κB pathway inhibitor PDTC, 25 μg/mL of SP600125 (MAPK/JNK pathway inhibitor), 2 μmol/L of LY294002 (inhibitor of PI3K) and 10 μmol/L SB203580 (MAPK/p38 pathway inhibitor) were used.
Total RNA was extracted from liver biopsy specimens for the relative p53 expression. For the RNA expression from HepG2 and HepG2.2.15 cells, 1 × 106 cells were plated in 6-well plate for the different experiments. For the mRNA expression of TLR4, different cell cycle regulators and the different TLR4 signaling molecules from HepG2/HepG2.2.15 cells, total RNA was treated with DNase and was reverse transcribed with random hexamers using Revert Aid first-strand cDNA synthesis kit (MBI Fermentas). Real time PCR was performed in ABI 7200 SDS (Applied Biosystems, Foster City, CA, United States) using Power SYBR Green (Applied Biosystems). The target mRNA was relatively quantified and was normalized to the internal control (GAPDH). The PCR cycle number (CT) at which the exponential growth in the fluorescence from the dye (SYBR Green) passes a certain threshold was used to calculate the relative gene expression. 2-ΔΔCT was calculated to represent the relative quantification of the gene, where ΔCT (CT-target gene - CT-GAPDH). ΔΔCT= ΔCT (Experiment) - ΔCT (Control). Real time PCR was performed in triplicates for all the samples.
Primer sequences used in the paper are as follows:
GAPDH: FP-5’-AATCCCATCACCATCTTCCA-3’
RP-5’-TGGACTCCACGACGTACTCA-3’
TLR4: FP-5’-AGGATGATGCCAGGATGATGTC-3’
RP-5’-TCAGGTCCAGGTTCTTGGTTGAG-3’
p53: FP-5’-CCCAAGCAATGGATGATTTGA-3’
RP-5’-GGCATTCTGGGAGCTTCATCT-3’
p21: FP-5’-GGACAGCAGAGGAAGAC-3’
RP-5’-GGCGTTTGGAGTGGTAGAAA-3’
Rb: FP-5’-CGGGAGTCGGGAGAGGACGG-3’
RP-5’-CGAGAGGCAGGTCCTCCGGG-3’
Cyclin D1: FP-5’-AGCTCCTGTGCTGCGAAGTGGAAAC-3’
RP-5’-AGTGTTCAATGAAATCGTGCGGGGT-3’
Cyclin E: FP-5’-CAGCACTTTCT TG AGCAACACCCTC-3’
RP-5’-TCTCTAT GTCGCACCACTGATACCC-3’
Cyclin B1: FP-5’-AAGAGCTTTAAACTTTGGTCTGGG-3’
RP-5’-CTTTGTAAGTCCTTGATTTACCATG-3’
Cyclin A: FP-5’GCATGTCACCGTTCCTCCTT-3’
RP-5’CAGGGCATCTTCACGCTCTAT-3’
MyD88: FP-5’-AAGTTATTT GTTTACAAACAGCGACCA-3’
RP-5’-GGA AGAATGGCAAATATCGGCT-3’
Traf 6: FP-5’-GCCCAGGCTGTTCATAGTTT-3’
RP-5’-CAAGGGAGGTGGCTGTCATA-3’
TRIF: FP-5’-ACGCCATAGACCACTCAGCTTTCA-3’
RP-5’-AGGTTGCTCATCATGGCTTGGTTC-3’
IRF3: FP-5’-TCTTCCAGCAGACCATCTCC-3’
RP-5’-TGCCTCACGTAGCTCATCAC-3’
IRF7: FP-5’-CAGATCCAGTCCCAACCAAG-3’
RP-5’-GTCTCTACTGCCCACCCGTA-3’
TBK1: FP-5’-AGCGGCAGAGTTAGGTGAAA-3’
RP-5’-CCAGTGATCCACCTGGAGAT-3’
JNK: FP-5’-GTACTTGTATGAAACCACCTTTCT-3’
RP-5’-AGCATCTCTTTCTGAATCTATGAAG-3’
p38: FP-5’-TCTGCTTACCCTTCACCTTTG-3’
RP-5’-CACATCCTCACTCTGCTAGAAAT-3’
PI3K: FP-5’-AAGGGTGCTAAAGAGGAACAC-3’
RP-5’-CATGAGGTACTGGCCAAAGAT-3’
NF-κB: FP-5’-CGCATCCAGACCAACAACA-3’
RP-5’-TGCCAGAGTTTCGGTTCAC-3’
For western blotting, cells were harvested and lysed with Laemelli buffer containing 120 mMTris-HCl (pH 6.8), 20% glycerol and 4% SDS. Equal amounts of protein was run in a SDS PAGE and transferred on Nitrocellulose membrane (Millipore). Following incubation with primary antibody (overnight at 4 °C) and HRP conjugated secondary antibody (3 h at room temperature), the blots were developed using chemiluminescent substrate (Millipore). Densitometry measurements of bands were used for quantification of each marker by integrating each peak in Image J software.
