Published online May 14, 2016. doi: 10.3748/wjg.v22.i18.4501
Peer-review started: January 10, 2016
First decision: January 28, 2016
Revised: January 30, 2016
Accepted: March 1, 2016
Article in press: March 2, 2016
Published online: May 14, 2016
Processing time: 115 Days and 10.8 Hours
AIM: To explore the role and potential mechanism of miR-30b regulation of autophagy in hepatic ischemia-reperfusion injury (IRI).
METHODS: An animal model of hepatic IRI was generated in C57BL/6 mice. For in vitro studies, AML12 cells were immersed in mineral oil for 1 h and then cultured in complete Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 to simulate IRI. Mice and cells were transfected with miR-30b agomir/mimics or antagomir/inhibitor to examine the effect of miR-30b on autophagy to promote hepatic IRI. The expression of miR-30b was measured by real-time polymerase chain reaction. Apoptotic cells were detected by terminal uridine nick-end labeling (TUNEL) staining, and cell viability was detected by methylthiazole tetrazolium assay. The expression of light chain 3, autophagy-related gene (Atg)12, Atg5, P62, and caspase-3 were detected by western blotting analysis.
RESULTS: miR-30b levels were significantly downregulated after hepatic IRI, and the numbers of autophagosomes were increased in response to IRI both in vivo and in vitro. These findings demonstrate that low levels of miR-30b could promote hepatic IRI. Furthermore, we found that miR-30b interacted with Atg12-Atg5 conjugate by binding to Atg12. Overexpression of miR-30b diminished Atg12 and Atg12-Atg5 conjugate levels, which promoted autophagy in response to IR. In contrast, downregulation of miR-30b was associated with increased Atg12-Atg5 conjugate levels and increased autophagy.
CONCLUSION: miR-30b inhibited autophagy to alleviate hepatic ischemia-reperfusion injury via decreasing the Atg12-Atg5 conjugate.
Core tip: miR-30b levels were significantly downregulated after hepatic ischemia-reperfusion injury (IRI) in mice. The number of autophagosomes was increased in response to IRI both in vivo and in vitro. Decreased levels of miR-30b could promote hepatic IRI, as revealed by reductions in cells viability in vitro. Overexpression of miR-30b diminished autophagy-related gene (Atg)12 and Atg12-Atg5 conjugate levels which promoted autophagy in response to hepatic IRI. Therefore, miR-30b inhibits autophagy to alleviate hepatic IRI via decreasing the Atg12-Atg5 conjugate.
- Citation: Li SP, He JD, Wang Z, Yu Y, Fu SY, Zhang HM, Zhang JJ, Shen ZY. miR-30b inhibits autophagy to alleviate hepatic ischemia-reperfusion injury via decreasing the Atg12-Atg5 conjugate. World J Gastroenterol 2016; 22(18): 4501-4514
- URL: https://www.wjgnet.com/1007-9327/full/v22/i18/4501.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i18.4501
Hepatic ischemia-reperfusion injury (IRI) is an important factor for the prognosis of surgical outcomes and patient survival as well as the protection of hepatic cells[1]. A key consideration regarding liver function was revealed from a recent study demonstrating that autophagy represents a principal component of hepatology[2]. Autophagy consists of a tightly regulated intracellular catabolic pathway involving lysosomal degradation of cytoplasmic organelles and proteins[3]. miRNAs are closely linked to virtually all fundamental biological pathways[4], and they play critical roles in a broad range of biological processes including proliferation, differentiation, apoptosis, and stress responses[5].
Recent findings have indicated some novel roles for miRNAs in the regulation of autophagy[6,7]. In this report, we focused on miRNAs with direct autophagic implications, as exerted either through putative core components of autophagy machinery or through less well-characterized mechanisms. The 3′-untranslated region (UTR) of the autophagy associated gene 12 (Atg12) contains the predicated target sites for miRNA-30b (miR-30b), which were identified by luciferase reporter gene assays. The possibility exists that miR-30b might contribute to alleviating IRI via modulating autophagy through targeting Atg12. In this study, we attempted to determine whether miR-30b modulates autophagy and thus alleviate hepatic IRI. Specifically, we upregulated or downregulated expression of miR-30b to examine the effects of miR-30b on Atg12 and Atg12-Atg5 conjugate levels that regulate autophagy in hepatic IRI. Our data indicate that miR-30b might serve as a novel therapeutic target regulating autophagy in hepatic IRI.
Male C57BL/6 mice (7-8-wk-old, 23 ± 3 g) were purchased from the experimental animal center of the PLA Military Medical Science Academy. All animals received humane care according to established standards and were maintained in an air-conditioned animal room at 25 °C with free access to water and food. All protocols conformed to the National Institute of Health (NIH) guidelines and all animals received care in compliance with the principles of laboratory animal care. The AML12 cell line (mouse hepatic cell) was purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States). The study was performed according to Tianjin Medical University Institutional Review Board guidelines, and the protocol was approved by the Institutional Review Board.
Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 medium and fetal bovine serum (FBS) were purchased from Gibco (Grand Island, NY, United States); miR-30b-5p mimics/agomir, miR-30b-5p inhibitor/antagomir, miR-NC, Atg12 siRNA and RiboFECTTM CP Reagent were purchased from RiboBio Co., Ltd. (Guangzhou, China); rapamycin and 3-MA were purchased from Selleck Inc (Houston, TX, United States). The In Situ Cell Death Detection Kit, TMR red and SYBR Green quantitative real time polymerase chain reaction (qRT-PCR) Master Mix were purchased from Roche Diagnostics GmbH (Mannheim, Germany); Trizol and Lipofectamine 2000 were obtained from Invitrogen (Carlsbad, CA, United States). Antibody Atg12, light chain 3 (LC3), P62, caspase-3, cleave caspase-3, poly ADP-ribose polymerase 1 (PARP1), β-actin, and horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Cell Signaling Technology Inc (Beverly, MA, United States).
The segmental (70%) hepatic ischemia model was performed as previously described[1]. There were six mice in the sham group, and the 24 mice in the IR group were divided by reperfusion times, consisting of 2, 6, 12, and 24 h. Mice in the miR-30b-5p agomir group (n = 6), miR-30b-5p antagomir group (n = 6), and miR-NC group (n = 6) received miR-30b-5p agomir (10 nmol/L), antagomir (10 nmol/L), or miR-NC (10 nmol/L), respectively, by tail intravenous injection 24 h prior to ischemia.
Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels of the mice in all groups were determined with use of a commercial assay kit (Nanjing Jiancheng Biological Technology, Nanjing, China). Enzyme activities were expressed as international units per liter (U/L).
Samples of liver were fixed in 4% mediosilicic isotonic formaldehyde for 24 h, dehydrated, and embedded in paraffin. Five micrometer-thick sections were cut from each paraffin embedded tissue and stained with hematoxylin and eosin (HE) to evaluate the degree of liver damage. In addition, liver and cell samples were placed in 1% glutaraldehyde and post-fixed with 2% osmium tetroxide. The cell pellets or sections were embedded in epon resin. The data were quantified by counting the number of autophagosomes per cross-sectioned cell.
AML12 cells were plated at a density of 2 × 105 cells/mL in 6-well plates and divided into seven groups[8]: (1) Control group, cells were cultured in DMEM/F12 without treatment; (2) IR group, cells were immersed in mineral oil (1 mL/well) for 1 h to simulate ischemia then cultured in DMEM/F12 for 12 h to simulate reperfusion; (3) miR-30b-5p mimics group, cells were transfected with 50 nmol/L miR-30b-5p mimics or miR-NC using RiboFECTTM CP Reagent for 24 h according to the manufacturer’s protocol, followed by reperfusion as described above; (4) miR-30b-5p inhibitor group, cells were transfected with 50 nmol/L miR-30b-5p inhibitor or miR-NC, followed by reperfusion; (5) Atg12 siRNA group, cells were treated with Atg12 siRNA or siRNA-NC for 24 h, followed by reperfusion; (6) Rapamycin group, cells were treated with 40 nmol/L rapamycin for 2 h, followed by reperfusion; and (7) 3-MA group, cells were treated with 60 μM 3-MA for 2 h, followed by reperfusion.
Cells were seeded onto 96-well plates (5 × 104 cells/well) and after culture for 24 h at 37 °C, subjected to reperfusion as described above. Fresh medium was then added to each well together with 20 μL methylthiazole tetrazolium solution (5 mg/mL), and the plate was incubated at 37 °C for 4 h. The medium was then removed, and 200 μL dimethylsulfoxide was added per well. The optical density of each well was determined with a test wavelength of 490 nm.
AML12 cells were cultured in 6-well plates to 60%-70% confluence. The cells were transfected with tandem GFP-RFP-LC3 adenovirus (Hanbio, Shanghai, China) according to the GFP-RFP-LC3 instruction manual to further confirm autophagy induction.
The streptavidin-peroxidase staining technique was used to detect protein following antigen retrieval by microwave treatment. After blocking endogenous peroxidase activity by incubating in 3% H2O2 for 15 min, specimens were incubated with antibodies [proliferating cell nuclear antigen (PCNA) and caspase-3] at 4 °C overnight. Specimens were incubated at room temperature for 1 h with the secondary antibody, then diaminobenzidine solution was used. Counterstaining was performed with hematoxylin.
Total RNA was isolated by Trizol and 1 μg of RNA for reverse transcription was prepared as described above. RT-PCR was performed in a total volume of 25 μL reaction mixture. U6 or GAPDH was used as an internal control and the expression levels was calculated using MxPro software (Version 4.0, Stratagene, La Jolla, CA, United States).
The miRWalk database was used to predict the binding site on the 3’-UTR of miR-30b, and this database combines several bioinformatic platforms including TargetScan 4.2, miRBase, and miRanda. Luciferase reporter gene assay was performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States) according to the manufacturer’s instructions[9-11]. Cells were transferred into 24-well plates at 3 × 104 cells/well. After 24 h, the cells were transiently co-transfected with pRL-TK plasmid (Promega), and various constructs containing different lengths of the Atg12 5’-flanking region or pGL3-Basic. The luciferase activities were measured according to the manufacturer’s instructions.
Protein samples were harvested from mice livers and AML12 cells. The proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to the nitrocellulose membranes. Membranes were probed with the antibodies to Atg12, LC3, P62, caspase-3, cleave caspase-3, RARP1 and β-actin. Bound antibodies were then visualized using an enhanced chemiluminescence (ECL) detection kit with an appropriate HRP-conjugated secondary antibody.
Terminal uridine nick-end labeling (TUNEL) reactions were performed using an In Situ Cell Death Detection Kit, TMR red. For quantification, the mean number of TUNEL-positive cells, as determined using 200 ×, in five different fields was determined.
All data are presented as mean ± SD. Differences among groups were analyzed using a one-way analysis of variance followed by the Student-Newman-Keuls post-hoc test. The SPSS software 19.0 (Armonk, NY, United States) was used for these analyses, where P < 0.05 was considered to be statistically significant.
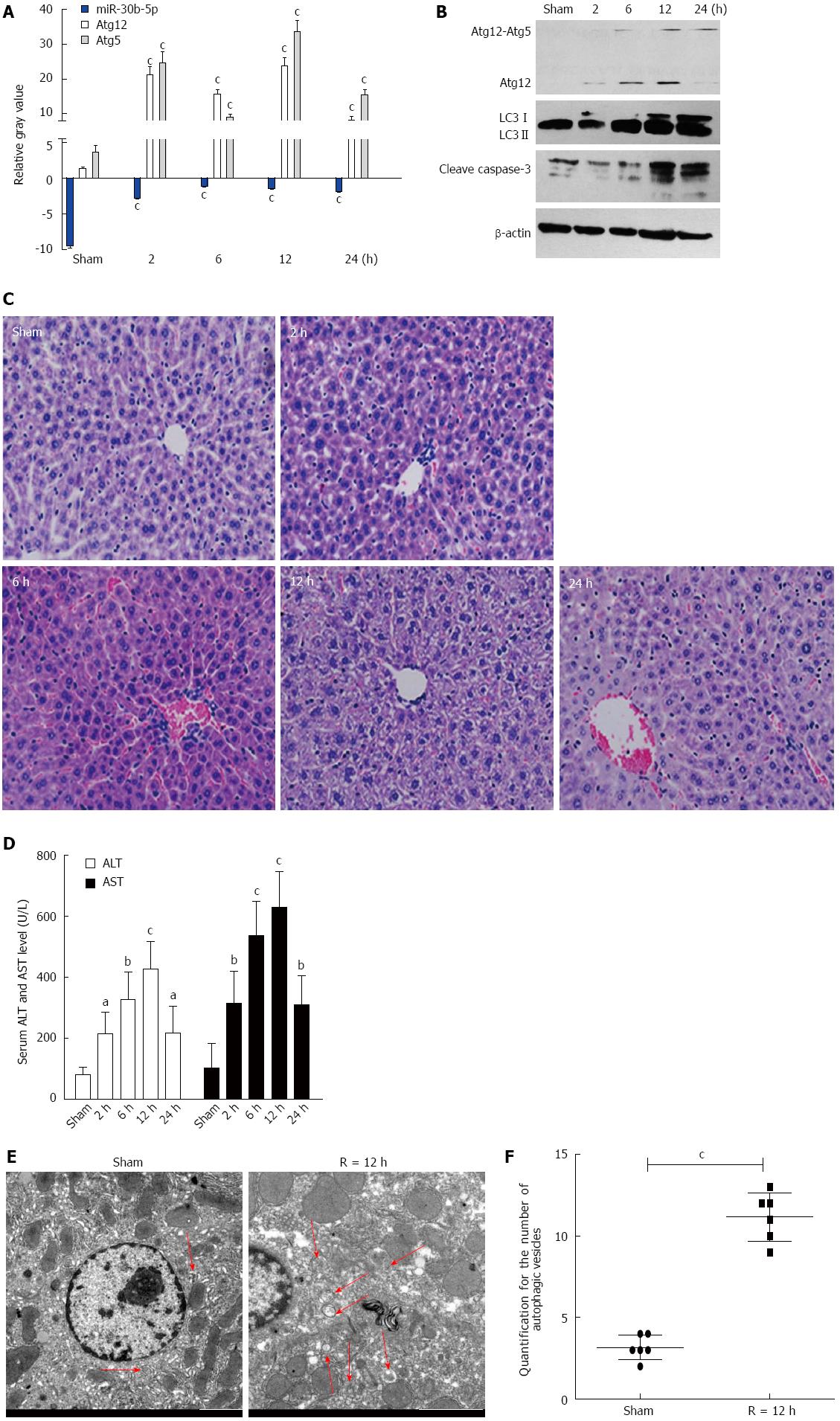
With the IR mouse model, miR-30b-5p expression levels gradually decreased (P < 0.05) after reperfusion, however, levels of Atg12 and Atg5 mRNA increased thereafter (P < 0.05), as compared with the Sham group (Figure 1A).
As shown in Figure 1B, the expression of Atg12, Atg12-Atg5 conjugate, and LC3II was upregulated as a function of time following reperfusion (P < 0.05). Meanwhile, cleave caspase-3 expression increased as a function of time after reperfusion.
Pathological analyses as presented in Figure 1C, revealed considerable hepatocyte edema, congestion and apoptosis at 6-24 h post-reperfusion as compared with the Sham group. The time-dependent changes in serum AST and ALT levels of mice are illustrated in Figure 1D. Serum AST and ALT levels gradually increased (P < 0.05), reaching a peak at 12 h (P < 0.001) following reperfusion. Next, we evaluated autophagic vacuoles using Transmission Electron Microscopy (TEM). Autophagosomes, which contained partially degraded cytoplasmic material, were clearly observed with TEM (Figure 1E). The basal number of autophagosomes within the IR group was increased relative to the Sham group (P < 0.001).
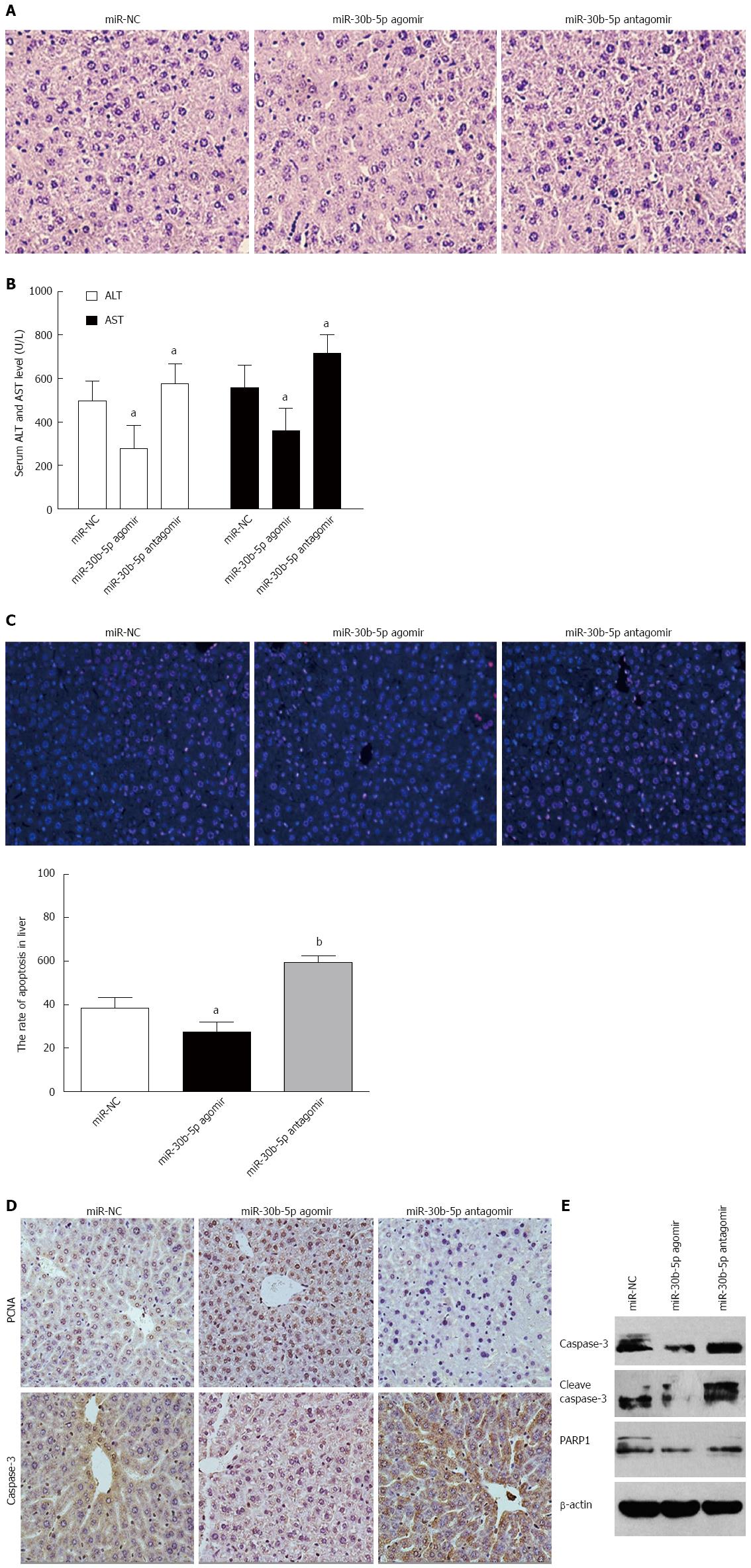
To clarify whether miR-30b can alleviate hepatic IRI, we either upregulated or downregulated the expression of miR-30b in mice after tail intravenous injection of miR-30b-5p agomir or antagomir, respectively, at 12 h following reperfusion. As shown in Figure 2A, we found that miR-30b-5p agomir significantly decreased the histopathologic changes of liver induced by IR treatment, but miR-30b-5p antagomir aggravated these changes. Compared with the miR-NC group, serum AST and ALT levels in the miR-30b-5p agomir group mice were decreased while those of the miR-30b-5p antagomir group mice were increased as a function of time following reperfusion (P < 0.05, Figure 2B). As illustrated in Figure 2C, the number of TUNEL-positive cells was significantly decreased compared with that of the miR-NC group (P < 0.05); however, miR-30b-5p antagomir increased the number of TUNEL-positive cells compared with that of the miR-NC group (P < 0.01). When compared with the miR-NC group at 12 h reperfusion, the miR-30b-5p agomir resulted in a significant increase in PCNA expression but a decrease in caspase-3, cleave caspase-3, and PARP1 expression. In contrast, the miR-30b-5p antagomir decreased in PCNA expression but increased in caspase-3, cleave caspase-3, and PARP1 expression (Figure 2D and E). These findings demonstrate that miR-30b can alleviate hepatic IRI.
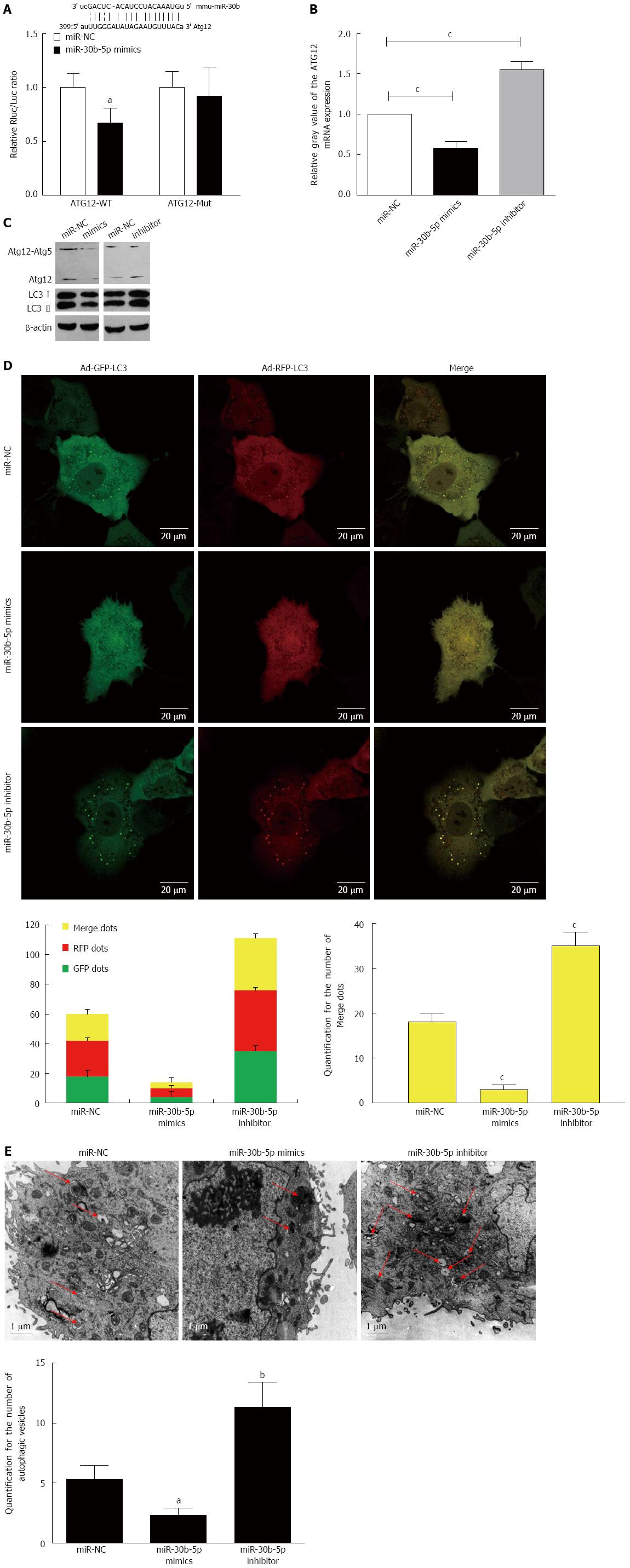
Based upon information contained within the bio-informatics database, we hypothesized that the miR-30b binding site was at the 3’-UTR of Atg12, and a luciferase reporter assay was performed to determine the effects of miR-30b on the 3′-UTR of Atg12 (Figure 3A). When examined at 12 h post-reperfusion, overexpression of miR-30b with miR-30b-5p mimics in AML12 cells significantly reduced Atg12 mRNA and protein levels compared with those cells transfected with the miR-NC (P < 0.05, Figure 3B). Moreover, levels of Atg12-Atg5 conjugate and LC3II decreased after transfection of miR-30b-5p mimics when compared with that of miR-NC (P < 0.05, Figure 3C). In contrast, the results in AML12 cells treated with miR-30b-5p inhibitor were opposite. Our findings indicate that miR-30b could decrease Atg12 and Atg12-Atg5 conjugate levels in AML12 cells.
We performed a Ad-GFP-RFP-LC3 to examine the potential role of miR-30b in autophagy after IRI in AML12 cells. The appearance of GFP- or RFP-LC3 dots within the cytoplasm reflects the recruitment of LC3 proteins to autophagosomes. Upon activation of autophagy, both GFP and RFP are expressed as yellow dots when merged, representing autophagosomes. When autophagosomes fuse with lysosomes and form autolysosomes, the GFP degrades in an acid environment, but the RFP-LC3 remains showing as red dots. A confocal immunofluorescence experiment was performed to demonstrate an increase of LC3. The Ad-GFP-RFP-LC3 was transfected into AML12 cells to confirm the induction of autophagy. As shown in Figure 3D, miR-30b inhibited significantly the induction of autophagosomes. We observed both fluorescent proteins were expressed after the infection with Ad-GFP-RFP-LC3. There were significant decreases in yellow dots with marginal elevations in red dots in the miR-30b-5p mimics group compared to the miR-NC or miR-30b-5p inhibitor groups (P < 0.001). Next, we evaluated autophagic vacuoles using TEM, an Autophagosomes were clearly visualized with TEM (Figure 3E). The basal number of autophagosomes within the miR-30b-5p mimics group was decreased relative to the miR-30b-5p inhibitor or miR-NC group (P < 0.001). Taken together, the results show that miR-30b inhibits autophagic flux by sequestering Atg12.
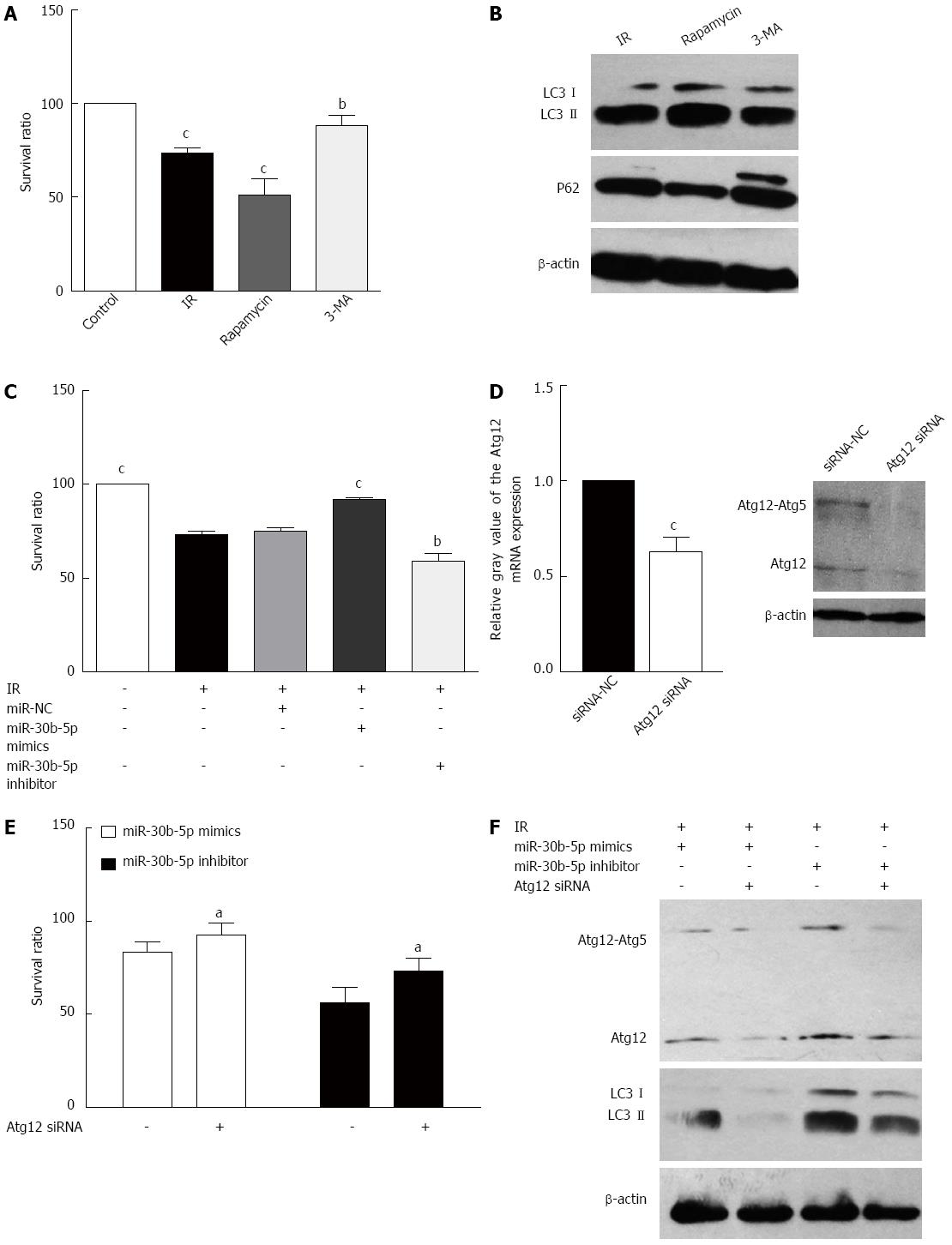
In order to assess whether cells become more resistant to IRI depending on autophagy status in a hepatic IRI model, we examined the effects of rapamycin and 3-MA, an activator and inhibitor of autophagy, respectively. Rapamycin decreased the survival ratio of AML12 cells induced by IR treatment, and 3-MA inhibited this change (P < 0.05, Figure 4A). Moreover, rapamycin increased the levels of LC3II and decreased P62 expression, whereas 3-MA decreased the levels of LC3II and increased P62 expression (Figure 4B). These data found that activating autophagy could aggravate hepatic IRI. miR-30b-5p mimics increased viability of IRI AML12 cells treated responding to IR (P < 0.05), but viability of IRI AML12 cells treated with miR-30b-5p inhibitor was decreased (P < 0.05, Figure 4C). As demonstrated in Figure 4D, Atg12 siRNAs significantly down-regulated Atg12 expression in AML12 cells. The viability of IRI AML12 cells treated with miR-30b-5p mimics or inhibitor (Figure 4E) was enhanced by siRNA knockdown of Atg12. AML12 cells were also treated with miR-30b-5p mimics/inhibitor or Atg12 siRNA to investigate potential interactions between the miR-30b and Atg12-Atg5 conjugate during IR. At 12 h post-reperfusion, siRNA-mediated knockdown of Atg12 contributed to Atg12-Atg5 conjugate and LC3II protein levels inhibited by miR-30b-5p mimics, while Atg12 siRNA decreased the levels of Atg12-Atg5 conjugate and LC3II induced by miR-30b-5p inhibitor (Figure 4F). These data suggest that miR-30b inhibits autophagy to alleviate hepatic IRI by targeting Atg12.
Autophagy plays a pivotal role in cellular homeostasis and adaptation to adverse environments[12,13], and although the regulation of this process remains incompletely understood[14], it is known to provide a cytoprotective role important for survival[15]. Autophagy is regarded as a natural and essential defense mechanism against inflammatory, damnification, and oncotherapy[16]. Hence, regulation of the autophagy pathway has been implicated in the pathogenesis of numerous human diseases. Recently, a growing number of studies on autophagy and liver diseases have focused on liver ischemia reperfusion[17-20]. miR-30b, a member of the miR-30 family, has been suggested to play a role in the differentiation of several cell types[21]. The miR-30 family is also involved in the control of structural changes in the extracellular matrix of the myocardium[22] and in the regulation of the apoptosis[23].
In this study, we discovered that miR-30b was downregulated in mice livers subjected to IR. In addition, the expression of Atg12 and LC3II was upregulated, and the numbers of autophagosomes observed in the IR group increased as a function of time following reperfusion. miR-30b expression was decreased in response to hepatic IRI and was accompanied by a corresponding activation of autophagy. The induction of autophagy represents an initial response to IR in mice livers, while Atg12 and Atg12-Atg5 conjugate expression decreased as a function of time following reperfusion. To clarify whether miR-30b can alleviate hepatic IRI, we upregulated or downregulated expression of miR-30b in mice after tail intravenous injection of miR-30b-5p agomir or antagomir, respectively. miR-30b-5p agomir significantly decreased the histopathologic changes of livers induced by IR treatment, and the number of TUNEL-positive cells were significantly decreased. However, downregulated expression of miR-30b could aggravate the histopathologic changes. PCNA, a subunit of the mammalian DNA polymerase delta, is synthesized primarily during the S phase of the cell cycle[24]. PCNA is a relay molecule that functions as a molecular integrator for proteins involved in the control of the cell cycle, DNA repair, and cell death[25]. Therefore, PCNA is a convincing marker to distinguish proliferating cells. The miR-30b-5p agomir resulted in a significant increase in PCNA expression but a decrease in caspase-3, cleave caspase-3 and PARP1 expression. In contrast, down-regulated expression of miR-30b decreased in PCNA expression but increase in caspase-3, Cleave caspase-3 and PARP1 expression. These findings demonstrate that miR-30b can alleviate hepatic ischemia-reperfusion injury.
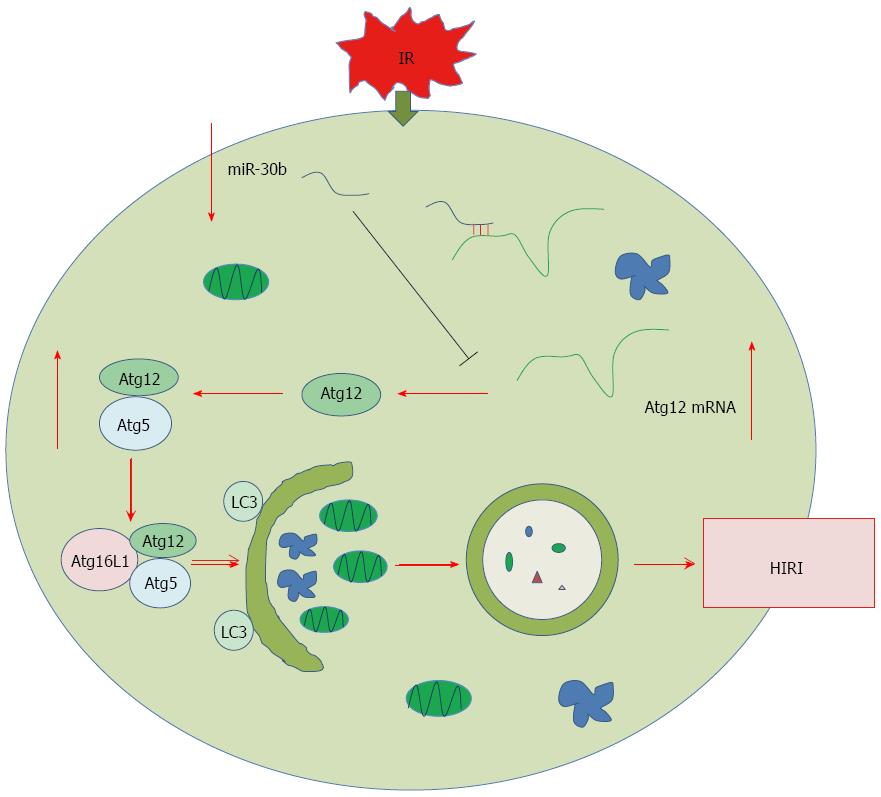
Autophagosome formation requires two ubiquitin-like conjugation systems, the Atg12 and LC3 systems[26]. The Atg12 system is located upstream of the LC3 system in the context of Atg protein organization. Atg12 is conjugated to Atg5, forming the irreversible Atg12-Atg5 complex, which strongly enhances the formation of LC3-phosphatidylethanolamine conjugation[27]. caveolin-1 also regulated Atg12-Atg5 conjugate during autophagosome formation, and caveolin-1 competitively interacts with the Atg12-Atg5 system to suppress the formation and function of the latter in lung epithelial cells[28]. Based upon miRNA target gene prediction, we identified the 3’-UTR area of the Atg12 gene as a match of miR-30b.
Our data showed that up-regulation of the expression of miR-30b significantly increased cell viability as a function of time following reperfusion, while the miR-30b inhibitor significantly decreased cell viability. Related to these findings, a significant decrease of LC3 dots in AML12 transfected with miR-30b-5p mimics compared with miR-NC group. The number of LC3 dots increased in the miR-30b inhibitor group relative to the miR-30b mimics group. These results indicate that miR-30b increased the viability of hepatocytes induced by IR via inhibiting autophagy. Over-expression of miR-30b significantly reduced Atg12 and Atg12-Atg5 conjugate protein levels after AML12 cells transfected with miR-30b mimics; and LC3II expression was decreased in these cells. The expression of Atg12 and Atg12-Atg5 conjugate protein levels increased after AML12 cells were transfected with the miR-30b inhibitor, while LC3II was also increased at the same time. Taken together, it seems clear that miR-30b can decrease Atg12 and Atg12-Atg5 conjugate expression, thereby down-regulating autophagy to alleviate hepatic IRI.
Autophagy is a self-digesting process that occurs in response to stress and plays important roles in the pathogenesis of a variety of diseases[15]. Autophagy functions mainly in a pro-survival capacity for cells to cope with nutrient starvation and anoxia[29,30], however, excessive levels of autophagy within impaired cells can contrarily induce cell death[31]. Oxidative stress may lead to autophagy which induces cell death in cisplatin-induced AKI[32]. In our study, we found that activation of autophagy aggravated hepatic IRI, an effect that was dependent on Atg12 and Atg12-Atg5 conjugate. We also treated AML12 cells with the miR-30b mimics, miR-30b inhibitor, or Atg12 siRNA to investigate whether any potential interaction may exist between miR-30b and Atg12 during IR. Our findings suggest that miR-30b mediated apoptosis to alleviate hepatic IRI, an effect that was dependent on Atg12 activity. As was shown in Figure 5, we found that miR-30b inhibited autophagy to alleviate hepatic IRI by decreasing the Atg12-Atg5 conjugate. This finding may serve as a guide to prevent hepatic IRI and provide a future strategy in research areas of IRI.
The authors would like to thank the members of the Key Laboratory of Organ Transplantation of Tianjin, Laboratory of Immunology and Inflammation of Tianjin Medical University, and Oriental Organ Transplant Center of Tianjin First Central Hospital for their technical support.
Hepatic ischemia reperfusion injury (IRI) represents an important clinical problem as related to liver resection or transplantation. miRNAs participate in various hepatic pathophysiological processes via regulating autophagy. miR-30b, a member of the miR-30 family, is involved in the control of structural changes in the regulation of the apoptosis. However, the importance and function of miR-30b and whether miR-30b regulate autophagy to alleviate hepatic IRI have not yet been elucidated.
It was reported that overexpression of miR-30b had an anti-apoptotic effect on the early phase of rat myocardial ischemia injury model through targeting Kirsten ras sarcoma (KRAS) and activating the Ras/Akt pathway. E2F1-regulated miR-30b suppressed Cyclophilin D to protect the heart from IRI and necrotic cell death.
This is the first study to investigate the role of miR-30b in inhibiting autophagy to alleviate hepatic IRI, and we found that miR-30b decreases the level of Atg12-Atg5 conjugate. This study provides the basis for a future research strategy in hepatic ischemia reperfusion injury.
This study provides insight into the role of miR-30b in the inhibition of autophagy to alleviate hepatic IRI by decreasing the autophagy-related gene Atg12-Atg5 conjugate. Understanding how miR-30b regulates autophagy during hepatic IRI may facilitate the design of new therapeutic approaches to prevent and cure hepatic IRI.
Atg12 ubiquitin-like conjugation systems are required in autophagosome formation, and the Atg12 system is located upstream of the light chain 3 (LC3) system in the context of Atg protein organization. Atg12 is conjugated to Atg5, forming the irreversible Atg12-Atg5 complex, which strongly enhances the formation of LC3-phosphatidylethanolamine conjugation.
This is an interesting paper, providing important information on the expression of miR-30b and its role in autophagy during hepatic IRI. This study found that the miR-30b inhibited autophagy to alleviate hepatic IRI via decreasing the Atg12-Atg5 conjugate. The result is very important for advanced research studies on hepatic IRI.
P- Reviewer: De Ponti F, Liu ZH S- Editor: Qi Y L- Editor: Filipodia E- Editor: Zhang DN
| 1. | Ji H, Shen X, Gao F, Ke B, Freitas MC, Uchida Y, Busuttil RW, Zhai Y, Kupiec-Weglinski JW. Programmed death-1/B7-H1 negative costimulation protects mouse liver against ischemia and reperfusion injury. Hepatology. 2010;52:1380-1389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 2. | Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 323] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 3. | Marchesi N, Osera C, Fassina L, Amadio M, Angeletti F, Morini M, Magenes G, Venturini L, Biggiogera M, Ricevuti G. Autophagy is modulated in human neuroblastoma cells through direct exposition to low frequency electromagnetic fields. J Cell Physiol. 2014;229:1776-1786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 4. | Kalla R, Ventham NT, Kennedy NA, Quintana JF, Nimmo ER, Buck AH, Satsangi J. MicroRNAs: new players in IBD. Gut. 2015;64:504-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 207] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 5. | Weiss JB, Eisenhardt SU, Stark GB, Bode C, Moser M, Grundmann S. MicroRNAs in ischemia-reperfusion injury. Am J Cardiovasc Dis. 2012;2:237-247. [PubMed] |
| 6. | Menghini R, Casagrande V, Marino A, Marchetti V, Cardellini M, Stoehr R, Rizza S, Martelli E, Greco S, Mauriello A. MiR-216a: a link between endothelial dysfunction and autophagy. Cell Death Dis. 2014;5:e1029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 7. | Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T, Zhang J, Peng C, Lin Y, Chen J. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013-7026. [PubMed] |
| 8. | Wang Y, Shen J, Xiong X, Xu Y, Zhang H, Huang C, Tian Y, Jiao C, Wang X, Li X. Remote ischemic preconditioning protects against liver ischemia-reperfusion injury via heme oxygenase-1-induced autophagy. PLoS One. 2014;9:e98834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 9. | Spenlehauer C, Gordon CA, Trkola A, Moore JP. A luciferase-reporter gene-expressing T-cell line facilitates neutralization and drug-sensitivity assays that use either R5 or X4 strains of human immunodeficiency virus type 1. Virology. 2001;280:292-300. [PubMed] |
| 10. | Himes SR, Shannon MF. Assays for transcriptional activity based on the luciferase reporter gene. Methods Mol Biol. 2000;130:165-174. [PubMed] |
| 11. | Fulton R, Van Ness B. Luminescent reporter gene assays for luciferase and beta-galactosidase using a liquid scintillation counter. Biotechniques. 1993;14:762-763. [PubMed] |
| 12. | Vucicevic L, Misirkic-Marjanovic M, Paunovic V, Kravic-Stevovic T, Martinovic T, Ciric D, Maric N, Petricevic S, Harhaji-Trajkovic L, Bumbasirevic V. Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy. 2014;10:2362-2378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 13. | Vlahakis A, Powers T. A role for TOR complex 2 signaling in promoting autophagy. Autophagy. 2014;10:2085-2086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Thapalia BA, Zhou Z, Lin X. Autophagy, a process within reperfusion injury: an update. Int J Clin Exp Pathol. 2014;7:8322-8341. [PubMed] |
| 15. | Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102-1109. [PubMed] |
| 16. | Chen ZH, Wu YF, Wang PL, Wu YP, Li ZY, Zhao Y, Zhou JS, Zhu C, Cao C, Mao YY. Autophagy is essential for ultrafine particle-induced inflammation and mucus hyperproduction in airway epithelium. Autophagy. 2016;12:297-311. [PubMed] |
| 17. | Matsumoto N, Ezaki J, Komatsu M, Takahashi K, Mineki R, Taka H, Kikkawa M, Fujimura T, Takeda-Ezaki M, Ueno T. Comprehensive proteomics analysis of autophagy-deficient mouse liver. Biochem Biophys Res Commun. 2008;368:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Zou H, Zhuo L, Han T, Hu D, Yang X, Wang Y, Yuan Y, Gu J, Bian J, Liu X. Autophagy and gap junctional intercellular communication inhibition are involved in cadmium-induced apoptosis in rat liver cells. Biochem Biophys Res Commun. 2015;459:713-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Yang J, Wang Y, Sui M, Liu F, Fu Z, Wang QX. Tri-iodothyronine preconditioning protects against liver ischemia reperfusion injury through the regulation of autophagy by the MEK/ERK/mTORC1 axis. Biochem Biophys Res Commun. 2015;467:704-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 20. | Cursio R, Colosetti P, Gugenheim J. Autophagy and liver ischemia-reperfusion injury. Biomed Res Int. 2015;2015:417590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 21. | Le Guillou S, Sdassi N, Laubier J, Passet B, Vilotte M, Castille J, Laloë D, Polyte J, Bouet S, Jaffrézic F. Overexpression of miR-30b in the developing mouse mammary gland causes a lactation defect and delays involution. PLoS One. 2012;7:e45727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170-18, 6p following 178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 685] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 23. | Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 281] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 24. | Melo RM, Martins YS, Luz RK, Rizzo E, Bazzoli N. PCNA and apoptosis during post-spawning ovarian remodeling in the teleost Oreochromis niloticus. Tissue Cell. 2015;47:541-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Cox LS. PCNA tightens its hold on the nucleus. Cell Cycle. 2015;14:2727-2728. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 26. | Murrow L, Debnath J. ATG12-ATG3 connects basal autophagy and late endosome function. Autophagy. 2015;11:961-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298-37302. [PubMed] |
| 28. | Chen ZH, Cao JF, Zhou JS, Liu H, Che LQ, Mizumura K, Li W, Choi AM, Shen HH. Interaction of caveolin-1 with ATG12-ATG5 system suppresses autophagy in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2014;306:L1016-L1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 29. | Guan X, Qian Y, Shen Y, Zhang L, Du Y, Dai H, Qian J, Yan Y. Autophagy protects renal tubular cells against ischemia / reperfusion injury in a time-dependent manner. Cell Physiol Biochem. 2015;36:285-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Zhang D, Li C, Zhou J, Song Y, Fang X, Ou J, Li J, Bai C. Autophagy protects against ischemia/reperfusion-induced lung injury through alleviating blood-air barrier damage. J Heart Lung Transplant. 2015;34:746-755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2886] [Cited by in RCA: 2761] [Article Influence: 184.1] [Reference Citation Analysis (0)] |
| 32. | Bolisetty S, Traylor AM, Kim J, Joseph R, Ricart K, Landar A, Agarwal A. Heme oxygenase-1 inhibits renal tubular macroautophagy in acute kidney injury. J Am Soc Nephrol. 2010;21:1702-1712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |