Published online Aug 7, 2016. doi: 10.3748/wjg.v22.i29.6573
Peer-review started: March 22, 2016
First decision: May 12, 2016
Revised: May 25, 2016
Accepted: June 15, 2016
Article in press: June 15, 2016
Published online: August 7, 2016
Processing time: 129 Days and 15.2 Hours
The use of direct-acting antivirals (DAAs) to treat chronic hepatitis C has resulted in a significant increase in rates of sustained viral response (around 90%-95%) as compared with the standard treatment of peginterferon/ribavirin. Despite this, however, the rates of therapeutic failure in daily clinical practice range from 10%-15%. Most of these cases are due to the presence of resistant viral variants, resulting from mutations produced by substitutions of amino acids in the viral target protein that reduce viral sensitivity to DAAs, thus limiting the efficacy of these drugs. The high genetic diversity of hepatitis C virus has resulted in the existence of resistance-associated variants (RAVs), sometimes even before starting treatment with DAAs, though generally at low levels. These pre-existing RAVs do not appear to impact on the sustained viral response, whereas those that appear after DAA therapy could well be determinant in virological failure with future treatments. As well as the presence of RAVs, virological failure to treatment with DAAs is generally associated with other factors related with a poor response, such as the degree of fibrosis, the response to previous therapy, the viral load or the viral genotype. Nonetheless, viral breakthrough and relapse can still occur in the absence of detectable RAVs and after the use of highly effective DAAs, so that the true clinical impact of the presence of RAVs in therapeutic failure remains to be determined.
Core tip: The use of direct-acting antivirals (DAAs) to treat chronic hepatitis C has resulted in a significant increase in rates of sustained viral response as compared with the standard treatment of peginterferon/ribavirin. The presence of resistance-associated variants (RAVs) can reduce viral sensitivity to DAAs, thus limiting the efficacy of these drugs . As well as the presence of RAVs, virological failure is generally associated with other factors related with a poor response. Nonetheless, therapeutic failure can still occur in the absence of detectable RAVs and after the use of highly effective DAAs, so that the true clinical impact of the RAVs remains to be determined.
- Citation: Jiménez-Pérez M, González-Grande R, España Contreras P, Pinazo Martínez I, de la Cruz Lombardo J, Olmedo Martín R. Treatment of chronic hepatitis C with direct-acting antivirals: The role of resistance. World J Gastroenterol 2016; 22(29): 6573-6581
- URL: https://www.wjgnet.com/1007-9327/full/v22/i29/6573.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i29.6573
The introduction of direct-acting antivirals (DAAs) for the treatment of chronic hepatitis C virus (HCV) has represented a qualitative leap of enormous importance in the approach to HCV infection. The high global efficacy of DAAs, with rates of sustained viral response (SVR) of around 90%-95%, their good tolerability, safety profile and applicability, enable the disease to be cured in a large number of patients, with the resulting positive impact on the natural history of the infection and the associated costs[1-3].
Any of the stages in the life cycle of the virus can theoretically be a potential therapeutic target of DAAs. Currently, four classes of DAAs have been approved for the treatment of HCV. These act on three therapeutic targets: non-structural NS3/4A protease inhibitors (telaprevir, boceprevir, simeprevir, paritaprevir, grazoprevir), NS5A replication complex inhibitors (daclatasvir, ledipasvir, ombitasvir, elbasvir) and nucleos(t)ide (sofosbuvir) and non-nucleoside (dasabuvir) NS5B RNA dependent polymerase inhibitors[4,5].
In everyday clinical practice the rate of therapeutic failure with these DAAs is estimated at 10%-15%[6-14], mainly presenting as recurrence after treatment and occasionally as viral breakthrough during treatment[15]. Most cases are associated with the presence of drug-resistant viral variants, resulting from mutations produced by amino acid substitutions in the target virus protein that reduce viral sensitivity to DAAs, limiting their efficacy[15-18]. The great genetic variability of HCV means there may be resistance-associated variants (RAVs) even before starting DAA treatment, though usually at low levels. The presence of the RAVs before treatment does not appear to affect the SVR, so testing for basal resistance before treatment is not recommended for näive patients, though testing is recommended in patients who have already experienced therapeutic failure with DAAs in order to determine the retreatment strategy[1,15].
There also exists a certain individual susceptibility concerning eradication of HCV infection. This explains cases of treatment failure with DAAs and can involve such factors as the degree of liver fibrosis, the response to previous therapy, the viral load, the viral genotype or suboptimum interaction of the DAAs with the therapeutic targets due to the presence of viral variants. Indeed, RAVs are generally associated with therapeutic failure when some of these factors are also present[18]. Nevertheless, viral breakthrough and recurrence can still occur in the absence of detectable RAVs and even with the use of highly effective DAAs[19]. Thus, the true clinical impact of the presence of RAVs in therapeutic failure remains to be determined[20].
The HCV is an RNA virus belonging almost solely to the genus Hepacivirus of the family Flaviviridae. It possesses a wide genetic variability, with 7 genotypes described (numbered from 1 to 7), each with its respective subtypes (identified by letters). In addition to this inter-individual diversity there also exist intra-individual variations. These quasispecies are a heterogeneous mixture of genomes with a homology greater than 98% that can be present in the same person and which is due to the viral replication capacity (1012 virions per day) and the defective corrective capacity of the viral polymerase (1 error each 10000-100000 nucleotides)[21-23]. The appearance of mutations during infection shows the capacity of the virus to adapt to the environment and is associated with resistance-associated variants (RAVs) that limit the efficacy of DAAs[15].
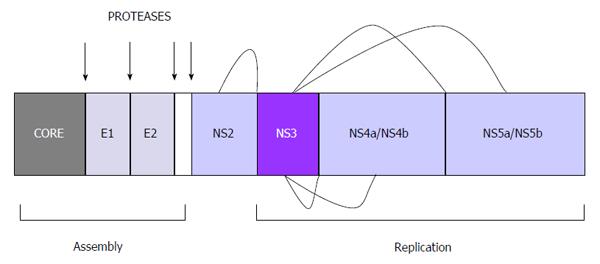
The virus itself is small (about 50 nm in diameter) and is composed of: (1) Coat or envelope. This is formed by two glycoproteins, called E1 and E2, linked to a lipid bilayer of the host cell and responsible for the pleomorphic structure of the virus[24]; (2) Nucleocapsid. Made of protein, it is formed by copies of the core protein, situated beneath the envelope and surrounding the genome; and (3) Genome. A positive sense single-stranded RNA formed by 9600 nucleotides. It possesses a single open reading frame (ORF) flanked by noncoding regions at the ends, called the 5’ untranslated region (UTR) and the 3´ UTR. These two regions of the virus are essential for RNA translation and replication. The 5’ end acts as the internal ribosome entry site (IRES) which initiates protein synthesis. The 3’ UTR also seems to participate directly in viral replication, though its mechanisms of action are less well known[25,26].
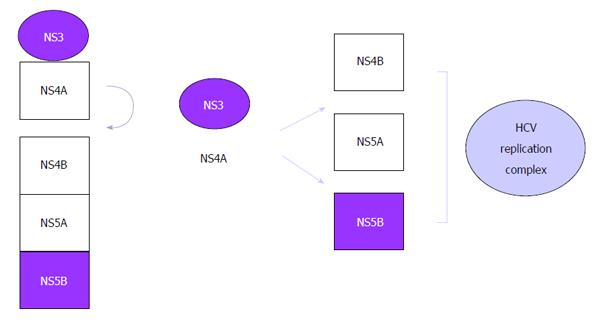
Translation of the reading frame originates a precursor polyprotein of some 3000 amino acids, which is processed by viral and host proteases giving rise to nonstructural and structural viral proteins, respectively[26]: (1) Three structural proteins: core (nucleocapsid), E1 and E2 (envelope). These constitute the main components of the virion; (2) p7 protein: viroporin that intervenes in the budding of virions by formation of pores in infected cell membranes; and (3) six nonstructural (NS) proteins: responsible for processing of the polyprotein and viral replication: (a) NS2 protein. Together with the N-terminal domain of NS3 it forms the NS2-NS3 protease, which processes the cleavage between NS2 and NS3; (b) NS3 and NS4A proteins. NS3 possesses a serine protease domain associated with the cofactor NS4A, forming the NS3-NS4 protease. It is one of the main targets of antiviral treatment, due to its crucial role in the replication cycle catalyzing the processing of the other nonstructural proteins at the sites NS3/NS4A, NS4A/NS4B, NS4B/NS5A and NS5A/NS5B. The products released are responsible for the formation of RNA (Figures 1 and 2); (c) NS4B protein. Essential for the formation of the replication complex; (d) NS5A protein. Metalloprotease with an important role in viral replication; and (e) NS5B protein. This is a viral polymerase, a central piece in the replication machinery. It is localized on the lipid membrane on the cytosolic side of the host endoplasmic reticulum.
Understanding the life cycle of the HCV has been essential in order to identify therapeutic targets and the subsequent development of antiviral agents. Ten years were necessary between the discovery of the virus and the time Lohmann et al[27] developed the first functional subgenomic replicons of HCV. Although these were unable to produce viral particles as they lacked structural proteins, they did enable understanding and characterization of the viral replication complexes, the development of new antiviral agents and the identification of resistance.
Later, use of replicon techniques enabled infecting viral particles to be obtained from the supernatant of cell cultures. Antiviral strategies advanced with production, with all chimeric virus genotypes highly replicating[26].
Although any step in the life cycle of the virus can in theory be a therapeutic target, it is the replication stage that has given the best therapeutic results so far. The commercialized drugs are inhibitors of these therapeutic targets that are essential for replication[28]: (1) NS3/4A protease. It contributes via proteolysis of the precursor polyprotein to the release of other proteins involved in replication; (2) viral NS5B polymerase. This is a central piece in replication and inhibitors have been developed of the active site of the polymerase as well as the allosteric sites (inducers of conformational changes in the enzyme that inactivate the replication complex); and (3) NS5A protein.
Viral resistance to treatment with DAAs is due to selection of viral variants that permit substitution of an amino acid in the region of the viral therapeutic target, making the virus less susceptible to the inhibitory activity of the drug[29]. Drug-resistant viral variants exist naturally prior to exposure to the drug, within the quasispecies already present in a patient. The great replication capacity of the HCV, with an estimated production and clearance of virions of 1010-1012 per day and a virion half-life of just 2-3 h together with errors in the viral replication mechanism, predispose to the constant appearance of a high number of genetically different viral variants that can confer resistance[30-32].
When a new virion is generated in a patient infected with a wild-type virus that is sensitive to a drug, this is estimated to have a 91% probability of having a non-mutated genome, an 8.7% probability of carrying one substitution, a 0.4% probability of carrying two substitutions, and a 0.013% probability of carrying three substitutions. It is also estimated that these variants can appear in 5%-20% of the total virus population as early as the second day after starting antiviral treatment. Of the 1012 virions that are generated daily, a mean of 8.7 × 1010 and 4.2 × 109 mutants are generated with a single or double site mutation, respectively[31].
Drug resistance can be measured in terms of the 50%-90% effective concentration (EC50 and EC90), that is the minimum concentration of the drug able to reduce values of the resistant variant by 50% or 90%. No agreement yet exists about the minimum value of increased concentration required to consider that a mutation confers resistance, as a viral variant can behave differently in vitro or in vivo, so results should be interpreted within a clinical context[11,29].
Viral resistance is usually associated with an “escape” pattern in which the mutation does not confer a loss of viral replication capacity in the presence of the drug and the viral variant maintains or rapidly recovers the pretreatment levels of replication.
Three levels of factors involved in the appearance of resistance to DAAs can be identified.
Not all mutations have the same likelihood of generating resistance. This depends on the quality or type of nucleotide alteration generating the resistance associated with the mutation[17,33].
Mutant variants associated with resistance generally have a lower replication capacity than wild strains, which are dominant. This makes it more difficult to detect them with usual techniques, requiring more sensitive techniques for their detection[34,35]. The selective pressure of a drug rapidly eliminates the wild variant, displacing it with the variant associated with resistance that then becomes the dominant strain. Persistent exposure to a drug can give rise to compensatory or secondary mutations that make replication in this mutant variant more effective, thus leading to reduced sensitivity to the drug[36].
The HCV subtype has been identified as a key determinant in the efficacy of DAAs. Patients with subtype 1a show a lower response to treatment than those infected with subtype 1b. Accordingly, subtyping is essential for the correct choice of therapy[17,37] .
Factors related with the HCV that have been associated with treatment failure after DAAs are: patients infected by genotype 1a receiving regimens with sofosbuvir plus simeprevir, sofusbuvir/ledipasvir or 3D regimen (ombitasvir/paritaprevir/ritonavir + dasabuvir); patients infected by genotype 3 receiving regimens with sofosbuvir plus ribavirin; the presence of variants associated with basal resistance to NS5A (only patients with cirrhosis); and the presence of the Q80K polymorphism (genotype 1a patients receiving regimens with simeprevir)[15].
These factors are determined by: 1. Genetic barrier to DAA resistance (type of nucleotide alteration (transition vs transversion) and number of mutations required to confer resistance to a particuar drug). DAAs with a low genetic barrier can require just transitions and/or one or two amino acid substitutions, whereas those with a high genetic barrier can require three or more nucleotide alterations and/or transversions 2. Drug exposure, defined as the minimum concentration of a drug required to produce inhibition of replication of 50%-90% of the virus (IC50-IC90)[33].
Clinically, therapeutic failure with DAAs has been associated with short-term regimens, poor adherence to treatment, and not giving ribavirin[15].
Factors involved in the immune response or the distribution of the drug to target cells, as well as genetic factors or the degree of liver fibrosis at the start of treatment, can all favor the appearance of resistance. One of the main causes of the appearance of viral escape variants and the resulting failure of antiviral therapy is specific CD8+ T cell exhaustion caused by continued exposure to the virus. Studies have shown that treatment with DAAs can restore the damage undergone by the specific CD8+ T cells, unlike interferon-based therapies. This would explain the success of DAAs in eradicating HCV infection[38,39].
Other factors associated with treatment failure after DAAs include male gender, the IL28B non CC genotype, the cirrhotic state, null responders to previous treatment with peginterferon/ribavirin (PEG/RBV), and previous failure to multiple DAAs[15,18].
The detection of RAVs is usually done with population sequencing techniques, with a detection sensitivity of about 20% among HCV quasispecies. More sensitive techniques, like clonal and deep sequencing, can improve the detection frequency of viral variants to 0.5%-1%[40]. Even so, no frequency cut-off has been established that is clinically relevant to predict treatment failure[18].
Currently approved NS3/4A inhibitors (telaprevir, boceprevir, simeprevir, paritaprevir, grazoprevir) posses a relatively low genetic barrier to resistance, with wide cross-resistance between them all; simple amino acid changes can be associated with an important loss of sensitivity. Grazoprevir, a second-generation NS3 inhibitor possesses a high genetic barrier, though a few RAVs (R155, A156, D168) have also been found to be associated with treatment failure[41]. Resistance to almost all protease inhibitors is due to substitutions at key points in NS3 (Arg 155, Ala 156, Asp 168)[17,42,43]. These variants usually have a low replication capacity, which explains the relatively low likelihood of detection as well as rapid replacement by the wild-type after treatment. Their natural frequency of appearance in genotype 1 patients is 0.1%-3.1%. The Q80K variant, however, differs in that it maintains its replication capacity and relatively high frequency associated with genotype 1a in certain populations (North America: 48%, South America: 9%, Europe: 19%)[44]. A few studies[45,46] have found loss of RAV detectability in 8-14 mo associated with boceprevir, telaprevir and simeprevir.
First-generation NS5A inhibitors approved so far (daclatasvir, ledipasvir, ombitasvir) posses both a low genetic barrier to resistance and wide cross- resistance. The second-generation NS5A inhibitor, elbasvir (pending approval), on the other hand, has shown a higher genetic barrier[47,48]. The most common mutations are associated with substitutions in Met 28, Gln 30, Leu 31, Pro 32, and Tyr 93[49,50]. Their natural frequency of presentation ranges from 0.3% to 2.8%, and they persist for 1-2 years in 85% of patients who have experienced treatment failure, maintaining their replication fitness[51-54].
Sofosbuvir, the only NS5B nucleos(t)ide inhibitor commercialized, has shown a high genetic barrier in all clinical studies with no appearance of breakthrough in regimens involving either monotherapy or in combination with other DAAs[55-59]. Nevertheless, substitutions have been identified in NS5B (S282T/G/C/R, M289L, I293L, C316N, V321A, I434M L159F/L320F, T179A) associated with resistance to sofosbuvir, as well as compensatory mutations such as M343T and H479P that can determine recovery of viral replication capacity in the presence of the main mutation S282T[60-62]. So far the natural presence of RAVs to sofosbuvir has not been detected, probably due to the marked loss of replication capacity of the mutation S282T, which would also explain why no breakthrough has been detected during treatment with sofosbuvir. Patients who experience viral recurrence after finishing treatment with sofosbuvir who are found to have S282T usually eliminate the mutation rapidly after just a few weeks[63,64].
Dasabuvir, the only non-nucleoside NS5B inhibitor so far approved, is however considered to have a low genetic barrier[65,66]. The pre-existence of RAVs associated with dasabuvir occurs in 0.2%-3.1% of genotype 1 patients. RAVs like C316N have a frequency of 10%-35% in genotype 1b. Overall, RAVs to dasabuvir remain detectable at 6 and 12 mo in 75% and 57% of patients, respectively[51].
The true impact of naturally-present resistant mutations in näive patients on treatment efficacy, as well as the impact of mutations resulting from earlier treatment on the efficacy of future therapy with the same or different DAAs are issues that remain to be clarified[16,20,67].
RAVs are not detected in all patients with virological failure to treatment. RAVs are almost always detected in patients who experience virological breakthrough, whereas in patients with viral recurrence the detection frequency of RAVs can vary from 53% to 91%, depending on the treatment regimen, the DAA used, and the duration of treatment. In addition, the sensitivity to the sequencing technique used, the low frequency of strains with RAVs, the low replication capacity of these or their rapid disappearance and reversion to wild-type, can all explain the lack of detectability of RAVs in many of these cases; in addition to the use of short antiviral regimens that may not completely eliminate the virus and account for recurrence by the predominantly wild-type variant[5,18,20,67].
Various studies[68-70] have shown the pre-existence of variants resistent to NS3/4A, NS5A and NS5B inhibitors in patients with no previous DAA treatment. At the moment basal determination of the Q80K polymorphism before treatment with triple therapy with simeprevir (PEG/RBV + simeprevir) is only recommended in näive genotype 1a patients, given the naturally high prevalence of this variant in patients with this genotype and its clear association with a low sustained viral response after treatment with this regimen[1,2,35,71,72]. Nonetheless, the presence of the Q80K variant in regimens using simeprevir plus sofosbuvir does not appear to influence the result significantly[73,74].
In general, no significant differences have been found concerning the pre-existence of mutant variants between responders and non-responders. The occasional study[75] did find that the basal presence de R155K/T/Q was associated with treatment failure in genotype 1a patients. Other studies[68], though, found no differences in the prevalence of DAA-resistant mutations between genotype 1a and genotype 1b responders.
It is not known how long mutant variants selected after previous treatment can remain in the viral population of the patient nor what their true impact is on future therapy. Some studies have shown the persistence over several years of resistant variants after treatment with telaprevir and boceprevir[52] and after combination therapy with NS5A inhibitors[76].
It remains to be determined whether the presence of DAA-resistant mutations with a high resistance barrier has important clinical relevance in treatment failure.
The best way to prevent the appearance of resistant variants is to achieve rapid and deep viral suppression with the first treatment, using a combination of various drugs that have a potent antiviral effect, high genetic barrier to resistance, different mechanisms of action and no cross-resistance[15] .
Current scientific evidence concerning the recommendations for retreatment of patients who have experienced failure to an earlier treatment with DAAs is still very limited. Generally, it would appear logical to give retreatment using regimens of two or three DAAs, including one with a high genetic barrier (in this case sofosbuvir) and without cross-resistance, during 12 wk accompanied by ribavirin, or 24 wk if it is not possible to give ribavirin[1,15] .
In patients who have experienced failure after triple therapy with boceprevir or telaprevir, retreatment with sofosbuvir plus daclatavir or sofosbuvir plus ledipasvir has achieved SVR rates of 94%-100%, without being influenced by the presence of NS3 RAVs[57,77].
Patients who have experienced failure to sofosbuvir plus ribavirin or sofosbuvir plus PEG/RBV can be retreated with sofosbuvir plus simeprevir if they are genotype 1 or 4, or with sofosbuvir plus daclatasvir for all genotypes, sofosbuvir plus ledipasvir for genotypes 1, 4, 5 and 6, or with the 3D regimen (ombitasvir/paritaprevir/ritonavir + dasabuvir) for genotype 1 and the 2D regimen (ombitasvir/paritaprevir/ritonavir) for genotype 4[1,2,15].
Patients who experience failure to PEG/RBV and simeprevir can be treated with sofosbuvir plus ledipasvir or daclatasvir (genotypes 1 and 4). Those who fail after sofosbuvir plus simeprevir can be retreated with sofosbuvir plus daclatasvir or ledipasvir (genotypes 1 and 4) whilst those who fail after sofosbuvir plus daclatasvir or ledipasvir can be retreated with sofosbuvir plus simeprevir (genotypes 1 and 4). The best strategy for failures to treatment for genotypes 2, 3, 5 and 6 who have already used sofosbuvir plus daclatasvir or ledipasvir is unclear; retreatment with the same regimen adding ribavirin or prolonging treatment for 24 wk may be an option[1,2,15].
Failures with the 3D combination can be retreated using a combination with sofosbuvir or adding PEG/RBV to the 3D regimen. The safety and efficacy of regimens combining three drugs (sofosbuvir, a protease inhibitor and a NS5A inhibitor) is not known[1,15].
Nonetheless, unless the need for retreatment is urgent one option may be to wait for better evidence and new drugs. No doubt this evidence will appear in the coming months.
Despite the great efficacy of therapies using DAAs, the high number of patients currently infected with HCV who are already receiving or will receive treatment, will lead to the appearance of a considerable number of patients with virological failure to these treatments. The use of regimens with drugs having a high antiviral activity and high genetic barrier for resistance means that basal resistance testing is not generally necessary prior to starting new treatment. It does, however, seem important to bear in mind factors that may predict the response, such as the genotype, subtype, histological damage, or prior treatments, in order to select a suitable therapeutic regimen and avoid possible therapeutic failure. Patients who have experienced virological failure to DAAs could warrant a resistance test prior to retreatment, and it is recommended to use other DAAs without cross-resistance, prolong the treatment, add ribavirin or even the joint use of peginterferon. Nonetheless, further studies are still required in order to define a suitable strategy in these cases.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Larrubia JR, Osna NA, Waheed Y S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
| 1. | European association for the study of the liver. EASL Recommendations on Treatment of Hepatitis C 2015. J Hepatol. 2015;63:199-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 877] [Cited by in RCA: 910] [Article Influence: 91.0] [Reference Citation Analysis (0)] |
| 2. | AASLD/IDSA HCV Guidance Panel. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus. Hepatology. 2015;62:932-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 912] [Cited by in RCA: 992] [Article Influence: 99.2] [Reference Citation Analysis (0)] |
| 3. | González-Grande R, Jiménez-Pérez M, González Arjona C, Mostazo Torres J. New approaches in the treatment of hepatitis C. World J Gastroenterol. 2016;22:1421-1432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 115] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (4)] |
| 4. | Vermehren J, Sarrazin C. New HCV therapies on the horizon. Clin Microbiol Infect. 2011;17:122-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 5. | Perales C, Quer J, Gregori J, Esteban JI, Domingo E. Resistance of Hepatitis C Virus to Inhibitors: Complexity and Clinical Implications. Viruses. 2015;7:5746-5766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Perales C, Quer J, Gregori J, Esteban JI, Domingo E. Resistance of Hepatitis C Virus to Inhibitors: Complexity and Clinical Implications. Viruses. 2015;7:5746-5766. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Bacon B, Dieterich D, Flamm S, Kowdley K, Lawitz E, Milligan S, Younossi Z Naoky Tsai. Efficacy of sofosbuvir and simeprevir-based regimens for 304 HCV treatment experienced patients in a reallife setting; data from the TRIO network. [Abstract 975] 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) Nov 7-11, 2014; Boston, MA. Hepatology. 2014;60 Suppl 1:S672. |
| 8. | Dieterich D, Bacon B, Flamm S, Kowdley K, Milligan S, Tsai N, Younossi Z, Lawitz E. Final evaluation of 955 HCV patients treated with 12 week regimens containing sofosbuvir/simeprevir in the TRIO network: academic and community treatment of a real-world, heterogeneous population. J Hepatol. 2015;62:S621. [DOI] [Full Text] |
| 9. | Reddy R, Lim JK, Kuo A, Di Bisceglie AM, Vargas HE, Galati JS, Morelli G, Everson GT, Kwo PY, Brown RS. Vainorius M, Peter JA, Nelson DR, Fried MW, Manns MP. All oral HCV therapy is safe and effective in patients with decompensated cirrhosis: interim report from the HCV-TARGET real world experience. J Hepatol. 2015;62:S193. [DOI] [Full Text] |
| 10. | Buggisch P, Sarrazin C, Mauss S, Hinrichsen H, Simon K, Vermehren J, Hueppe D, Petersen J. Sofosbuvir-based treatment under real life conditions in Germany (The SOFGER trial). J Hepatol. 2015;62:S622. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 11. | Sulkowski MS, Vargas HE, Di Bisceglie AM, Kuo A, Reddy KR, Lim JK, Morelli G, Feld J, Brown RS, Frazier LM. Safety and efficacy of sofosbuvir (SOF) in combination with simeprevir (SIM) ribavirin (RBV) in patients with genotype 1: interim results of a prospective, observational study. [Abstract 955] 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) Nov 7-11, 2014; Boston, MA. Hepatology. 2014;60 Suppl 1:S660. |
| 12. | Pol S, Bourliere M, Lucier S, De Ledinghen V, Zoulim F, Dorival-Mouly C, Métivier S, Larrey D, Tran A, Hezode C. Daclatasvir-Sofosbuvir in HCV genotype 1-mono-infected patients from the French observational cohort ANRS CO22 HEPATHER. J Hepatol. 2015;62:S258. [DOI] [Full Text] |
| 13. | Saxena V, Koraishy F, Sise M, Lim J, Chung R, Liapakis A, Nelson DR, Schmidt M, Fried NW, Terrault N. HCV-TARGET. Safety and efficacy of sofosbuvir-containing regimens in hepatitis C infected patients with reduced renal function: real-word experience from HCV-TARGET. J Hepatol. 2015;62:S267. [RCA] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Buggisch P, Petersen J, Wursthorn K, Atanasov P, Gauthier A. Real-world effectiveness of ledipasvir/sofosbuvir 8 weeks chronic hepatitis C treatment. J Hepatol. 2015;62:S280. [DOI] [Full Text] |
| 15. | Buti M, Riveiro-Barciela M, Esteban R. Management of direct-acting antiviral agent failures. J Hepatol. 2015;63:1511-1522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 16. | Halfon P, Sarrazin C. Future treatment of chronic hepatitis C with direct acting antivirals: is resistance important? Liver Int. 2012;32 Suppl 1:79-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 17. | Sarrazin C, Zeuzem S. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology. 2010;138:447-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 412] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 18. | Sarrazin C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol. 2016;64:486-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 341] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 19. | Wyles DL, Rodriguez-Torres M, Lawitz E, Shiffman ML, Pol S, Herring RW, Massetto B, Kanwar B, Trenkle JD, Pang PS. All-oral combination of ledipasvir, vedroprevir, tegobuvir, and ribavirin in treatment-naïve patients with genotype 1 HCV infection. Hepatology. 2014;60:56-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 20. | Lontok E, Harrington P, Howe A, Kieffer T, Lennerstrand J, Lenz O, McPhee F, Mo H, Parkin N, Pilot-Matias T. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology. 2015;62:1623-1632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 239] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 21. | Lindenbach BD, Rice CM. Flaviviridae: the viruses and their replication. Fields Virology. 5th Edition. Philadelphia: Lippincott-Raven Publishers 2012; 1101-1152. |
| 22. | Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol. 2009;510:33-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 23. | Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225-3229. [PubMed] |
| 24. | Merz A, Long G, Hiet MS, Brügger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018-3032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 285] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 25. | Milam SB, Magnuson VL, Steffensen B, Chen D, Klebe RJ. IL-1 beta and prostaglandins regulate integrin mRNA expression. J Cell Physiol. 1991;149:173-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 26. | Koutsoudakis G, Forns X, Pérez-Del-Pulgar S. [The molecular biology of hepatitis C virus]. Gastroenterol Hepatol. 2013;36:280-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2294] [Cited by in RCA: 2251] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 28. | Chueca Porcuna N, Alvarez Estévez M, Parra Ruiz J, Hernández Quero J, García García F. [Update on hepatitis C therapy. New drugs, treatment response monitoring and emergence of resistance]. Enferm Infecc Microbiol Clin. 2013;31 Suppl 1:40-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 29. | Pawlotsky JM. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53:1742-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 257] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 30. | Pawlotsky JM. Hepatitis C virus genetic variability: pathogenic and clinical implications. Clin Liver Dis. 2003;7:45-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 31. | Rong L, Dahari H, Ribeiro RM, Perelson AS. Rapid emergence of protease inhibitor resistance in hepatitis C virus. Sci Transl Med. 2010;2:30ra32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 307] [Cited by in RCA: 288] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 32. | Herrmann E, Neumann AU, Schmidt JM, Zeuzem S. Hepatitis C virus kinetics. Antivir Ther. 2000;5:85-90. [PubMed] |
| 33. | Powdrill MH, Tchesnokov EP, Kozak RA, Russell RS, Martin R, Svarovskaia ES, Mo H, Kouyos RD, Götte M. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc Natl Acad Sci USA. 2011;108:20509-20513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 34. | Lataillade M, Chiarella J, Yang R, Schnittman S, Wirtz V, Uy J, Seekins D, Krystal M, Mancini M, McGrath D. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naïve subjects in the CASTLE study. PLoS One. 2010;5:e10952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 99] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 35. | Verbinnen T, Van Marck H, Vandenbroucke I, Vijgen L, Claes M, Lin TI, Simmen K, Neyts J, Fanning G, Lenz O. Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing. J Virol. 2010;84:11124-11133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Soriano V, Perelson AS, Zoulim F. Why are there different dynamics in the selection of drug resistance in HIV and hepatitis B and C viruses? J Antimicrob Chemother. 2008;62:1-4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 70] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | McCown MF, Rajyaguru S, Kular S, Cammack N, Nájera I. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob Agents Chemother. 2009;53:2129-2132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 97] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 38. | Martin B, Hennecke N, Lohmann V, Kayser A, Neumann-Haefelin C, Kukolj G, Böcher WO, Thimme R. Restoration of HCV-specific CD8+ T cell function by interferon-free therapy. J Hepatol. 2014;61:538-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 194] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 39. | Larrubia JR, Moreno-Cubero E, Miquel J, Sanz-de-Villalobos E. Hepatitis C virus-specific cytotoxic T cell response restoration after treatment-induced hepatitis C virus control. World J Gastroenterol. 2015;21:3480-3491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 40. | Dietz J, Schelhorn SE, Fitting D, Mihm U, Susser S, Welker MW, Füller C, Däumer M, Teuber G, Wedemeyer H. Deep sequencing reveals mutagenic effects of ribavirin during monotherapy of hepatitis C virus genotype 1-infected patients. J Virol. 2013;87:6172-6181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 41. | Howe AY, Black S, Curry S, Ludmerer SW, Liu R, Barnard RJ, Newhard W, Hwang PM, Nickle D, Gilbert C. Virologic resistance analysis from a phase 2 study of MK-5172 combined with pegylated interferon/ribavirin in treatment-naive patients with hepatitis C virus genotype 1 infection. Clin Infect Dis. 2014;59:1657-1665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Thompson AJ, Locarnini SA, Beard MR. Resistance to anti-HCV protease inhibitors. Curr Opin Virol. 2011;1:599-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 43. | Wyles DL. Antiviral resistance and the future landscape of hepatitis C virus infection therapy. J Infect Dis. 2013;207 Suppl 1:S33-S39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 44. | Sarrazin C, Lathouwers E, Peeters M, Daems B, Buelens A, Witek J, Wyckmans Y, Fevery B, Verbinnen T, Ghys A. Prevalence of the hepatitis C virus NS3 polymorphism Q80K in genotype 1 patients in the European region. Antiviral Res. 2015;116:10-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 45. | Sullivan JC, De Meyer S, Bartels DJ, Dierynck I, Zhang EZ, Spanks J, Tigges AM, Ghys A, Dorrian J, Adda N. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin Infect Dis. 2013;57:221-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 46. | Lenz O, Verbinnen T, Fevery B, Tambuyzer L, Vijgen L, Peeters M, Buelens A, Ceulemans H, Beumont M, Picchio G. Virology analyses of HCV isolates from genotype 1-infected patients treated with simeprevir plus peginterferon/ribavirin in Phase IIb/III studies. J Hepatol. 2015;62:1008-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 47. | Yeh WW, Lipardi C, Jumes P, De Lepeleire IM, Van den Bulk N, Caro L, Huang X, Mangin E, Nachbar RB, Gane EJ. MK-8742, a HCV NS5A inhibitor with a broad spectrum of HCV genotypic activity, demonstrates potent antiviral activity in genotype-1 and -3 HCV infected patients. [Abstract 479] 64th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) Nov 1-5, 2013; Washington DC. Hepatology. 2013;58 Suppl 1:438a-439a. |
| 48. | Lahser F, Liu R, Bystol K, Xia E, Raubertas R, Asante-Appiah E. A combination containing MK-5172 (HCV NS3 protease inhibitor) and MK-8742 (HCV NS5A inhibitor) demonstrates high barrier to resistance in HCV replicons. [Abstract 87] 63rd Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) Nov 9-13, 2012; Boston, MA. Hepatology. 2012;56:236a. |
| 49. | Gao M. Antiviral activity and resistance of HCV NS5A replication complex inhibitors. Curr Opin Virol. 2013;3:514-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 113] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 50. | Nakamoto S, Kanda T, Wu S, Shirasawa H, Yokosuka O. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J Gastroenterol. 2014;20:2902-2912. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 108] [Cited by in RCA: 103] [Article Influence: 9.4] [Reference Citation Analysis (1)] |
| 51. | Krishnan P, Tripathi R, Schnell G, Reisch T, Beyer J, Dekhtyar T, Irvin M, Xie W, Larsen L, Podsadecki T. Long term follow-up of treatment-emergent resistance-associated variants in NS3, NS5A and NS5B with paritaprevir/r, ombitasvir- and dasabuvir-based regimens. J Hepatol. 2015;62:S220-S220. [DOI] [Full Text] |
| 52. | Susser S, Vermehren J, Forestier N, Welker MW, Grigorian N, Füller C, Perner D, Zeuzem S, Sarrazin C. Analysis of long-term persistence of resistance mutations within the hepatitis C virus NS3 protease after treatment with telaprevir or boceprevir. J Clin Virol. 2011;52:321-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 53. | Thomas XV, de Bruijne J, Kieffer TL, Sullivan JC, Rebers SP, deVries M, Reesink HW, Weegink CJ, Molenkamp R, Schinkel J. Long-term follow-up of chronic hepatitis C infected patients treated with telaprevir: evaluation of persistence of resistant variants by ultra-deep sequencing. J Hepatol. 2011;54:S490-S491. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 54. | McPhee F, Hernandez D, Yu F, Ueland J, Monikowski A, Carifa A, Falk P, Wang C, Fridell R, Eley T. Resistance analysis of hepatitis C virus genotype 1 prior treatment null responders receiving daclatasvir and asunaprevir. Hepatology. 2013;58:902-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 55. | Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Symonds WT, Hindes RG, Berrey MM. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N Engl J Med. 2013;368:34-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 564] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 56. | Afdhal N, Zeuzem S, Kwo P, Chojkier M, Gitlin N, Puoti M, Romero-Gomez M, Zarski JP, Agarwal K, Buggisch P. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med. 2014;370:1889-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1365] [Article Influence: 124.1] [Reference Citation Analysis (0)] |
| 57. | Afdhal N, Reddy KR, Nelson DR, Lawitz E, Gordon SC, Schiff E, Nahass R, Ghalib R, Gitlin N, Herring R. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med. 2014;370:1483-1493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1065] [Cited by in RCA: 1064] [Article Influence: 96.7] [Reference Citation Analysis (0)] |
| 58. | Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, Shiffman ML, Schiff E, Ghalib R, Ryan M. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med. 2014;370:1879-1888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 911] [Cited by in RCA: 928] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 59. | Lawitz EJ, Rodriguez-Torres M, Denning J, Mathias A, Mo H, Gao B, Cornpropst MT, Berrey MM, Symonds WT. All-oral therapy with nucleotide inhibitors sofosbuvir and GS-0938 for 14 days in treatment-naive genotype 1 hepatitis C (nuclear). J Viral Hepat. 2013;20:699-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 60. | Lam AM, Espiritu C, Bansal S, Micolochick Steuer HM, Niu C, Zennou V, Keilman M, Zhu Y, Lan S, Otto MJ. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother. 2012;56:3359-3368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 205] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 61. | Tong X, Le Pogam S, Li L, Haines K, Piso K, Baronas V, Yan JM, So SS, Klumpp K, Nájera I. In vivo emergence of a novel mutant L159F/L320F in the NS5B polymerase confers low-level resistance to the HCV polymerase inhibitors mericitabine and sofosbuvir. J Infect Dis. 2014;209:668-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 62. | Childs-Kean LM, Hand EO. Simeprevir and sofosbuvir for treatment of chronic hepatitis C infection. Clin Ther. 2015;37:243-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 63. | Hedskog C, Dvory-Sobol H, Gontcharova V, Martin R, Ouyang W, Han B, Gane EJ, Brainard D, Hyland RH, Miller MD. Evolution of the HCV viral population from a patient with S282T detected at relapse after sofosbuvir monotherapy. J Viral Hepat. 2015;22:871-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 64. | Svarovskaia ES, Dvory-Sobol H, Parkin N, Hebner C, Gontcharova V, Martin R, Ouyang W, Han B, Xu S, Ku K. Infrequent development of resistance in genotype 1-6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis. 2014;59:1666-1674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 65. | Krishnan P, Tripathi R, Schnell G, Reisch T, Beyer J, Irvin M, Xie W, Larsen L, Podsadecki T, Pilot-Matias T. Pooled analysis of resistance in patients treated with ombitasvir/ABT-450/r and dasabuvir with or without ribavirin in Phase 2 and Phase 3 clinical trials. [Abstract 1936] 65th Annual Meeting of the American Association for the Study of Liver Diseases (AASLD) Nov 7-11, 2014; Boston, MA. Hepatology. 2014;60 Suppl 1:1134a-1135a. |
| 66. | Kati W, Koev G, Irvin M, Beyer J, Liu Y, Krishnan P, Reisch T, Mondal R, Wagner R, Molla A. In vitro activity and resistance profile of dasabuvir, a nonnucleoside hepatitis C virus polymerase inhibitor. Antimicrob Agents Chemother. 2015;59:1505-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 67. | Vermehren J, Sarrazin C. The role of resistance in HCV treatment. Best Pract Res Clin Gastroenterol. 2012;26:487-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 68. | Margeridon-Thermet S, Le Pogam S, Li L, Liu TF, Shulman N, Shafer RW, Najera I. Similar prevalence of low-abundance drug-resistant variants in treatment-naive patients with genotype 1a and 1b hepatitis C virus infections as determined by ultradeep pyrosequencing. PLoS One. 2014;9:e105569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 69. | Paolucci S, Fiorina L, Mariani B, Gulminetti R, Novati S, Barbarini G, Bruno R, Baldanti F. Naturally occurring resistance mutations to inhibitors of HCV NS5A region and NS5B polymerase in DAA treatment-naïve patients. Virol J. 2013;10:355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 70. | Suzuki F, Sezaki H, Akuta N, Suzuki Y, Seko Y, Kawamura Y, Hosaka T, Kobayashi M, Saito S, Arase Y. Prevalence of hepatitis C virus variants resistant to NS3 protease inhibitors or the NS5A inhibitor (BMS-790052) in hepatitis patients with genotype 1b. J Clin Virol. 2012;54:352-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 71. | McCloskey RM, Liang RH, Joy JB, Krajden M, Montaner JS, Harrigan PR, Poon AF. Global origin and transmission of hepatitis C virus nonstructural protein 3 Q80K polymorphism. J Infect Dis. 2015;211:1288-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 72. | Ruggiero T, Proietti A, Boglione L, Milia MG, Allice T, Burdino E, Orofino G, Bonora S, Di Perri G, Ghisetti V. Predominance of hepatitis C virus Q80K among NS3 baseline-resistance-associated amino acid variants in direct-antiviral-agent-naïve patients with chronic hepatitis: single-centre experience. Arch Virol. 2015;160:2881-2885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 73. | Jacobson IM, Dore GJ, Foster GR, Fried MW, Radu M, Rafalsky VV, Moroz L, Craxi A, Peeters M, Lenz O. Simeprevir with pegylated interferon alfa 2a plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-1): a phase 3, randomised, double-blind, placebo-controlled trial. Lancet. 2014;384:403-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 357] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 74. | Manns M, Marcellin P, Poordad F, de Araujo ES, Buti M, Horsmans Y, Janczewska E, Villamil F, Scott J, Peeters M. Simeprevir with pegylated interferon alfa 2a or 2b plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-2): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2014;384:414-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 291] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 75. | Chatterji U, Lim P, Bobardt MD, Wieland S, Cordek DG, Vuagniaux G, Chisari F, Cameron CE, Targett-Adams P, Parkinson T. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J Hepatol. 2010;53:50-56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 76. | Yoshimi S, Imamura M, Murakami E, Hiraga N, Tsuge M, Kawakami Y, Aikata H, Abe H, Hayes CN, Sasaki T. Long term persistence of NS5A inhibitor-resistant hepatitis C virus in patients who failed daclatasvir and asunaprevir therapy. J Med Virol. 2015;87:1913-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 77. | Sulkowski MS, Gardiner DF, Rodriguez-Torres M, Reddy KR, Hassanein T, Jacobson I, Lawitz E, Lok AS, Hinestrosa F, Thuluvath PJ. Daclatasvir plus sofosbuvir for previously treated or untreated chronic HCV infection. N Engl J Med. 2014;370:211-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 911] [Article Influence: 82.8] [Reference Citation Analysis (0)] |