Antibodies used are as follows:
Anti-H3K27me3 - 1:500(ACTIVE MOTIF: 39155)
Anti-H3K39me3 - 1:1000 (ACTIVE MOTIF: 39161)
Anti-H3K4me3 - 1:500 (ACTIVE MOTIF: 39159)
Anti-H3K36me3 - 1:500 (ABCAM: ab9050)
Anti-H3K9Ac - 1:1000 (MILLIPORE, 07-352)
Anti-H3K18Ac - 1:500(ACTIVE MOTIF: 39587)
Anti-H3 - 1:10000 (ABCAM: ab10799)
Anti-p53 - 1:5000 (SANTA CRUZ: SC-126)
Anti-NF-κB - 1:1000 (e-Bioscience14-6731-81)
Anti-β-actin - 1:2000 (Sigma: A2228-100UL)
Anti-Rabbit IgG-HRP - 1:10000 (SIGMA: A1949
Anti-Mouse IgG-HRP - 1:5000 (PROMEGA: W402B)
HepG2.2.15 cells were grown on coverslips and treated with 4 μg/mL of LPS-B5 Ultrapure as stated earlier. The cells were washed with phosphate buffered saline (PBS) three times and then fixed with 4% Paraformaldehyde (Sigma) in PBS for 10 min at room temperature (RT). Cells were washed thrice with PBS and permeabilized with 1% Triton X-100 in PBS for 10 min at RT, washed with PBS thrice followed by blocking with 3% BSA in PBS for 1 h at RT. Cells were incubated with Anti-NF-κB [1:1000 (e-Bioscience14-6731-81) for 1 h at RT, washed thrice with PBST (PBS + 0.05% Tween20) and then incubated with secondary antibody (Alexa-Fluor anti-rabbit 488 (1:1000), Invitrogen) for 1 h in dark at RT, followed by three washes with PBST. Coverslips were then mounted with mounting media containing DAPI (Sigma Aldrich). Fluorescence for Alexa and DAPI was visualized with Nikon Ti-E confocal microscope with A1RMP scanner head equipped with Nikon imaging software (NIS).
For cell cycle analysis, the cells were harvested and re-suspended in phosphate buffered saline (PBS). The cells were then fixed by adding double volume of chilled 70% ethanol (Merck) drop wise, with continuous vortexing. After incubating the mixture overnight at -20 °C, it was spinned and the cells were resuspended in 500 μL of PBS. The cells were then incubated with RNaseA (0.2 mg/mL) for 30 min followed by Propidium Iodide (50 μg/mL) (Sigma) at 37 °C for 1 h. Flow cytometric data acquisition was performed on BD FACS Calibur platform for 10000 events of each sample.
Cell proliferation was measured by MTT assay. HepG2.2.15 cells were plated in a 6 well format with a seeding density of 1 × 106 cells/well and grown in RPMI 1640 supplemented with 10% FBS. After 24 h, the media was removed and replaced with fresh media and treated with LPS-B5 Ultrapure. The plates were incubated at 37 °C in a humidified atmosphere of 5% CO2 for 72 h. At indicated time-points, relative cell numbers were determined by incubating cells with MTT3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide. The absorbance was taken at 570 and 650 nm. All experiments were performed in triplicates.
Cytokines were screened from the supernatant of the HepG2.2.15 cells treated with 4 μg/mL of LPS-B5 Ultrapure (seeded in 6 well plate with a seeding density of 1 × 106 cells/well) using cytokine bead array (CBA, BD Biosciences). The assay was conducted using 25 μL of sample and using a 10-point standard curve (ranging from 0 to 5000 pg/mL) was included for each cytokine measured (IL-6, IL-8, IL-10, IL-1β, IL-12p70 and Human TNF). The samples were analyzed using a BD Accuri C6 flow cytometer (BD Bioscience). FCAP Array software (BD version 3.1) was used to create the standard curves for each cytokine and convert the fluorescent MFI values into cytokine concentrations.
All data are expressed as mean ± standard deviation (SD) from at least three separate experiments. The differences between groups were analyzed using unpaired 2-tailed Student’s t-test. Differences were estimated at a statistical significance of P < 0.05.
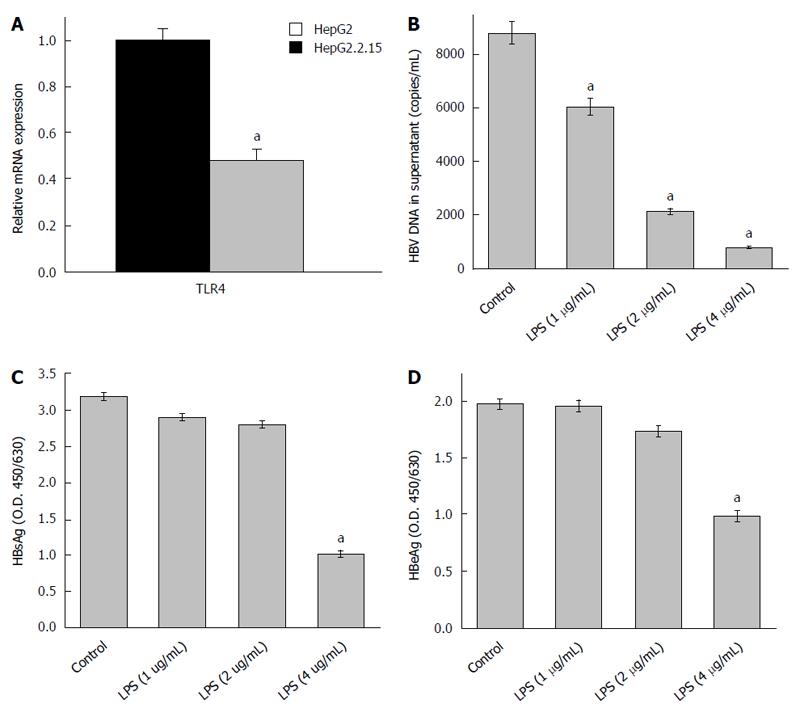
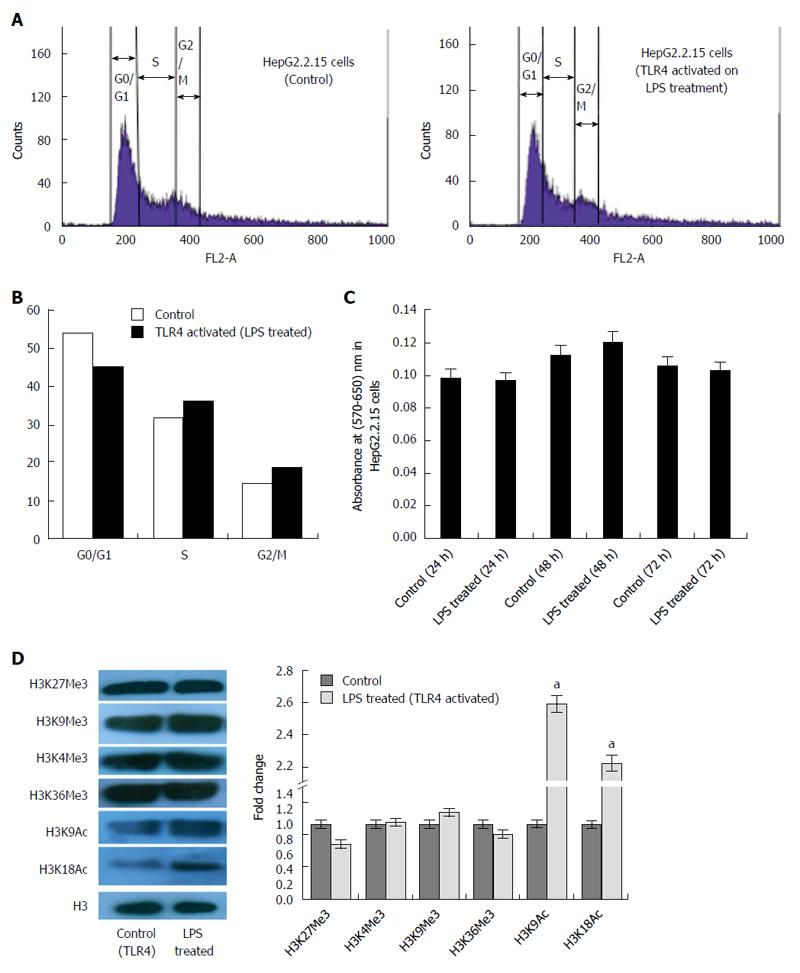
Previous reports have shown that TLR4 expression remains down regulated in HepG2.2.15 cells compared to HepG2 cells[14]. This was re-affirmed in our study (Figure 1A). TLR4 activation using LPS significantly decreased the HBV DNA viral load and proteins (HBsAg and HBeAg) in a dose dependent manner. The most effective dose of LPS, which repressed HBV replication, was observed to be 4 μg/mL (Figure 1B-D). Since, there was a marked reduction in HBV viremia and viral proteins (HBsAg and HBeAg) on stimulating the TLR4 pathway; we expected to observe changes in host cell cycle stages. It has already been well established that HBV affects host cell cycle stages. It has been earlier observed that HepG2.2.15 undergoes slow proliferation and approximately 80% of cells remain arrested in G1 phase due to high HBV DNA load[15]. It was thus expected that triggering TLR4 pathway would partially release the G1/S arrest due to viral suppression. Interestingly, TLR4 stimulation induces a release in G1/S arrest (Figure 2A) and percentage distribution of cells in the S-phase clearly indicated a G1 escape (Figure 2B). The effect of the ligand in causing cytotoxicity was next assessed by MTT assay. In the ligand concentration used in our assays, no significant cell death could be monitored (Figure 2C).
Recent studies have shown that some viruses subvert cellular epigenetic mechanisms and recruits host transcription factors to their advantage and modulate chromatin structure ensuring its successful existence in the host[16]. In order to identify the role of TLR4 affecting transcription of the chromatin template, global alterations of histone modifications was observed in HepG2.2.15 cells after triggering the TLR4 pathway. The transcription activation signatures (H3K9Ac and H3K18Ac) were significantly upregulated on stimulating the TLR4 signaling cascade. However, H3K27Me3, H3K4Me3, H3K9Me3 and H3K36Me3 histone marks did not show significant changes (Figure 2D). Thus, HBV infection represses the host transcription, which is observed through epigenetic changes. Stimulating TLR4 pathway with LPS, clearly rescues the transcription halt, by upregulating histone activation signatures. Thus HBV induced alteration of the epigenetic signatures; highlight the role of LPS in affecting cellular transcription profile of host cells.
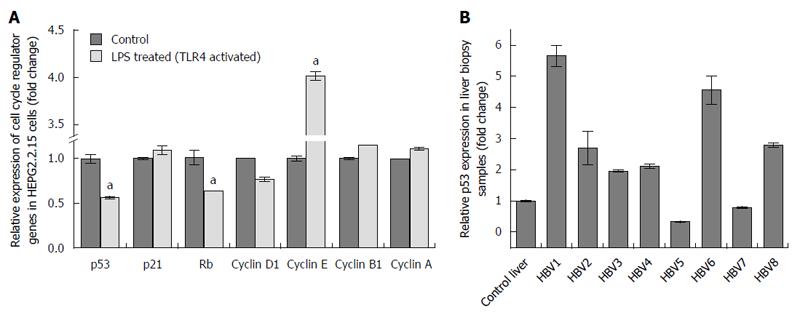
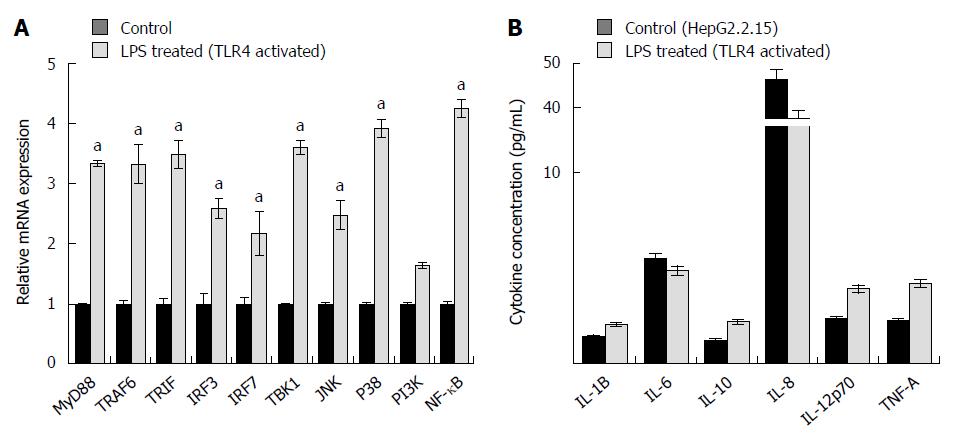
Transition through the different cell cycle stages were subsequently corroborated with the expression of various cell cycle regulators. Viral elimination imposed due to activation of TLR4 pathway, abolishes the G1/S arrest. Significantly, TLR4 activation up regulates the expression of Cyclin E. Similarly, the RNA expression of Cyclin D1, which remains upregulated in HepG2.2.15 cells, gets partially downregulated on stimulation with LPS followed by viral elimination. Retinoblastoma (Rb) gene playing an important role in G1/S arrest is markedly downregulated on TLR4 activation. However, Cyclin B1 and Cyclin A, involved in late S to G2 phase entry and G2/M transition respectively, are not significantly modulated showing the late S, G2 and M phases are not largely affected on viral elimination (Figure 3A). p53 considered as the “guardian of genome” is a tumor suppressor protein, which plays a crucial role in G1/S arrest as a result of DNA damage, oxidative stress, viral infections, nutrient depletion, etc. Though p53 RNA expression gets repressed on TLR4 activation compared to control HepG2.2.15 cells as a result of viral repression, the protein expression is partially upregulated (Figures 3A and 4A). However the p21 levels did not show significant changes. Interestingly this observation was corroborated in patient liver biopsy samples as well. Patients with CHB infection had a higher p53 expression compared to control liver sample. Liver biopsy samples HBV1, 2, 3, 4, 6 and 8 had significant upregulated p53 expression compared to that from control liver sample (Figure 3B). This showed that viral persistence directly affected the p53 levels. Modulation of TLR4 expression significantly affects the cell cycle regulators. We monitored the alteration of other host factors including cytokines to understand the immune response pathways which were triggered upon TLR4 activation. TLR4 activates a MyD88- dependent and independent signaling pathways. The chief players in the MyD88 signaling pathway including TRAF6 and TBK1 showed an increased RNA expression on triggering TLR4 with LPS. TRIF, IRF3 and IRF7, which are important signaling molecules in the MyD88-independent signaling pathway also showed elevated RNA expression (Figure 5A). Further, the principal downstream regulators that are triggered during activated host cell transcription including MAPKs (JNK and p38), NF-κB and PI3K showed upregulated RNA expression (Figure 5A). TLR4 activation triggers an important innate immune signaling pathway. The above-mentioned downstream regulators activate different transcription factors, which in turn upregulates the expression of relevant inflammatory cytokines. Cytokine bead array analysis showed that TLR4 activation turns-on the expression of a couple of inflammatory cytokines including IL-1B, IL-10, IL-12p70 and TNF-A (Figure 5B). These pro-inflammatory cytokines are important mediators that may play crucial role in viral elimination.
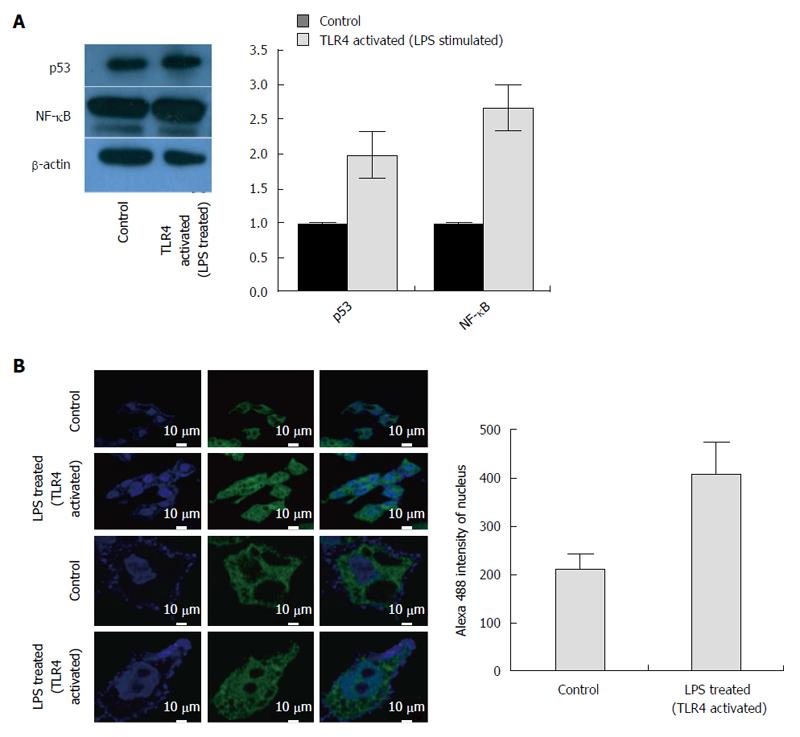
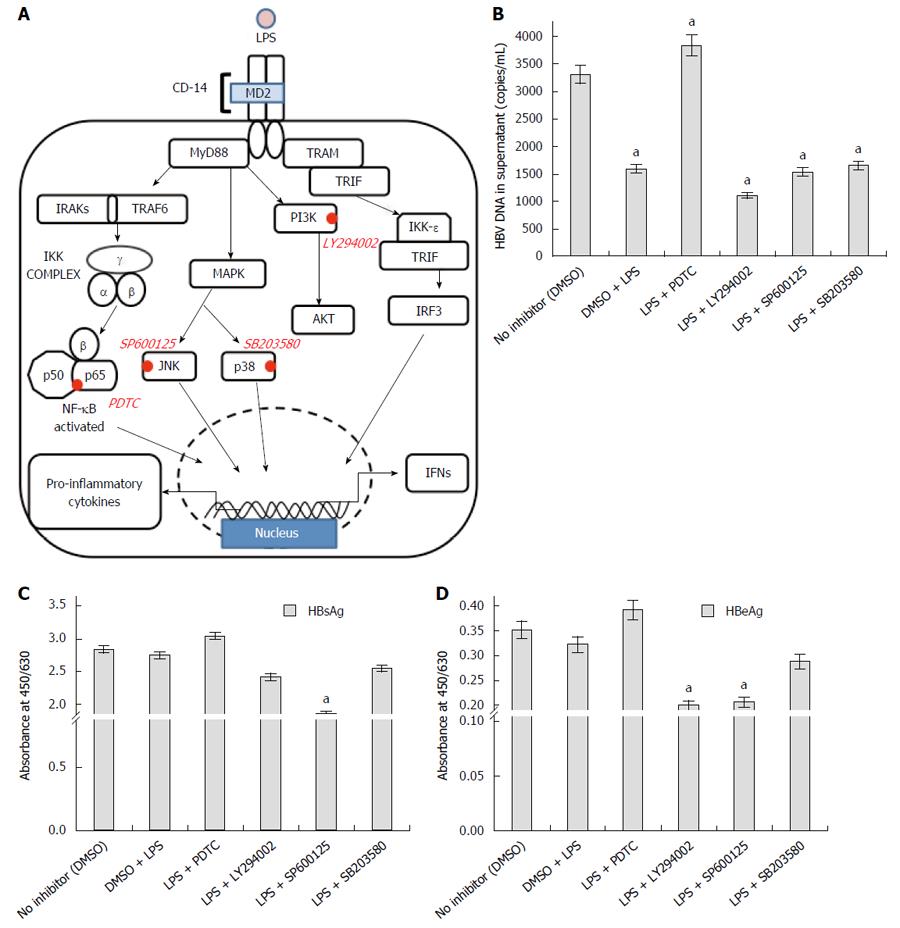
Subsequently we monitored the expression levels of the transcription factor p53 and NF-κB. Following a reduction in transcript level, the protein expression of p53 was slightly upregulated (Figure 4A). In comparison to the RNA expression, the change in NF-κB protein level was subtle (Figure 4A). However, TLR4 activation using LPS promoted the proteolytic degradation of Inhibitor-κB (I-κB) and allowed nuclear translocation of (NF-κB) p65 subunit (Figure 4B). The p65 subunit was expected to bind different parts of the DNA and affect host transcription. Thus, TLR4 activation and subsequent commencement of the signaling cascade was confirmed on LPS treatment. Since TLR4 activation, reduced the viral replication, we wanted to identify the pathway responsible for viral elimination. The TLR4 signaling cascade is observed in the figure (Figure 6A) and the important protein blockers targeting chief downstream regulators are marked. In an attempt to identify the pathway that is responsible for viral elimination, different chemical inhibitors (PDTC for NF-κB, LY294002 for PI3K, SP600125 for JNK and SB203580 for p38) were used, after which LPS was used to stimulate the TLR4 signaling pathway. These chemical blockers block MAPKs and NF-κB activation specifically, and have been used in several studies previously for pathway analysis[14,17]. Interestingly, on blocking NF-κB with its inhibitor eliminated the anti-viral role of TLR4. The HBV DNA load was maximal on blocking this pathway (Fig 6B). Further, the viral protein (HBsAg and HBeAg) production was greatly increased on inhibiting NF-κB pathway with PDTC (Figure 6C and D). Thus, it was concluded that the anti-viral action of TLR4 was probably operated through the NF-κB pathway.
In the present study, we have seen that stimulating the TLR4 pathway in HepG2.2.15 cells reduces Hepatitis B viral DNA and protein titre in a dose dependent manner. It was seen that stimulating TLR4 pathway with its specific ligand (LPS), activated a MyD88-dependent and independent pathway. The viral suppression on TLR4 activation possibly takes place through the NF-κB pathway and thus blocking it, impedes the antiviral action of TLR4. Further, it has already been seen that HBV affects the host cell cycle stages and induces a G1/S arrest. Interestingly, viral elimination induced by TLR4 activation, partially released the arrest after a G1 escape. The cell cycle status was further confirmed with the expression of different cell-cycle regulators. HBV plays an active role in upregulating the expression of p53. This was confirmed in liver biopsy samples and in HepG2.2.15 cells. The p53 expression gets repressed on viral elimination, following TLR4 activation. Since, there were significant changes in the host gene expression on TLR4 stimulation; we analyzed the status of host transcription by looking into the epigenetic modifications. Important host active signatures, H3K9Ac and H3K18Ac showed significant upregulation on pathway stimulation, highlighting the ability of LPS to affect the cellular micro-environment.
Previous studies have shown that HepG2.2.15 cells show slow proliferation compared to HepG2 cells. Results showed that Hepg2.2.15 cells undergo slower proliferation due to G1 arrest and not due to increase in apoptosis[15,18]. The present study shows that TLR4 stimulation modulates a couple of important host factors. It was also seen that HBV affects the host cell cycle stages. Stimulation of TLR4 pathway resulted in elimination of the virus, which in turn facilitated a G1 escape. The study was corroborated with the expression of different cell cycle markers. p53 along with the cyclins play a crucial role in G1 to S phase transition. The virus increases the level of p53 and induces a G1 arrest. It was noteworthy, that p53 expression was upregulated in the liver biopsy samples of CHB patients compared to control liver sample. However, no significant change of p21 expression was observed on TLR4 activation. Cyclins and the Retinoblastoma (Rb) gene play a vital role in establishing a G1/S control[19]. It has been shown earlier that Cyclin D1 levels are high during G1 phase to initiate DNA synthesis, but eventually it is suppressed to low levels in S-phase to ensure efficient DNA synthesis[20]. Down regulation of Cyclin D1 levels on TLR4 activation, was thus instrumental in G1 escape. Rb is one of the key regulators in G1/S transition. Rb can sequester different transcription factors that are essential for the progress of cell cycle. Studies show that at the late G1 restriction point, Cyclin E: Cdk2 complex inactivates Rb by hyperphosphorylating it at different sites and driving cells to the next phase of cell cycle[21]. Our studies also show that Cyclin E is significantly upregulated and Rb gene gets downregulated on viral elimination as a sequel to TLR4 activation. Thus probably a p53 independent pathway plays a potential role in G1/S escape on TLR4 stimulation resulting in viral evasion. Since there was no marked change in G2/M phases on the presence or absence of virus, the regulators in these phases showed no significant modification.
Recently, there has been a significant dynamics of virus-host chromatin interaction in determination of establishment of viral infection. Further, there are strong evidences of interplay of viral encoded proteins in host chromatin modulation[16]. Several viruses on host cell entry elicit an epigenetic reprogramming of the host chromatin by changing methylation and acetylation status and subverting chromatin-associated enzymes which leads to a change in host chromatin structure and gene expression[22]. Modulation of host epigenetic landscape upon virus infection has been observed in several cases. Adenoviral as well as SV40 virus infection leads to hypoacetylation of H3K18Ac. A recent report shows that H3K18Ac is downregulated upon HBV infection[23]. Thus LPS treatment which further restores the H3K18Ac status could be an ideal futuristic anti-viral therapy. It can also be observed that LPS treatment leads to significant increase of H3K9Ac mark in human genome. Thus epigenetic therapy to hyperacetylate H3K9 or H3K18 at gene promoters, which leads to a classical gene activation scenario, can be adopted as a novel strategy to eliminate HBV infection. The epigenetic reprogramming and modifications in gene expression promotes a permissive nuclear environment for the viral existence. Previous reports show that viruses including human γ-herpesviruses, Kaposi’s sarcoma-associated virus (KSHV or HHV8), Epstein-Barr virus (EBV), etc. epigenetically manipulate host genes involved in senescence, cell cycle progression, survival, immunity and inflammation to ensure its survival[24]. Interestingly, TLR4 activation showed no significant change in the histone methylation marks. However, the acetylation signatures (H3K9Ac and H3K18Ac), which are transcription activation marks showed increased levels on TLR4 activation. The virus induces host epigenetic modifications that would probably silence specific host gene clusters that are detrimental for viral infection. Thus TLR4 stimulation induces transcription activation which upregulates the expression of different genes that probably encode antiviral factors that lead to HBV elimination. The virus induced host modifications raise the possibility of a combination therapy, which can target multiple epigenetic restrictions to limit HBV infection.
It has been seen that TLR4 levels are altered in HBV infection and plays a very crucial role in its pathogenesis[9].A recent study showed that TLR4 plays a tumor promoting role in HBV- related HCC cells associated with regulation of ERK 1/2 activation and interaction with HBx[25]. Investigations show[26] that TLR4 on stimulation activates an inflammatory pathway and a host of signaling molecules are triggered that is responsible for production of inflammatory cytokines[27]. Previous reports show that TLR4 expression is greatly impaired in PBMCs from chronic HBV-infected patients in a Chinese population[28]. Studies have showed that supernatant from TLR4 stimulated Kupffer cells from C57/BL6 wild-typemice are effective in suppressing HBV replication[29]. It was observed in this study that a MyD88 dependent and independent pathway is triggered on TLR4 activation using its specific ligand. TLR4 activation enhanced the production of a number of pro-inflammatory cytokines, among which TNF-A, IL-10 and IL-1β is noteworthy. These cytokines probably play a major role in viral elimination and suppression of viral replication. A recent study showed that IL-1 and TNF-A have anti-viral mechanisms against HBV infection in hepatocytes cell lines, through the NF-κB pathway. The study further showed that the effect of the cytokines was mediated by induction of activation- induced cytidinedeaminase (AID). This is a relevant work in context of a link between pro-inflammatory cytokines and development of innate anti-viral defense[30]. Another study showed that IL-1 and TNF-A is produced spontaneously during interferon-alpha treatment of hepatocytes, suggesting their therapeutic potency during HBV infection[31]. It was investigated in our study that the viral elimination of TLR4 takes place through the NF-κB. Previous studies had shown that PI3K/AKT and MAPK/ERK pathways are responsible for viral suppression through TLR2[3]. However, the present study has been limited to NF-κB and MAPK signaling pathways. In the future we plan to look into additional pathways that may play role in viral repression through TLR4.
The study thus holds a promising immune-therapeutic approach for viral elimination based on triggering the TLR4 pathway. Though, HepG2.2.15 carrying HBV genome is the only cell line used for this study, but this is an extremely relevant model and sufficient work has been done on host-virus interaction[26,32,33] . However, we plan to investigate our work in other relevant cell lines and primary human hepatocytes in future.
DD is a recipient of Inspire Fellowship, DST, Government of India. We thank Mr. Chinmay Mondal for his exceptional technical assistance.
Hepatitis B virus (HBV) infection is a major health problem, worldwide. Despite the availability of different vaccines and anti-virals, HBV continues to be a massive threat globally. Immunotherapeutic approaches, are evolving to combat the infection. In this article, we look into the role of toll like receptor 4 (TLR4) in HBV infection and a possible mode of invading the infection by triggering the receptor, with its specific ligand.
TLRs are fundamental sensors of innate immunity and development of TLR-targeted therapies in the form of agonists and antagonists offers exciting and thrilling new possibilities for the prevention of virus-induced infectious diseases and management of virus induced harmful inflammatory responses. Different TLR agonists have been clinically assessed for treatment of chronic viral infections like HBV, hepatitis C virus (HCV) and human immunodeficiency virus (HIV).
Previous studies have showed that TLRs are potential targets in combating HBV infection. Recently it has been shown that TLR7 agonist GS-9620 has significant anti-HBV effects and is now in clinical trials. An earlier study showed that TLR8 agonist ssRNA40 could selectively activate liver-resident innate immune cells and could trigger the production of anti-viral cytokine IFN-γ in chronically HBV- and HCV- infected livers. This is the first study showing the effective role of TLR4 in HepG2.2.15 cells and its effect on a different range of host factors.
The present study looks into the role of TLR4 in HBV infection. It has been observed here that stimulating TLR4 represses the viral infection. The study opens up broader opportunities and scope to develop immune-therapeutic approaches by utilizing TLR4 to combat HBV infection.
Toll like receptors are modulators of innate immune response and is responsible for triggering elimination of different viral infections. The present study shows that TLR4 stimulation with its specific ligand/agonist lipopolysaccharide modulates a couple of host factors and represses HBV infection.
The aim of the manuscript is to focus on the role of TLR4 in HBV infection to further investigate a new therapeutic approach against HBV by immune-targeting TLR4. The main results of the present study showed that TLR4 activation repressed HBV infection and several changes in host factors, such as release in G1/S cell cycle arrest and modifications in host epigenetic marks. It was also observed that anti-viral action of TLR4 took place through NF-κB pathway.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: India
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chen CJ, Grasa L, Schvoerer E, Senapati S S- Editor: Yu J L- Editor: A E- Editor: Wang CH
| 1. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1700] [Cited by in RCA: 1712] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 2. | Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6034] [Cited by in RCA: 6318] [Article Influence: 300.9] [Reference Citation Analysis (0)] |
| 3. | Zhang X, Ma Z, Liu H, Liu J, Meng Z, Broering R, Yang D, Schlaak JF, Roggendorf M, Lu M. Role of Toll-like receptor 2 in the immune response against hepadnaviral infection. J Hepatol. 2012;57:522-528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 4. | Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol. 2005;79:7269-7272. [PubMed] |
| 5. | Gane EJ, Lim YS, Gordon SC, Visvanathan K, Sicard E, Fedorak RN, Roberts S, Massetto B, Ye Z, Pflanz S. The oral toll-like receptor-7 agonist GS-9620 in patients with chronic hepatitis B virus infection. J Hepatol. 2015;63:320-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 161] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 6. | Thompson AJ, Colledge D, Rodgers S, Wilson R, Revill P, Desmond P, Mansell A, Visvanathan K, Locarnini S. Stimulation of the interleukin-1 receptor and Toll-like receptor 2 inhibits hepatitis B virus replication in hepatoma cell lines in vitro. Antivir Ther. 2009;14:797-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Paik YH, Schwabe RF, Bataller R, Russo MP, Jobin C, Brenner DA. Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology. 2003;37:1043-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 509] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 8. | Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47:571-579. [PubMed] |
| 9. | Zare-Bidaki M, Tsukiyama-Kohara K, Arababadi MK. Toll-like receptor 4 and hepatitis B infection: molecular mechanisms and pathogenesis. Viral Immunol. 2014;27:321-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Zhang RN, Pan Q, Zhang Z, Cao HX, Shen F, Fan JG. Saturated Fatty Acid inhibits viral replication in chronic hepatitis B virus infection with nonalcoholic Fatty liver disease by toll-like receptor 4-mediated innate immune response. Hepat Mon. 2015;15:e27909. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Soares JB, Pimentel-Nunes P, Afonso L, Rolanda C, Lopes P, Roncon-Albuquerque R, Gonçalves N, Boal-Carvalho I, Pardal F, Lopes S. Increased hepatic expression of TLR2 and TLR4 in the hepatic inflammation-fibrosis-carcinoma sequence. Innate Immun. 2012;18:700-708. [PubMed] |
| 12. | Roberts BJ, Dragon JA, Moussawi M, Huber SA. Sex-specific signaling through Toll-Like Receptors 2 and 4 contributes to survival outcome of Coxsackievirus B3 infection in C57Bl/6 mice. Biol Sex Differ. 2012;3:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Chandra PK, Banerjee A, Datta S, Chakravarty R. G1862T mutation among hepatitis B virus-infected individuals: association with viral genotypes and disease outcome in Kolkata, Eastern India. Intervirology. 2007;50:173-180. [PubMed] |
| 14. | Sarkar N, Panigrahi R, Pal A, Biswas A, Singh SP, Kar SK, Bandopadhyay M, Das D, Saha D, Kanda T. Expression of microRNA-155 correlates positively with the expression of Toll-like receptor 7 and modulates hepatitis B virus via C/EBP-β in hepatocytes. J Viral Hepat. 2015;22:817-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Wang T, Zhao R, Wu Y, Kong D, Zhang L, Wu D, Li C, Zhang C, Yu Z, Jin X. Hepatitis B virus induces G1 phase arrest by regulating cell cycle genes in HepG2.2.15 cells. Virol J. 2011;8:231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Simões M, Rino J, Pinheiro I, Martins C, Ferreira F. Alterations of Nuclear Architecture and Epigenetic Signatures during African Swine Fever Virus Infection. Viruses. 2015;7:4978-4996. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Zhou H, Huang X, Cui H, Luo X, Tang Y, Chen S, Wu L, Shen N. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885-5894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 205] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 18. | Ozer A, Khaoustov VI, Mearns M, Lewis DE, Genta RM, Darlington GJ, Yoffe B. Effect of hepatocyte proliferation and cellular DNA synthesis on hepatitis B virus replication. Gastroenterology. 1996;110:1519-1528. [PubMed] |
| 20. | Yang K, Hitomi M, Stacey DW. Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Div. 2006;1:32. [PubMed] |
| 21. | Narasimha AM, Kaulich M, Shapiro GS, Choi YJ, Sicinski P, Dowdy SF. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife. 2014;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 327] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 22. | Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol. 2010;64:61-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 23. | Pandey V, Kumar V. Stabilization of SIRT7 deacetylase by viral oncoprotein HBx leads to inhibition of growth restrictive RPS7 gene and facilitates cellular transformation. Sci Rep. 2015;5:14806. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Knipe DM, Lieberman PM, Jung JU, McBride AA, Morris KV, Ott M, Margolis D, Nieto A, Nevels M, Parks RJ. Snapshots: chromatin control of viral infection. Virology. 2013;435:141-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 25. | Wang Y, Cai J, Zeng X, Chen Y, Yan W, Ouyang Y, Xiao D, Zeng Z, Huang L, Liu A. Downregulation of toll-like receptor 4 induces suppressive effects on hepatitis B virus-related hepatocellular carcinoma via ERK1/2 signaling. BMC Cancer. 2015;15:821. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 26. | Wu S, Kanda T, Nakamoto S, Jiang X, Nakamura M, Sasaki R, Haga Y, Shirasawa H, Yokosuka O. Cooperative effects of hepatitis B virus and TNF may play important roles in the activation of metabolic pathways through the activation of NF-κB. Int J Mol Med. 2016;38:475-481. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2377] [Cited by in RCA: 2484] [Article Influence: 112.9] [Reference Citation Analysis (0)] |
| 28. | Chen Z, Cheng Y, Xu Y, Liao J, Zhang X, Hu Y, Zhang Q, Wang J, Zhang Z, Shen F. Expression profiles and function of Toll-like receptors 2 and 4 in peripheral blood mononuclear cells of chronic hepatitis B patients. Clin Immunol. 2008;128:400-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 29. | Wu J, Lu M, Meng Z, Trippler M, Broering R, Szczeponek A, Krux F, Dittmer U, Roggendorf M, Gerken G. Toll-like receptor-mediated control of HBV replication by nonparenchymal liver cells in mice. Hepatology. 2007;46:1769-1778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 231] [Article Influence: 12.8] [Reference Citation Analysis (1)] |
| 30. | Watashi K, Liang G, Iwamoto M, Marusawa H, Uchida N, Daito T, Kitamura K, Muramatsu M, Ohashi H, Kiyohara T. Interleukin-1 and tumor necrosis factor-α trigger restriction of hepatitis B virus infection via a cytidine deaminase activation-induced cytidine deaminase (AID). J Biol Chem. 2013;288:31715-31727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 31. | Daniels HM, Meager A, Eddleston AL, Alexander GJ, Williams R. Spontaneous production of tumour necrosis factor alpha and interleukin-1 beta during interferon-alpha treatment of chronic HBV infection. Lancet. 1990;335:875-877. [PubMed] |
| 32. | Zhu X, Xie C, Li YM, Huang ZL, Zhao QY, Hu ZX, Wang PP, Gu YR, Gao ZL, Peng L. TMEM2 inhibits hepatitis B virus infection in HepG2 and HepG2.2.15 cells by activating the JAK-STAT signaling pathway. Cell Death Dis. 2016;7:e2239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Li L, Lei QS, Zhang SJ, Kong LN, Qin B. Suppression of USP18 Potentiates the Anti-HBV Activity of Interferon Alpha in HepG2.2.15 Cells via JAK/STAT Signaling. PLoS One. 2016;11:e0156496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |