Published online Jun 14, 2016. doi: 10.3748/wjg.v22.i22.5173
Peer-review started: February 19, 2016
First decision: March 31, 2016
Revised: April 26, 2016
Accepted: May 4, 2016
Article in press: May 4, 2016
Published online: June 14, 2016
Processing time: 105 Days and 18.6 Hours
AIM: To elucidate the molecular mechanisms leading to development of functionally impaired dendritic cells (DCs) in chronic hepatitis C (CHC) patients infected with genotype 3 virus.
METHODS: This prospective study was conducted on the cohorts of CHC individuals identified as responders or non-responders to antiviral therapy. Myeloid DCs were isolated from the peripheral blood of each subject using CD1c (BDCA1)+ DC isolation Kit. Monocytes from healthy donor were cultured with DC growth factors such as IL-4 and GM-CSF either in the presence or absence of hepatitis C virus (HCV) viral proteins followed by LPS stimulation. Phenotyping was done by flowcytometry and gene expression profiling was evaluated by real-time PCR.
RESULTS: Non-responders [sustained virological response (SVR)-ve] to conventional antiviral therapy had significantly higher expression of genes associated with interferon responsive element such as IDO1 and PD-L1 (6-fold) and negative regulators of JAK-STAT pathway such as SOCS (6-fold) as compared to responders (SVR+ve) to antiviral therapy. The down-regulated genes in non-responders included factors involved in antigen processing and presentation mainly belonging to major histocompatibility complex (MHC) Class-II family as HLA-DP, HLA-DQ (2-fold) and superoxide dismutase (2-fold). Cells grown in the presence of HCV viral proteins had genes down-regulated for factors involved in innate response, interferon signaling, DC maturation and co-stimulatory signaling to T-cells, while the genes for cytokine signaling and Toll-like receptors (4-fold) were up-regulated as compared to cells grown in absence of viral proteins.
CONCLUSION: Underexpressed MHC class-II genes and upregulated negative regulators in non-responders indicate diminished capacity to present antigen and may constitute mechanism of functionally defective state of DCs.
Core tip: The study was aimed to understand the mechanisms of dendritic cells dysfunction during chronic hepatitis C (CHC) infection. The findings highlight the association between different immune response genes and viral persistence in non-responders to antiviral therapy. Up regulation of negative regulators and down-regulation of molecules involved with antigen presentation seems to associate with non-responsiveness to antiviral therapy. Some novel pathways can be targeted to achieve better management of CHC patients.
- Citation: Tomer S, Chawla YK, Duseja A, Arora SK. Dominating expression of negative regulatory factors downmodulates major histocompatibility complex Class-II expression on dendritic cells in chronic hepatitis C infection. World J Gastroenterol 2016; 22(22): 5173-5182
- URL: https://www.wjgnet.com/1007-9327/full/v22/i22/5173.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i22.5173
Hepatitis C virus (HCV), a positive sense single stranded RNA virus, infecting around 180 million people worldwide, is becoming a significant global health problem[1,2]. Transmitted through infected blood and body fluids, it is responsible for chronic hepatitis, which ultimately leads to life threatening liver diseases like fibrosis, cirrhosis, steatosis and finally causing hepatocellular carcinoma (HCC), thus a need for liver transplant[3]. Genotype 3 of HCV being more prevalent in South Asia, accounts for more than 50% of all the genotypes[4]. Although more patients infected with genotype 3 respond successfully to therapy as compared to genotype 1, yet approximately 25%-30% patients fail to achieve sustained virological response (SVR) and are considered as non-responders (NR) to antiviral therapy containing IFN-α and ribavirin, offered till recently[5,6].
Dendritic cells (DC) are professional Antigen- presenting cells (APC) having the unique property to induce a primary immune response[7]. Antigen uptake, processing and presentation to naïve T-cells for activation of the immune system are the main functions carried out by DC. There have been studies reported from our laboratory which showed that DC are numerically, functionally and phenotypically dysfunctional in patients infected with CHC[8,9]. The study also showed that functionally defective monocyte-derived dendritic cells (moDC) from CHC patients who did not achieve SVR, failed to reconstitute the capacity to mature, indicating that the dysfunctional status of DC in CHC patients was directly associated with the persistence of the virus.
The expression of many genes is responsible for regulation of DC maturation. They involve genes associated with antigen processing and presentation[10], interferon α, β response[11,12], cytokine signaling[13], adhesion and migration[14], phagocytosis[15], interferon responsive elements[16], anti-inflammatory process[17], negative regulators of JAK-STAT pathway[18] and genes involved in TLR mediated signaling[19].
Effective cellular immune response directed against HCV is mediated through T-cell and DC crosstalks[20]. A subdued adaptive immune response in chronic HCV patients might be due to the suboptimal antigen presentation and signaling via impaired DCs in these individuals. So, in the proposed study, we wanted to find out whether this non-responsiveness to standard anti-viral therapy in a proportion of CHC patients is associated with the virus-modulated expression of certain genes, which may culminate into dysfunctional status of DC (maturation as well as functional defects). So, we planned to investigate the expression levels of a set of selected genes in the myeloid dendritic cells (MDC) of CHC patients with the hypothesis that, an analysis of the association of immune response genes and non-responsiveness to therapy may reveal molecular mechanisms of DC dysfunction in the non-responders.
The study was approved by the Institute Ethics Committee of the PGIMER, Chandigarh (Reg. No. NKG/947). An informed written consent was obtained from all the subjects before taking blood samples.
A total of 20 CHC patients were recruited for the study. Patients were divided into two groups on the basis of response to therapy in terms of SVR i.e., HCV RNA negative at 24 wk after cessation of antiviral therapy. Patients achieving SVR (SVR+ve) were considered as “Responders” whereas those who failed to achieve SVR (SVR-ve) were termed as “non-responders”. Responders (n = 10, MDC-R) and Non-responders (n = 10, MDC-NR) were recruited on the basis of inclusion and exclusion criteria. Inclusion criteria included patients positive for anti-HCV antibodies and serum HCV RNA, HCV RNA genotype 3 only, no prior history of any treatment for HCV, negative for auto-antibodies (ANA, SMA, LKM, AMA and PCA) and non-viral factors (alcoholism, inherited metabolic disorders). Exclusion criteria included patients with HBV, HCV genotype 1, 2 or 4, HIV and other co-infections, patients with regular use of hepato-toxic drugs and alcohol intake and any evidence of auto-immune or metabolic disease. Venous blood was taken in heparin vacutainer vials (BD) from each recruited patient in the hepatology clinic of PGIMER, Chandigarh. Age and sex matched healthy volunteers were recruited as control subjects (HC; n = 10). Inclusion criteria for HC included those subjects who had normal liver function tests with no history of jaundice or viral hepatitis infection in the past.
Plasma was stored at -80 °C before isolation. From heparinised blood, peripheral blood mononuclear cells (PBMCs) were isolated by ficoll-hypaque density gradient centrifugation using Hisep (Himedia, Mumbai, India). MDC enrichment was performed by using CD1c (BDCA-1)+ Dendritic Cell Isolation Kit (MiltineyiBiotec, Germany) following manufacturer’s instructions. Briefly, the procedure included two steps: In the 1st step, CD1c (BDCA-1) expressing B cells labeled with CD19 magnetic microbeads got depleted by separation over a MACS column placed in a magnetic field of a MACS Separator. In the second step, CD1c (BDCA-1)+ MDC labeled with CD1c-Biotin and Anti-biotin microbeads in B cell depleted flow-through fraction were retained within the column and eluted after removing the column from magnetic field. These cells (MDC) were used for further experiments.
PBMCs and MDCs (10 μL each) were stained with fluorochrome-labeled antibodies (2 μL): Allophycocyanin (APC)-conjugated anti-HLA-DR, Fluorescein isothiocyanate (FITC)-conjugated Lineage Cocktail 1 (Lin1: CD3, CD14, CD16, CD19, CD20, CD56) and Phycoerythrin-Cy5 (PE-Cy5) - conjugated anti-CD11c from BD Biosciences (San Jose, CA, United States) for 15 min in the dark. Cells washed with staining buffer for 5 min at 1400 rpm were re-suspended in buffer for acquisition on Flowcytometer (FACS Calibur, BD, United States). Percent purity was calculated.
Monocyte-derived dendritic cells (moDCs) were derived according to the method described by Romani et al[21] and modified in our laboratory[8]. Cells were cultured in the presence (moDC-Ag) or absence (moDC-N) of HCV viral proteins. Briefly the PBMCs were isolated from venous blood as described above. Cells were suspended in RPMI 1640 medium (Sigma-Aldrich) and monocytes were made to adhere for 2 h at 37 °C (Plate adherence method). After incubation, non-adherent cells were removed. Adherent cells were cultured in the DC culture medium (DCCM) consisting of RPMI 1640 supplemented with: 2 mmol/L L-glutamine, 5 mmol/L HEPES buffer, 100 IU/mL penicillin and 100 μg/mL streptomycin, 10% fetal bovine serum (GIBCO), 20 ng/mL recombinant human GM-CSF (Peprotech Asia) and 20 ng/mL recombinant human IL-4 (Peprotech Asia) at 37 °C in a humidified incubator with CO2 volume fraction, 50 mL/L CO2 for six days. The cells were cultured in different sets as: either in presence or absence of viral proteins: core, NS3, NS4 and NS5 (Peprotech Asia). At the end of six days, these moDCs were stimulated with bacterial lipopolysaccharide (LPS) and further cultured for 48 h in maturation cocktail which comprised of DCCM with LPS (500 ng/mL). On the 8th day, moDCs were harvested and gene expression studies were carried out.
MDCs and moDCs were centrifuged and dissolved in 1 mL TRIzol (Sigma, United States). RNA was extracted and reverse transcribed to cDNA using the RT2 First Strand Kit (Qiagen, Germany) according the manufacturer’s protocol, and cDNA was stored at -20 °C till further use. A custom PCR array (RT2 Custom Profile PCR Array Human, Qiagen) was designed which included a panel of immune-stimulatory genes (ISGs) and genes involved in DC functioning (Table 1). Real-time PCR was undertaken using RT SYBR Green Master Mix (Qiagen, Germany) in a 96-well PCR plate pre-dispensed with primers in a Light Cycler 480 (Roche, Germany). Values were normalized against housekeeping genes (GAPDH, β-actin) in the same sample. Each experiment included positive PCR control (PPC), reverse transcription control (RTC) and human genomic DNA contamination (HGDC) control. Ct values were obtained for calculation of delta-CtCt and further analysis.
| CD209 | CSF1R | ADAMDEC1 | PDCD1 | HLA-DPB1 | HLA-DQA1 | HLA-DQB1 | HMOX1 | ITGB2 | CD40 | CD80 | CD86 |
| CD83 | LY75 | LAMP3 | ARHGDIB | CCL5 | CCL8 | TLR2 | CCL22 | CCR7 | CXCR3 | CXCR4 | CXCL6 |
| CXCL9 | CXCL10 | CXCL11 | CXCL12 | CXCL16 | ITGAX | ICAM1 | VCL | TLR8 | NFKB1 | NFKB2 | CD1A |
| CD1B | CD1C | CD52 | S100A4 | RELB | IDO1 | CD274 | IFNAR1 | IFNAR2 | IRF1 | IRF3 | CD44 |
| IRF7 | IRF9 | STAT1 | STAT2 | ADAR | EIF2AK2 | IFI6 | IFI27 | IFI35 | OAS1 | OAS2 | OAS3 |
| PRKRA | SOD2 | MX1 | MX2 | ISG15 | ISG20 | IFIT1 | FAS | LITAF | IFIT3 | IFITM1 | ITIH2 |
| GBP1 | GBP2 | PIAS1 | PIAS2 | SOCS1 | SOCS2 | SOCS3 | SOCS4 | SOCS5 | IL28B | TRIM22 | RARRES3 |
| TAP1 | TAP2 | RELA | TLR3 | TLR4 | TLR7 | TLR9 | GAPDH | ACTB | HGDC | RTC | PPC |
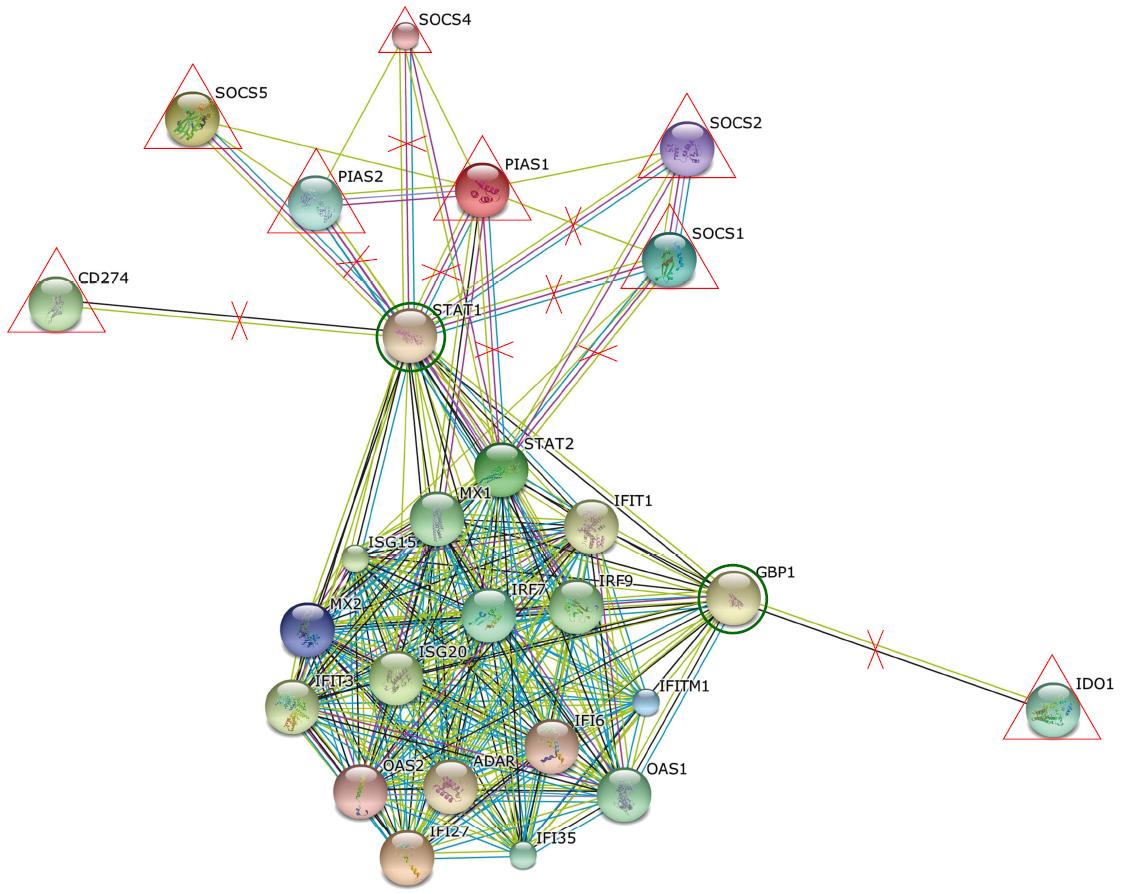
Statistical analysis for viral load (baseline and 4 wk) and other clinical features were done using GraphPad Prism software v 5.03 statistical package. Parametric and non-parametric t-tests were carried out and P < 0.05 was considered significant. For flow cytometry results, Cellquest software (BD Biosciences, United States) was used. Analysis of up-regulated and down-regulated genes was done using web based online software RT2 Profiler PCR array data analysis version 3.5 software. To check interactions and associations between different genes, string software available online was used.
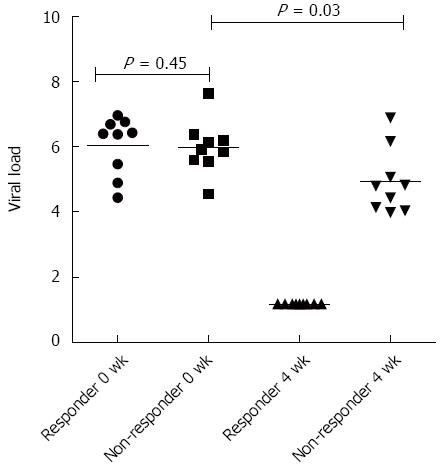
A total of 20 patients were recruited for the study. Their clinical and demographic parameters like gender, age, genotype, liver enzyme (AST-ALT) levels, total bilirubin/conjugated bilirubin, Alkaline Phosphatase (ALP) levels were recorded (Table 2). At the baseline, there was no significant difference in the viral loads of responders vs non-responders, but when compared between baseline vs 4 wk (at RVR - rapid virological response) the viral load became undetectable in responders, while remained detectable in non-responders although was significantly decreased (Figure 1). Also, the degree of liver fibrosis (LSM - liver stiffness measurements) which provides useful information in prognostication, therapeutic planning, and assessment of the impact of treatment in chronic liver diseases, was significantly increased in non-responders (P < 0.05), which suggests that the persistence of virus in the liver leads to cirrhosis of the liver.
| Parameter | Responder | Non-responder |
| Mean viral load (IU/mL) | 6.09 ± 0.29 | 5.97 ± 0.81 |
| Mean age (yr) | 42.0 ± 2.8 | 47 ± 2.9 |
| Male/Female | 6/4 | 8/2 |
| Mean TB/CB (mg/dL) | 0.75 ± 0.10 | 1.25 ± 0.20 |
| Mean AST (U/L) | 94.77 ± 23.40 | 102.3 ± 12.7 |
| Mean ALT (U/L) | 155.4 ± 45.2 | 111.2 ± 18.3 |
| Mean AP (U/L) | 94.50 ± 8.20 | 160.00 ± 24.44 |
| Mean A/G (mg/dL) | 1.25 ± 0.10 | 1.74 ± 0.10 |
| (Fibrosis) Median LSM (kPa) | 6.10 | 23.50 (P = 0. 01) |
For phenotyping and purity of the isolated MDC, the cells negative for Lineage (CD3, CD14, CD16, CD19, CD20, CD56) and dual positive for CD11c and HLA-DR were gated. Percent enrichment of MDC was 65% after magnetic sorting as compared to 10% in PBMCs before sorting.
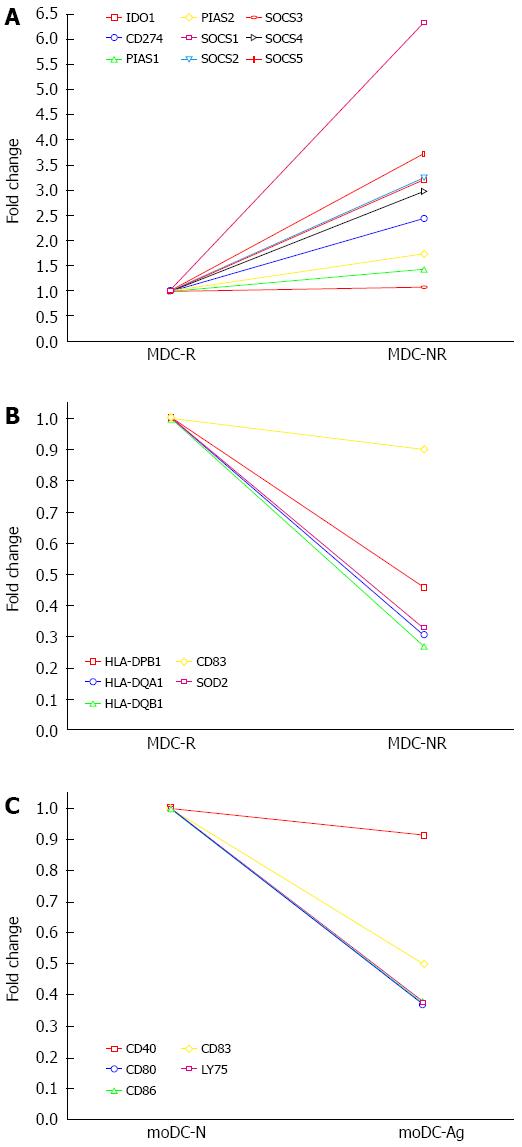
The gene expression profiles of the selected genes (as in Table 1) using custom-designed PCR array are shown in the heat map of genes indicating the differentially expressed genes (Figure 2). The genes upregulated or down-regulated are shown in Figure 3.
Non-responders (MDC-NR) vs Responders (MDC-R) group: Genes involved in negative signaling of JAK-STAT pathway, such as suppressor of cytokine signaling (SOCS1; six-fold, SOCS2, SOCS4 and SOCS5 all two-fold) and genes involved with down-modulation of immune response such as Indoleamine 2,3-Dioxygenase (IDO1) and Programmed death-ligand 1 (PD-L1) were found to be significantly upregulated (two-fold or more) in non-responders as compared to responders to therapy (Figure 4).
Further, the genes for antiviral innate response such as TLR, ISGs and JAK/STAT pathway were also found to be up-regulated in non-responders as compared to responders to antiviral therapy.TLR3 which is activated by viral RNA (HCV RNA) was four-fold up-regulated whereas TLR4 and TLR7 showed two-fold up-regulation in non-responders. The genes for Interferon regulatory factors (IRF 7 and IRF 9) and the Interferon stimulatory genes (ISG15 and ISG20) were also found to be significantly (six-fold) upregulated in non-responders. Further, the genes involved in JAK-STAT signaling, the STAT1 and STAT2, showed two-fold upregulation along with the increased expression of IFN-induced proteins with tetratricopeptide repeats (IFIT1; six-fold, IFIT3; two-fold), IFN-Inducible transmembrane family (IFITM1; two-fold), Interferon-induced GTP-binding protein encoding gene (MX1, MX2; both two-fold), 2′,5′-oligoadenylate synthetase (OAS1; six-fold, OAS2; two-fold), IFN-inducible genes (IFI6; two-fold, IFI27; two-fold, IFI35; four-fold), Adenosine deaminase acting on RNA (ADAR; two-fold) and eukaryotic translation initiation factor 2-alpha kinase 2 (EIF2AK2; two-fold). Also gene associated with apoptosis such as Fas cell surface death receptor (FAS) showed increased expression (two-fold) in non-responders to antiviral therapy.
moDC from healthy donor differentiated in presence (moDC-Ag) or absence (moDC-N) of viral proteins: Sixteen genes were upregulated in the moDC differentiated from monocytes grown in presence of HCV viral proteins as compared to the cells grown in absence of proteins. Amongst these, included the chemokine and their receptor genes (CXCR3, CXCR6, CXCL12, CCL8; all two-fold) and Toll-like receptor genes (TLR2, TLR4, TLR9; all two-fold).
Non-responders (MDC-NR) vs Responders (MDC-R) group: The genes downregulated in non-responders as compared to responders included the genes belonging to MHC-Class II family (HLA-DPB1, HLA-DQA1, HLA-DQB1) and Superoxide dismutase (SOD), the enzyme involved in transforming toxic superoxide anion radicals into hydrogen peroxide and oxygen for protecting DNA from oxidative stress showed two-fold reduced expression in non-responders as compared to responders (Figure 4).
DCs from healthy donor grown with (moDC-Ag) or without (moDC-N) viral proteins: A decreased expression of 21 genes in the cells grown in presence of viral proteins was observed as compared to the cells grown in absence of proteins. The genes found down-regulated (two-fold) include the ones involved in innate response and Interferon signaling (EIF2AK2, IFI27, OAS1, OAS2, MX1, IFIT1, IFIT3, GBP1, GBP2, ISG20); the genes involved with DC maturation (CD83, LY75, LAMP3) and genes involved in delivering co-stimulatory signals to T-cells (CD40, CD80, CD86) (Figure 4).
Non-responsiveness to antiviral therapy has been linked to defective phenotype of MDC by previous reports including our laboratory[8,9]. This indicates a direct association of immune defects with response to treatment in CHC, which could be attributed to many reasons such as (1) defect in IFN-α interactions with its receptors on MDCs; (2) defect in signal transduction machinery after this interaction; and (3) abnormal expression of certain transcription factors and immune response genes which are involved with the activation and maturation of DCs.
Although it has already been reported that there are functional and maturation defects in MDCs during CHC infection, yet the molecular mechanisms involved have not been fully elucidated[8]. The present study was designed with a view to understand these mechanisms and the role of different immune response genes that are involved in regulation of DC functions and may be associated with non-responsiveness to therapy and viral persistence during CHC. In order to achieve the objectives, gene expression profiles were studied in MDC isolated from the peripheral blood of CHC patients put on standard anti-viral treatment consisting of Type 1 IFN and ribavirin, some of those who achieved SVR were termed “responders” and those who did not achieve SVR were termed “non-responders”. The differentially expressed genes were identified after analysis of the expression profile results. Further, these findings were confirmed in an ex vivo moDC model where gene expression profiles were analyzed in monocytes from a healthy donor, differentiated to DC, either in presence or absence of some HCV proteins, using same custom-designed PCR array. Interestingly results from both these experiments although not exactly overlapping, yet revealed the set of genes down-regulated in “non-responders” or in cells grown in presence of viral proteins were those, which are involved with DC maturation and function. Similarly the genes that were found to be up-regulated in these cells were mainly of the negative regulators of DC functions suggesting that the continuous presence of virus or viral proteins in individuals infected with HCV, would facilitate the development of functionally defective phenotype of DCs in these individuals.
The monocytes cultured and differentiated in the presence of HCV viral proteins to dendritic cells in a culture system ex vivo, in the present study, induced the development of a defective phenotype of DCs with hampered maturation capabilities in a similar manner and also confirmed the findings from CHC patient experiments as described above. The analysis of gene expression profiles of these cells revealed downregulated expression of some important genes associated with DC maturation (LAMP3 and LY75), co-stimulatory signaling (CD80 and CD86) and many immune-stimulatory genes, the ISGs (EIF2AK2, IFI27, OAS1, OAS2, MX1, ISG20, IFIT3, IFIT1, GBP1 and GBP2), which are the first line of defense in innate antiviral immunity, suggesting that persistence of HCV down-modulates the host defense mechanisms and make conditions favorable for its own survival. These results are consistent with the earlier reports, which also indicated that different HCV viral proteins disrupt the host IFN signaling and ISGs to establish chronic infection[22].
Entry of the virus in the host results in up-regulation of many TLRs like TLR2, TLR3, TLR4, TLR7, TLR8 and TLR9 on PBMCs and monocytes[23]. The expression of TLR3, TLR4 and TLR7 was also found to be increased on the MDC of non-responders in our study, suggesting the immune activation due to the constant presence of viral RNA. Besides, RARRES (Retionic acid receptor responder protein 1), which is also activated by viral RNA, was also upregulated in non-responders. The TLR7 and RARRES cause IRF7 activation and induction of Type1 IFN gene, leading to activation of JAK-STAT signaling and upregulated expression of STAT1, STAT2 and IRF9, which further lead to enhanced expression of ISGs namely IFIT1, IFIT3, ISG15, ISG20, ADAR, GBP1, PRKRA, EIF2AK2 (PKR), IFITM1, MX1, MX2, OAS1, OAS2, IFI16, IFI27 and IFI35. The administration of exogenous IFN and upregulation of ISGs may be effective to a certain limit because virus replication and copy number gets significantly reduced from baseline to week 4 (RVR) in these patients, but complete removal was not achieved as viral load was still detectable, which suggests that there are other factors that are associated with the persistence of the virus.
The possible reasons may be attributed to dampening of the immune response (functionally impaired immature CD4+ cells) by Type 1 IFN, which leads to impaired T-cell immunity as evident in these patients. Moreover the genes important for optimal antigen presentation like MHC-II (HLA-DP, HLA-DQ) and co-stimulatory molecules (CD80, CD86) as well as the homing receptors like CCR7 were all found to be down-regulated in MDC from non-responder patients as compared to responders in our study, which again supports the hypothesis that such maturation defective DCs with hampered antigen-presenting and migration capabilities would be responsible for the generation of functionally impaired set of immature T-cells, which are incapable of clearing the virus in CHC. Also, it has been reported earlier that excessive or prolonged IFN-αβ signaling is associated with severe disease in HIV infection and favors the replication of virus in the host[23]. Although HCV has different mechanisms to down-regulate Type 1 IFN production, still ISGs are induced in infected hepatocytes in most of the chronically infected CHC patients. Earlier reports suggest that HCV patients having high pre-existing levels of ISGs are less likely to respond to IFN-α therapy in comparison to those with lower levels[24].
Besides, the genes for factors such as SOCS1, SOCS2, SOCS4 and SOCS5 which negatively regulate the inflammatory pathways such as JAK-STAT signaling[25]; PDL1 responsible for exhaustion of T-cell function and its blocking on DCs shown to enhance T-cell activation[26,27]; IDO1 which alters DC by decreasing its APC function and capable of suppressing local T-cell immune responses and promoting systemic tolerance[28], were all up-regulated in non-responders. Our findings corroborate the study reported earlier that up-regulation of PD-1 and SOCS-1 inhibitory molecules mediates functional impairment of the early immune response during HCV infection[29].
The expression of CD209 (DC-SIGN), a molecule expressed more on immature DCs and involved in innate immune responses also plays a critical role in viral pathogenesis, was also upregulated in non-responders[30,31]. Immature DCs bind more strongly to E1 and E2 (HCV Envelope proteins) through DC-SIGN with a difference in internalization pathway. These HCV viral like particles are targeted to non-lysosomal compartment in immature DCs and are protected from lysosomal degradation[32]. Thus, HCV may use DC-SIGN as an entry portal and facilitate viral infection of nearby hepatocytes and also use these DCs as reservoirs resulting in establishment of viral infection. HIV gp120 also binds to DC-SIGN and results in horizontal and vertical transfer and also helps in spreading the virus in the host[33].
HCV induces chronic increase in hepatic oxidative stress which plays an important role in pathogenesis of HCV[34]. Expression of genes of the factors involved with stress conditions such as Heme Oxygenase (HMOX1) was up-regulated in non-responders, suggesting higher oxidative stress in these patients. Since the HMOX1 catabolizes heme to bilirubin, this might be responsible for significantly higher bilirubin levels observed in non-responders as compared to responders. The HMOX1 being mainly expressed in immature DCs and its overexpression induces down-regulation of co-stimulatory molecules on DCs, might be responsible for inhibition of T-cell proliferation in CHC[35]. On the other hand, SOD responsible for transforming toxic superoxide anion radicals, showed down-regulated expression in non-responders resulting in increased levels of reactive oxygen species (ROS) and oxidative stress in these patients, might be responsible for increased apoptosis of cells, which is also supported by up-regulated expression of FAS gene observed in these patients in our study. Thus, these patients are unable to destroy superoxide anion radicals, which are normally produced within the cells and are toxic to biological systems. Earlier report has also shown increased levels of HMOX1 and decreased levels of SOD2 in PBMC of patients with CHC[36].
In summary, our study indicates that there is up-regulation of negative regulators and down-regulation of molecules involved with maturation and antigen-presentation on DCs of non-responders. This imbalanced state, possibly modulated by the continuous replication of HCV, results in the generation of maturation-defective phenotype of DCs which are not capable of presenting the viral antigens to the naïve T-cells and lead to the generation of functionally defective immature T-cells incapable of clearing the virus (Figure 5). Whether this defective state of DCs in these patients is the cause or effect of viral persistence, is not really clear, but possibly this vicious cycle might be the cause of non-responsiveness to anti-viral therapy. Never the less, the study points to some novel pathways that may be targeted to achieve better management of this chronic disease.
In patients infected with hepatitis C, it had already been reported that dendritic cells are numerically, functionally and phenotypically dysfunctional. Also functionally defective monocyte-derived dendritic cells (DCs) from chronic hepatitis C (CHC) patients who did not achieve sustained virological response (SVR) failed to reconstitute the capacity to mature, indicating the dysfunctional status of DC in CHC patients, however the molecular mechanisms regulating this defect have not been elucidated.
Previous experiments have indicated that CHC patients having dysfunctional dendritic cells led to therapy non-responsiveness in these patients.
The expression profile of selected genes related to hepatitis C virus (HCV) infection have been studied in various hepatocytes cell lines but, the role of dendritic cells in non-responsiveness to antiviral therapy has not been elucidated as yet.
The molecular profile of dendritic cells on their role to standard therapy may be used in better prognosis and will help in designing newer therapeutic modalities that might help in better management of non-responders who do not respond to extended regimens to antiviral therapy.
SVR - HCV RNA negative 24 wk after cessation of treatment. It is the best predictor of a long-term response to treatment.
The paper of Tomer S et al discusses the effects of HCV on DCs isolated from PBMC of IFNα-treated HCV patients. The comparisons in gene expression have been done between the responders and non-responders to treatment vs DC from healthy donors exposed or not to HCV proteins. This is an interesting attempt to elucidate the role of HCV-infection in impairment of DC function and to link this situation to non-responsiveness to IFNα treatment, which provides a promising connection to translational research.
P- Reviewer: Grant M, Osna NA S- Editor: Gong ZM L- Editor: A E- Editor: Zhang DN
| 1. | Chevaliez S, Pawlotsky JM. Hepatitis C Viruses: Genomes and Molecular Biology. Tan SL, editor. Norfolk (UK): Horizon Bioscience 2006; Chapter 1. |
| 2. | Farci P, Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome [Science 1989; 244: 359-362]. J Hepatol. 2002;36:582-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4996] [Cited by in RCA: 4657] [Article Influence: 129.4] [Reference Citation Analysis (0)] |
| 3. | Hiroishi K, Ito T, Imawari M. Immune responses in hepatitis C virus infection and mechanisms of hepatitis C virus persistence. J Gastroenterol Hepatol. 2008;23:1473-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology. 2015;61:77-87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1077] [Cited by in RCA: 1146] [Article Influence: 114.6] [Reference Citation Analysis (0)] |
| 5. | Chen CH, Yu ML. Evolution of interferon-based therapy for chronic hepatitis C. Hepat Res Treat. 2010;2010:140953. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2320] [Cited by in RCA: 2242] [Article Influence: 140.1] [Reference Citation Analysis (1)] |
| 7. | Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4895] [Cited by in RCA: 4820] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 8. | Rana D, Chawla YK, Duseja A, Dhiman R, Arora SK. Functional reconstitution of defective myeloid dendritic cells in chronic hepatitis C infection on successful antiviral treatment. Liver Int. 2012;32:1128-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 9. | Rana D, Chawla Y, Arora SK. Success of antiviral therapy in chronic hepatitis C infection relates to functional status of myeloid dendritic cells. Indian J Med Res. 2013;138:766-778. [PubMed] |
| 10. | Krishnadas DK, Ahn JS, Han J, Kumar R, Agrawal B. Immunomodulation by hepatitis C virus-derived proteins: targeting human dendritic cells by multiple mechanisms. Int Immunol. 2010;22:491-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Biron CA. Interferons alpha and beta as immune regulators--a new look. Immunity. 2001;14:661-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 550] [Cited by in RCA: 542] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 12. | Santini SM, Di Pucchio T, Lapenta C, Parlato S, Logozzi M, Belardelli F. A new type I IFN-mediated pathway for the rapid differentiation of monocytes into highly active dendritic cells. Stem Cells. 2003;21:357-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Zekri AR, Ashour MS, Hassan A, Alam El-Din HM, El-Shehaby AM, Abu-Shady MA. Cytokine profile in Egyptian hepatitis C virus genotype-4 in relation to liver disease progression. World J Gastroenterol. 2005;11:6624-6630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 39] [Cited by in RCA: 48] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 14. | Cicinnati VR, Kang J, Sotiropoulos GC, Hilgard P, Frilling A, Broelsch CE, Gerken G, Beckebaum S. Altered chemotactic response of myeloid and plasmacytoid dendritic cells from patients with chronic hepatitis C: role of alpha interferon. J Gen Virol. 2008;89:1243-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Pöhlmann S, Zhang J, Baribaud F, Chen Z, Leslie GJ, Lin G, Granelli-Piperno A, Doms RW, Rice CM, McKeating JA. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J Virol. 2003;77:4070-4080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 301] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 16. | Sedeño-Monge V, Santos-López G, Rocha-Gracia RC, Meléndez-Mena D, Ramírez-Mata A, Vallejo-Ruiz V, Reyes-Leyva J. Quantitative analysis of interferon alpha receptor subunit 1 and suppressor of cytokine signaling 1 gene transcription in blood cells of patients with chronic hepatitis C. Virol J. 2010;7:243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Abdalla MY, Mathahs MM, Ahmad IM. Reduced heme oxygenase-1 expression in steatotic livers infected with hepatitis C virus. Eur J Intern Med. 2012;23:649-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Longman RS, Braun D, Pellegrini S, Rice CM, Darnell RB, Albert ML. Dendritic-cell maturation alters intracellular signaling networks, enabling differential effects of IFN-alpha/beta on antigen cross-presentation. Blood. 2007;109:1113-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 19. | McCoy CE, O’Neill LA. The role of toll-like receptors in macrophages. Front Biosci. 2008;13:62-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Zhao L, Shields J, Tyrrell DL. Functional changes, increased apoptosis, and diminished nuclear factor-kappaB activity of myeloid dendritic cells during chronic hepatitis C infection. Hum Immunol. 2010;71:751-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Romani N, Gruner S, Brang D, Kämpgen E, Lenz A, Trockenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83-93. [PubMed] |
| 22. | Horner SM, Gale M. Intracellular innate immune cascades and interferon defenses that control hepatitis C virus. J Interferon Cytokine Res. 2009;29:489-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 23. | Davidson S, Maini MK, Wack A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res. 2015;35:252-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 148] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 24. | Heim MH, Thimme R. Innate and adaptive immune responses in HCV infections. J Hepatol. 2014;61:S14-S25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 224] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 25. | Akhtar LN, Benveniste EN. Viral exploitation of host SOCS protein functions. J Virol. 2011;85:1912-1921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 26. | Shen T, Chen X, Chen Y, Xu Q, Lu F, Liu S. Increased PD-L1 expression and PD-L1/CD86 ratio on dendritic cells were associated with impaired dendritic cells function in HCV infection. J Med Virol. 2010;82:1152-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | Brown JA, Dorfman DM, Ma FR, Sullivan EL, Munoz O, Wood CR, Greenfield EA, Freeman GJ. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J Immunol. 2003;170:1257-1266. [PubMed] [DOI] [Full Text] |
| 28. | Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1666] [Cited by in RCA: 1769] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 29. | Zhang Y, Ma CJ, Ni L, Zhang CL, Wu XY, Kumaraguru U, Li CF, Moorman JP, Yao ZQ. Cross-talk between programmed death-1 and suppressor of cytokine signaling-1 in inhibition of IL-12 production by monocytes/macrophages in hepatitis C virus infection. J Immunol. 2011;186:3093-3103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 30. | Neumann AK, Thompson NL, Jacobson K. Distribution and lateral mobility of DC-SIGN on immature dendritic cells--implications for pathogen uptake. J Cell Sci. 2008;121:634-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823-829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 665] [Cited by in RCA: 673] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 32. | Ludwig IS, Lekkerkerker AN, Depla E, Bosman F, Musters RJ, Depraetere S, van Kooyk Y, Geijtenbeek TB. Hepatitis C virus targets DC-SIGN and L-SIGN to escape lysosomal degradation. J Virol. 2004;78:8322-8332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 110] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 33. | da Silva RC, Segat L, Crovella S. Role of DC-SIGN and L-SIGN receptors in HIV-1 vertical transmission. Hum Immunol. 2011;72:305-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 34. | Houldsworth A, Metzner M, Shaw S, Kaminski E, Demaine AG, Cramp ME. Polymorphic differences in SOD-2 may influence HCV viral clearance. J Med Virol. 2014;86:941-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 35. | Chauveau C, Rémy S, Royer PJ, Hill M, Tanguy-Royer S, Hubert FX, Tesson L, Brion R, Beriou G, Gregoire M. Heme oxygenase-1 expression inhibits dendritic cell maturation and proinflammatory function but conserves IL-10 expression. Blood. 2005;106:1694-1702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 278] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 36. | Paracha UZ, Fatima K, Alqahtani M, Chaudhary A, Abuzenadah A, Damanhouri G, Qadri I. Oxidative stress and hepatitis C virus. Virol J. 2013;10:251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |