Published online Apr 28, 2016. doi: 10.3748/wjg.v22.i16.4250
Peer-review started: January 21, 2016
First decision: February 18, 2016
Revised: March 1, 2016
Accepted: March 14, 2016
Article in press: March 14, 2016
Published online: April 28, 2016
Processing time: 89 Days and 20.6 Hours
AIM: To examine the predictive effects of baseline serum bilirubin levels and UDP-glucuronosyltransferase (UGT) 1A1*28 polymorphism on response of colorectal cancer to irinotecan-based chemotherapy.
METHODS: The present study was based on a prospective multicenter longitudinal trial of Chinese metastatic colorectal cancer (mCRC) patients treated with irinotecan-based chemotherapy (NCT01282658). Baseline serum bilirubin levels, including total bilirubin (TBil) and unconjugated bilirubin (UBil), were measured, and genotyping of UGT1A1*28 polymorphism was performed. Receiver operating characteristic curve (ROC) analysis was used to determine cutoff values of TBil and UBil. The TBil values were categorized into > 13.0 or ≤ 13.0 groups; the UBil values were categorized into > 4.1 or ≤ 4.1 groups. Combining the cutoff values of TBil and UBil, which was recorded as CoBil, patients were classified into three groups. The classifier’s performance of UGT1A1*28 and CoBil for predicting treatment response was evaluated by ROC analysis. Associations between response and CoBil or UGT1A1*28 polymorphism were estimated using simple and multiple logistic regression models.
RESULTS: Among the 120 mCRC patients, the serum bilirubin level was significantly different between the UGT1A1*28 wild-type and mutant genotypes. Patients with the mutant genotype had an increased likelihood of a higher TBil (P = 0.018) and a higher UBil (P = 0.014) level compared with the wild-type genotype. Patients were stratified into three groups based on CoBil. Group 1 was patients with TBil > 13.0 and UBil > 4.1; Group 2 was patients with TBil ≤ 13.0 and UBil > 4.1; and Group 3 was patients with TBil ≤ 13.0 and UBil ≤ 4.1. Patients in Group 3 had more than a 10-fold higher likelihood of having a response in the simple (OR = 11.250; 95%CI: 2.286-55.367; P = 0.003) and multiple (OR = 16.001; 95%CI: 2.802 -91.371; P = 0.002) analyses compared with the Group 1 individuals. Patients carrying the UGT1A1*28 (TA)7 allele were 4-fold less likely to present with a response compared with the individuals harboring a homozygous (TA)6 genotype in the simple (OR = 0.267; 95%CI: 0.100-0.709; P = 0.008) and multiple (OR = 0.244; 95%CI: 0.088-0.678; P = 0.007) analyses. Classifier’s performance of CoBil and UGT1A1*28 were comparable.
CONCLUSION: CoBil and UGT1A1*28 are both independent biomarkers for predicting the treatment response of mCRC patients to irinotecan-based chemotherapy. After validation, CoBil, an easily determinable index in the clinic, might be helpful in facilitating stratification of mCRC patients for individualized treatment options.
Core tip: Serum bilirubin was reported to be associated with irinotecan-induced toxicity. The current study evaluated whether baseline bilirubin levels could predict treatment response of metastatic colorectal cancer patients given irinotecan-based chemotherapy in a Chinese population and found that a lower bilirubin level was an independent predictor of irinotecan treatment response.
- Citation: Yu QQ, Qiu H, Zhang MS, Hu GY, Liu B, Huang L, Liao X, Li QX, Li ZH, Yuan XL. Predictive effects of bilirubin on response of colorectal cancer to irinotecan-based chemotherapy. World J Gastroenterol 2016; 22(16): 4250-4258
- URL: https://www.wjgnet.com/1007-9327/full/v22/i16/4250.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i16.4250
Colorectal cancer (CRC) is the third most common cancer and a major cause of cancer related death worldwide[1]. More than 40% of CRC patients will eventually develop metastases and require palliative chemotherapy[2,3]. Irinotecan-based therapy is one of the most important fundamental chemotherapy regimens for metastatic CRC (mCRC)[4,5]. As a first-line treatment, the response rate to irinotecan-based therapy is approximately 35%-55%[6,7]. Patients who do not benefit from first-line therapy will miss the best opportunity for disease palliation or control. To date, no prospective tool has been validated for selecting the best therapy for an individual patient. Biomarkers with predictive effects on therapeutic efficacy warrant further investigations[8].
Circulating levels of 7-ethyl-10-hydroxycamptothecin (SN-38), the active metabolite of irinotecan, is associated with irinotecan-treatment efficacy[9]. Through the glucuronidation pathway, SN-38 is converted to an inactive form and eliminated in the bile[10]. UGT1A1 is the most important enzyme that metabolizes SN-38, and it is the only physiological enzyme that converts bilirubin to water-soluble glucuronides[11]. Pharmacological studies demonstrate that glucuronidation rates of both SN-38 and bilirubin are decreased when the TATAA box of the gene contains an extra TA insertion (UGT1A1*28)[12]. Additionally, UGT1A1*28 is involved in the pathogenesis of Gilbert syndrome (GS), which is an inherited disorder of hepatic bilirubin metabolism characterized by unconjugated hyperbilirubinemia[13]. Even in populations without GS, UGT1A1*28 has a strong impact on serum bilirubin levels[14]. Although it is reported that UGT1A1*28 is linked to SN-38 glucuronidation and irinotecan-related toxicity, the predictive role of the UGT1A1*28 polymorphism regarding treatment outcome of irinotecan-based therapy has been conflicting[15-18].
In the current study, we investigated the association between serum bilirubin levels, UGT1A1*28 polymorphism and the therapeutic response in a prospective series of patients with mCRC undergoing irinotecan-based first-line chemotherapy to determine whether serum bilirubin levels and UGT1A1*28 polymorphism could be predictors of therapeutic response.
Study design and patients: The study was based on a prospective longitudinal Chinese clinical trial sponsored by Huazhong University of Science and Technology. Patients treated with irinotecan-based therapy were consecutively recruited between November 2010 and December 2014 from the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology and 5 other cancer centers in south-central China.
Eligibility criteria were as follows: histologically confirmed adenocarcinoma of the colon or rectum; unresectable metastases; age from 18 to 75 years; measurable disease defined according to the Response Evaluation Criteria In Solid Tumors version 1.1 (RECIST1.1)[19]; no prior chemotherapy for metastatic disease (adjuvant chemotherapy except for irinotecan was allowed); Eastern Cooperative Oncology Group Performance Status Scale (PS) ≤ 2 or Karnofsky index of performance status (KPS) > 60%; total bilirubin ≤ 1.5 times the upper limit of normal (ULN); aspartate aminotransferase (AST) and alanine aminotransferase (ALT) ≤ 2.5 times ULN (≤ 5 times ULN if liver metastases present); creatinine clearance > 50 mL/min or serum creatinine ≤ 1.5 times ULN; and no history of Gilbert’s syndrome.
This study was approved by the Ethical Committee of Huazhong University of Science and Technology under reference number NCT01282658 (registered at http://www.clinicaltrials.gov). Written informed consent was required, and blood samples were obtained.
In the current study, only participants recruited from Tongji Hospital or Tongji Medical College were included because they were provided with recorded numerical values of baseline serum bilirubin.
Baseline clinical information, including demographics, KPS, tumor-related details and medical history, was collected prior to the commencement of chemotherapy. Total bilirubin (TBil) and conjugated bilirubin levels were measured in the participants recruited from Tongji Hospital. The unconjugated bilirubin (UBil) level was calculated by subtracting the conjugated bilirubin level from the TBil level. Reference value ranges were 3.4-20.5 μmol/L and 0.00-6.84 μmol/L for TBil and conjugated bilirubin, respectively.
Objective tumor response was categorized using computed tomography or magnetic resonance imaging every 6-8 weeks according to RECIST1.1. The disease was considered to be stable only if the duration of stabilization was at least 2 mo. Patients who received fewer than 3 cycles of chemotherapy were not evaluated for tumor response, except for those with rapid progression. Evaluations were performed blindly with respect to biochemical markers.
Genomic DNA was extracted from peripheral blood samples using the QIAGEN DNA Blood Mini Kit (Qiagen, Valencia, CA). TA repeats in the UGT1A1 promoter (UGT1A1*28) was genotyped by fragment sizing. Polymerase chain reaction (PCR) was performed in a total volume of 20 μL containing template DNA (80 ng/μL) according to the manufacturer’s instructions (2 × Taq PCR MasterMix; Tiangen Biotech, Beijing, China). A forward primer that was modified by adding a 5′ fluorescent label FAM (FAM_F: 5′-GAACTCCCTGCTACCTTT-3′) and an unlabeled reverse primer (R: 5′-GAACTCCCTGCTACCTTT-3′) were used. The amplification was performed with a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA, United States), started with initial denaturation at 95 °C for 3 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 30 s. Then, the mixtures of the PCR product (TA6, 242 bp; TA7, 244 bp) and Hi-Di formamide (containing the internal size standard Genescan 500 [Applied Biosystems]) were run in the ABI 3730 Genetic Analyzer (Applied Biosystems). Fragment sizes were determined by comparison with Genescan 500 using the local Southern algorithm and analyzed by GeneMapper 3.2 (Applied Biosystems). For quality control purposes, heterozygous and homozygous sequenced samples were included in each run. Genotypes were determined based on the number of TA repeats in each allele (i.e., [TA]6/6, [TA]6/7 and [TA]7/7). Repeat genotyping of 25% of the samples was 100% concordant with original results.
Statistical analyses were performed using SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, United States). Continuous variables are presented as the mean with SD and range and compared using two-sided t-tests or Wilcoxon tests, when appropriate. Categorical variables are expressed as the frequencies and percentages and compared using Pearson’s χ2 test. The cutoff values for TBil and UBil were determined using receiver operating curve (ROC) analysis based on optimal Youden Index. The TBil values were categorized into two groups: > 13.0 or ≤ 13.0; and the UBil values were categorized into two groups: > 4.1 or ≤ 4.1. The parameter combining the TBil and UBil cutoff values (CoBil) was used to further stratify patients. Associations between the objective tumor response and bilirubin indicators or UGT1A1*28 polymorphism were estimated using simple and multiple logistic regression models. Multiple logistic regression analyses were adjusted for age, sex, KPS (≥ 80% or < 80%), histology (glandular or others), primary tumor site (left- or right-side) and treatment regimens. The classifier’s performance of CoBil and UGT1A1*28 was evaluated using a ROC analysis. All tests were two-sided, and P≤ 0.05 indicated statistical significance. The statistical review of the study was performed by a biomedical statistician.
A total of 120 Han Chinese patients with recorded serum bilirubin data and who were available for response assessment were enrolled. Follow-up information was updated in April 2015 when 58% of the patients were deceased. The mean duration of follow-up is 26 mo (range 5-55 mo).
The baseline patient characteristics and tumor biological factors are shown in Table 1. The median age by the time of diagnosis was 50 years (range 18-72 years); 59.2% were males; 20.0% of patients had a KPS less than 80%; and 74.2% of patients were characterized as having a glandular histology. The primary tumors that were proximal or distal to the splenic flexure were categorized as right-sided (n = 33) or left-sided (n = 87), respectively, as described by Loupakis et al[20]. One hundred and nine (90.8%) patients received the FOLFIRI regimen[21], 7 (5.8%) patients received the mXILIRI regimen[21], and the rest (3.3%), who could not bear combined chemotherapy, were treated with irinotecan alone[22].
| Characteristics | Value |
| Total | 120 (100) |
| Age, yr | |
| mean ± SD | 49.6 ± 10.5 |
| Median (range) | 18-72 |
| Gender | |
| Male | 71 (59.2) |
| Female | 49 (40.8) |
| KPS | |
| ≥ 80% | 96 (80.0) |
| 70% | 20 (16.7) |
| 60% | 4 (3.3) |
| Primary tumor | |
| Right-sided | 33 (27.5) |
| Left-sided | 87 (72.5) |
| Histology | |
| Glandular | 89 (74.2) |
| Mucinous | 13 (10.8) |
| Signet-ring cell | 3 (2.5) |
| Mixed | 10 (8.3) |
| Unfixed | 5 (4.2) |
| First-line chemotherapy | |
| FOLFIRI | 109 (90.8) |
| mXELIRI | 7 (5.8) |
| Irinotecan | 4 (3.3) |
| Serum total bilirubin, μmol/L | |
| mean ± SD | 10.2 ± 4.3 |
| Range | 3.1-26.9 |
| Serum unconjugated bilirubin, μmol/L | |
| mean ± SD | 7.2 ± 3.1 |
| Range | 2.5-19.2 |
| UGT1A1*28 genotype1 | |
| (TA)6/6 | 84 (70.0) |
| (TA)6/7 | 34 (28.3) |
| (TA)7/7 | 2 (1.7) |
The mean ± SD of TBil was 10.2 ± 4.3 (range 3.1-26.9), and the mean ± SD of UBil was 7.2 ± 3.1 (range 2.5-19.2). Eighty-four (70%) patients harbored the UGT1A1*28 (TA)6/6 genotype, and 36 (30%) patients harbored variant genotypes, including 34 (28.3%) (TA)6/7 and 2 (1.7%) (TA)7/7 genotypes.
As shown in Table 2, there were no significant differences between patients who had an objective tumor response [complete response (CR) + partial response (PR)] and those who had stable disease or progressive disease (PD) with respect to sex, KPS, primary tumor site, histology and treatment regimen. The mean age was slightly different between the two groups classified by objective response (P = 0.047). Patients with older age tended to have an increased likelihood of objective response (P = 0.066).
| Characteristics | Objective response | P value | P value1 | |
| No | Yes | |||
| Age, yr (mean ± SD) | 48.2 ± 10.9 | 52.1 ± 9.0 | 0.0472 | 0.066 |
| Gender | ||||
| Male | 44 (62.0) | 27 (38.0) | 0.4023 | 0.492 |
| Female | 34 (69.4) | 15 (30.6) | ||
| KPS | ||||
| ≥ 80% | 64 (66.7) | 32 (33.3) | 0.4443 | 0.42 |
| < 80% | 14 (58.3) | 10 (41.7) | ||
| Primary tumor | ||||
| Right-sided | 22 (66.7) | 11 (33.3) | 0.8143 | 0.885 |
| Left-sided | 56 (64.4) | 31 (35.6) | ||
| Histology | ||||
| Glandular | 55 (61.8) | 34 (38.2) | 0.2133 | 0.407 |
| Other | 23 (74.2) | 8 (25.8) | ||
| First-line chemotherapy | ||||
| FOLFIRI | 71 (65.1) | 38 (34.9) | 0.8824 | 0.832 |
| mXELIRI | 4 (57.1) | 3 (42.9) | ||
| Irinotecan | 3 (75.0) | 1 (25.0) | ||
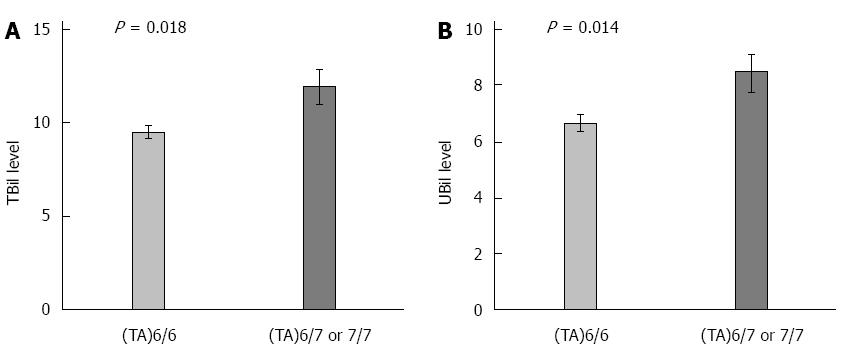
As shown in Figure 1, patients with UGT1A1*28 (TA)6/7 or (TA)7/7 genotype had an increased likelihood of having a higher TBil (P = 0.018, t tests) and a higher UBil (P = 0.014, t tests) level compared with the (TA)6/6 genotype. The mean and SD of TBil was 11.9 ± 5.5 for (TA)6/7 or (TA)7/7 patients, and 9.5 ± 3.4 for (TA)6/6 patients. The mean and SD of UBil was 8.5 ± 3.9 for (TA)6/7 or (TA)7/7 patients, and 6.7 ± 2.5 for (TA)6/6 patients.
As shown in Table 3, the objective response was observed in 42 (35.0%) of 120 patients, including 4 CRs (3.3%) and 38 PRs (31.7%). Stable disease was observed in 52 (43.3%) patients, and PD was observed in 26 (21.7%) patients.
| UGT1A1*28 genotype | Complete response | Partial response | Stable disease | Progressive disease | Objective response2 | Simple analysis | Multiple analysis3 | |||||
| n | Yes | OR | 95%CI | P value | OR | 95%CI | P value | |||||
| (TA)6/6 | 3 (3.6) | 33 (39.3) | 34 (40.5) | 14 (16.7) | 84 | 36 (42.9) | 1.000 (ref.) | 1.000 (ref.) | ||||
| (TA)6/7 or (TA)7/71 | 1 (2.8) | 5 (13.9) | 18 (50.0) | 12 (33.3) | 36 | 6 (16.7) | 0.267 | 0.100-0.709 | 0.008 | 0.244 | 0.088-0.678 | 0.007 |
Patients harboring the minor allele of UGT1A1*28 (TA)6>(TA)7 had a reduced likelihood of objective response compared with the wild-type genotype (OR = 0.267; 95%CI: 0.100-0.709; P = 0.008). In the multiple logistic regression model, the (TA)6/7 or (TA)7/7 genotype remained significantly associated with a decreased objective response rate (OR = 0.244; 95%CI: 0.088-0.678; P = 0.007) compared with the (TA)6/6 genotype.
The optimal Youden Index-based cut-off points were 13.0 μmol/L and 4.1 μmol/L for TBil and UBil, respectively. As shown in Table 4, in the simple analysis, patients with TBil ≤ 13.0 had an increased likelihood of an objective response to treatment compared with the TBil > 13.0 individuals (OR = 3.276; 95%CI: 1.038-10.333; P = 0.043). The association remained significant in multiple analyses adjusted for age, sex, KPS, histology, primary tumor site and treatment regimen (OR = 3.874; 95%CI: 1.127-13.319; P = 0.032). Patients with UBil ≤ 4.1 had approximately a 5-fold higher likelihood of having an objective response compared with the UBil > 4.1 individuals (OR = 5.045; 95%CI: 1.450-17.561; P = 0.011) in the simple analyses. The multiple analysis also showed a significantly increased likelihood of an objective response in patients with UBil ≤ 4.1 compared with the UBil > 4.1 individuals (OR = 5.923; 95%CI: 1.561-22.479; P = 0.011).
| Objective response1 | Simple analysis | Multiple analysis2 | ||||
| n | Yes (%) | OR (95%CI) | P value | OR (95%CI) | P value | |
| Serum total bilirubin | ||||||
| > 13.0 | 24 | 4 (16.7) | 1.000 (ref.) | 1.000 (ref.) | ||
| ≤ 13.0 | 96 | 38 (39.6) | 3.276 (1.038-10.333) | 0.043 | 3.874 (1.127-13.319) | 0.032 |
| Serum unconjugated bilirubin | ||||||
| > 4.1 | 107 | 33 (30.8) | 1.000 (ref.) | 1.000 (ref.) | ||
| ≤ 4.1 | 13 | 9 (69.2) | 5.045 (1.450-17.561) | 0.011 | 5.923 (1.561-22.479) | 0.009 |
Based on the combined TBil and UBil values (CoBil), patients were classified into three groups: Group 1 patients harbored TBil > 13.0 and UBil > 4.1; Group 2 patients harbored TBil ≤ 13.0 and UBil > 4.1; and Group 3 patients harbored TBil ≤ 13.0 and UBil ≤ 4.1. Compared with Group 1, Group 2 had a trend towards an increased likelihood of an objective response in the simple (OR = 2.685; 95%CI: 0.838-8.604; P = 0.096) and multiple (OR = 3.215; 95%CI: 0.918-11.255; P = 0.068) analyses, and Group 3 had more than a 10-fold increase in the likelihood of an objective response in the simple (OR = 11.250; 95%CI: 2.286-55.367; P = 0.003) and multiple (OR = 16.001; 95%CI: 2.802 -91.371; P = 0.002) analyses (Table 5).
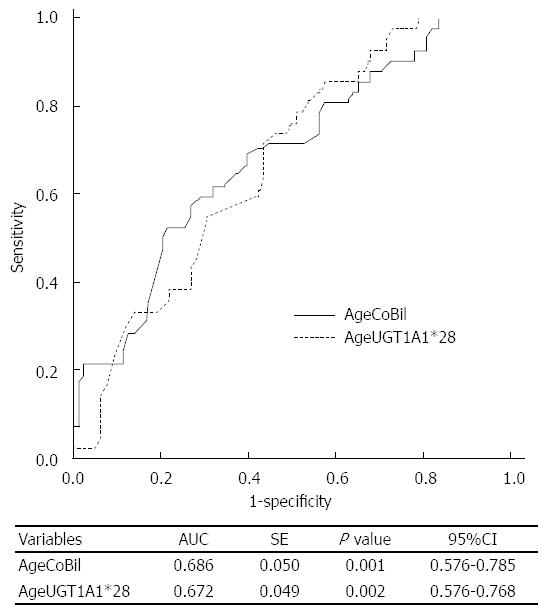
To compare the classifier’s performance of CoBil and UGT1A1*28, an ROC analysis was used. Considering that age may be a potential predictor of objective response, we drew ROC curves of age + CoBil (AgeCoBil) and age + UGT1A1*28 (AgeUGT1A1*28), as shown in Figure 2. The areas under the curves (AUCs) were 0.686 (95%CI: 0.587-0.785) and 0.672 (95%CI: 0.576-0.768) for AgeCoBil and AgeUGT1A1*28, respectively. Therefore, CoBil had a comparable performance for predicting objective response to UGT1A1*28.
This study evaluated whether serum bilirubin levels and UGT1A1*28 polymorphism could predict treatment response of mCRC patients treated with irinotecan-based chemotherapy in a Chinese population. We found that patients with TBil ≤ 13.0 and UBil ≤ 4.1 had a 16-fold higher likelihood of objective response compared with the TBil > 13.0 and UBil > 4.1 individuals; patients carrying the UGT1A1*28 (TA)7 allele were 4-fold less likely to present with an objective response compared with the individuals harboring the homozygous (TA)6 genotype.
Consistent with previous observations, we also found that the UGT1A1*28 variant genotypes were correlated with higher serum TBil and UBil levels[13,14,21,23]. Patients harboring the UGT1A1*28 variant genotypes have a decreased ability to glucuronidate bilirubin, resulting in relatively elevated levels of unconjugated bilirubin as well as total bilirubin[24,25]. At the same time, the metabolism of SN-38 is reduced, leading to higher SN-38 exposures in those patients[26,27].
However, either in patients with the higher bilirubin levels or in patients harboring the UGT1A1*28 variant genotypes, the response rate to irinotecan-based therapy was lower than in the others. The reduced response rate is not due to the toxicity-related dose-reduction because no significant difference in terms of dose-reduction and chemotherapy cycles was observed between the groups classified by the UGT1A1*28 genotypes or CoBil (data shown in the Supplementary Material).
It is pharmacologically plausible that the UGT1A1*28 genotype is associated with irinotecan treatment efficacy. However, published data from different clinical studies are inconsistent[16,28]. Our result was in agreement with the observations by McLeod et al[29] that the homozygous UGT1A1*28 patients showed a trend toward a decreased response rate, but our result was opposite to the findings by Toffoli et al[30] that the homozygous UGT1A1*28 patients had a higher response rate than the wild genotype patients. These inconsistencies may be due to different study populations, distinct study designs and diverse schedules of irinotecan treatment used.
Data with respect to bilirubin and irinotecan-based therapy were limited. Two articles based on previous clinical trials suggested that baseline bilirubin was predictive of grade 3 to 4 neutropenia but not severe diarrhea or therapeutic efficacy[31,32]. These trials were conducted in mCRC patients administered with single-agent irinotecan as second-line chemotherapy, and the analyses were performed retrospectively. In contrast, the current study was conducted prospectively in a Chinese patient population treated with irinotecan-based first-line chemotherapy. The treatment response in first-line therapy is less likely to be confounded by other regimens than that in second-line therapy, and prospective data has a higher level of evidence than retrospective data.
It is proverbial that initial treatment provides the best opportunity for disease palliation or control. The response rate could be reduced from 56% in first-line chemotherapy to 4% in second-line chemotherapy[7]. Hence, biomarkers that could improve therapeutic efficacy by stratifying patients using their predictive effects are of great importance to practitioners and patients alike[33]. For the inconsistency of UGT1A1*28 as a predictor for therapeutic efficacy, UGT1A1*28 genotyping has not been used as a routine test in clinic. However, serum bilirubin level is detected prior to the commencement of each cycle of chemotherapy. The predictive value of baseline bilirubin in tumor response to irinotecan-based therapy is a novel finding in Chinese mCRC patients. As predictors of irinotecan-treatment response, CoBil has a comparable classifier’s performance to UGT1A1*28, and the predictive power is more significant. With validation, CoBil could facilitate stratification of patients for optimal first-line therapy options without extra examinations.
There are some limitations in the current study. First, the study is limited by the restricted number of samples, and requires confirmation in independent external patient cohorts. Second, the result is derived from a unicentral population. Although the research is based on a multicenter clinical trial, only participants at Tongji Hospital are equipped with complete records of baseline bilirubin. Finally, for the longitudinal design, not all patients received standardized treatment schedules as strict as those described in international clinical trials. However, from another perspective, the longitudinal study may represent the treatment experience of patients in a more literal way.
In conclusion, CoBil, as a parameter combining TBil and UBil values, might be clinically useful for predicting treatment response in mCRC patients treated with irinotecan and may help clinicians make informed decisions about first-line treatment selection without extra examinations. We also found that UGT1A1*28 polymorphism is a predictor of treatment response. However, the results need to be validated in other independent prospective studies, and the classifiers for bilirubin warrant verification in cohorts with sufficient power to detect their predictive accuracy.
We thank Guohui Pan from NYU Langone Medical Center for biostatistics assistance and statistical review.
Irinotecan-based therapy is one of the most important fundamental chemotherapy regimens for metastatic colorectal cancer (mCRC). More than 40% of CRC patients will eventually develop metastases and possibly receive irinotecan treatment during the course of the disease. As a first-line treatment, the response rate to irinotecan-based therapy is approximately 35%-55%. No biomarker has been validated for predicting the individual likelihood of response to irinotecan treatment in mCRC patients. The current study was designed to evaluate the predictive effects of baseline serum bilirubin levels and UDP-glucuronosyltransferase (UGT)1A1*28 polymorphism on treatment response of mCRC to irinotecan-based chemotherapy.
Baseline bilirubin has been reported to be associated with irinotecan-induced toxicity. Pharmacological studies demonstrate that SN-38 (the active metabolite of irinotecan) and bilirubin are glucuronidated in the same pathway, and the activity of glucuronidation is influenced by UGT1A1*28 polymorphism.
Literature suggested a connection between circulating bilirubin, UGT1A1*28 polymorphism and irinotecan-induced toxicity. However, the predictive role of bilirubin and UGT1A1*28 regarding treatment outcome of irinotecan-based therapy has been conflicting. For the first time, the authors found that CoBil (an indicator combining the cutoff values of total bilirubin and unconjugated bilirubin) and UGT1A1*28 were both independent biomarkers for predicting the treatment response of mCRC to irinotecan-based chemotherapy in a Chinese population.
After validation, CoBil, an easily determinable index in the clinic, might be helpful in facilitating stratification of mCRC patients for individualized treatment options.
TBil and UBil are total bilirubin and unconjugated bilirubin for short, respectively. CoBil is an indicator combining the cutoff values of TBil and UBil. SN-38 is 7-ethyl-10- hydroxycamptothecin for short. SN-38 is the active metabolite of irinotecan and associated with irinotecan-treatment efficacy.
The authors have performed a good study, and the manuscript is interesting and has clinical relevance.
P- Reviewer: Demir Y S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Ma S
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21370] [Article Influence: 2137.0] [Reference Citation Analysis (3)] |
| 2. | McPhail S, Johnson S, Greenberg D, Peake M, Rous B. Stage at diagnosis and early mortality from cancer in England. Br J Cancer. 2015;112 Suppl 1:S108-S115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 205] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 3. | Yoo PS, Lopez-Soler RI, Longo WE, Cha CH. Liver resection for metastatic colorectal cancer in the age of neoadjuvant chemotherapy and bevacizumab. Clin Colorectal Cancer. 2006;6:202-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 4. | André T, Louvet C, Maindrault-Goebel F, Couteau C, Mabro M, Lotz JP, Gilles-Amar V, Krulik M, Carola E, Izrael V. CPT-11 (irinotecan) addition to bimonthly, high-dose leucovorin and bolus and continuous-infusion 5-fluorouracil (FOLFIRI) for pretreated metastatic colorectal cancer. GERCOR. Eur J Cancer. 1999;35:1343-1347. [PubMed] |
| 5. | Fuchs CS, Marshall J, Mitchell E, Wierzbicki R, Ganju V, Jeffery M, Schulz J, Richards D, Soufi-Mahjoubi R, Wang B. Randomized, controlled trial of irinotecan plus infusional, bolus, or oral fluoropyrimidines in first-line treatment of metastatic colorectal cancer: results from the BICC-C Study. J Clin Oncol. 2007;25:4779-4786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 546] [Cited by in RCA: 598] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 6. | Saltz LB, Cox JV, Blanke C, Rosen LS, Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta N. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med. 2000;343:905-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2273] [Cited by in RCA: 2222] [Article Influence: 88.9] [Reference Citation Analysis (0)] |
| 7. | Tournigand C, André T, Achille E, Lledo G, Flesh M, Mery-Mignard D, Quinaux E, Couteau C, Buyse M, Ganem G. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: a randomized GERCOR study. J Clin Oncol. 2004;22:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2282] [Cited by in RCA: 2200] [Article Influence: 104.8] [Reference Citation Analysis (1)] |
| 8. | De Divitiis C, Nasti G, Montano M, Fisichella R, Iaffaioli RV, Berretta M. Prognostic and predictive response factors in colorectal cancer patients: between hope and reality. World J Gastroenterol. 2014;20:15049-15059. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 55] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (2)] |
| 9. | Cai X, Cao W, Ding H, Liu T, Zhou X, Wang M, Zhong M, Zhao Z, Xu Q, Wang L. Analysis of UGT1A1*28 genotype and SN-38 pharmacokinetics for irinotecan-based chemotherapy in patients with advanced colorectal cancer: results from a multicenter, retrospective study in Shanghai. J Cancer Res Clin Oncol. 2013;139:1579-1589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ. Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes. J Clin Invest. 1998;101:847-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 496] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 11. | Bosma PJ, Seppen J, Goldhoorn B, Bakker C, Oude Elferink RP, Chowdhury JR, Chowdhury NR, Jansen PL. Bilirubin UDP-glucuronosyltransferase 1 is the only relevant bilirubin glucuronidating isoform in man. J Biol Chem. 1994;269:17960-17964. [PubMed] |
| 12. | Iyer L, Hall D, Das S, Mortell MA, Ramírez J, Kim S, Di Rienzo A, Ratain MJ. Phenotype-genotype correlation of in vitro SN-38 (active metabolite of irinotecan) and bilirubin glucuronidation in human liver tissue with UGT1A1 promoter polymorphism. Clin Pharmacol Ther. 1999;65:576-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 247] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 13. | Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet. 1996;347:578-581. [PubMed] |
| 14. | Chen YH, Hung SC, Tarng DC. Serum bilirubin links UGT1A1*28 polymorphism and predicts long-term cardiovascular events and mortality in chronic hemodialysis patients. Clin J Am Soc Nephrol. 2011;6:567-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Cecchin E, Innocenti F, D’Andrea M, Corona G, De Mattia E, Biason P, Buonadonna A, Toffoli G. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27:2457-2465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 16. | Liu X, Cheng D, Kuang Q, Liu G, Xu W. Association between UGT1A1*28 polymorphisms and clinical outcomes of irinotecan-based chemotherapies in colorectal cancer: a meta-analysis in Caucasians. PLoS One. 2013;8:e58489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Xu JM, Wang Y, Ge FJ, Lin L, Liu ZY, Sharma MR. Severe irinotecan-induced toxicity in a patient with UGT1A1 28 and UGT1A1 6 polymorphisms. World J Gastroenterol. 2013;19:3899-3903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 15] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Wang Y, Shen L, Xu N, Wang JW, Jiao SC, Liu ZY, Xu JM. UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J Gastroenterol. 2012;18:6635-6644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15860] [Cited by in RCA: 21625] [Article Influence: 1351.6] [Reference Citation Analysis (1)] |
| 20. | Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, Maus MK, Antoniotti C, Langer C, Scherer SJ. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst. 2015;107:pii dju427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 357] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 21. | Liu CY, Chen PM, Chiou TJ, Liu JH, Lin JK, Lin TC, Chen WS, Jiang JK, Wang HS, Wang WS. UGT1A1*28 polymorphism predicts irinotecan-induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer. 2008;112:1932-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 22. | Fuchs CS, Moore MR, Harker G, Villa L, Rinaldi D, Hecht JR. Phase III comparison of two irinotecan dosing regimens in second-line therapy of metastatic colorectal cancer. J Clin Oncol. 2003;21:807-814. [PubMed] |
| 23. | Sugatani J, Yamakawa K, Yoshinari K, Machida T, Takagi H, Mori M, Kakizaki S, Sueyoshi T, Negishi M, Miwa M. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun. 2002;292:492-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 157] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 24. | Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP. The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert’s syndrome. N Engl J Med. 1995;333:1171-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1077] [Cited by in RCA: 1004] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 25. | Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci USA. 1998;95:8170-8174. [PubMed] |
| 26. | Iyer L, Das S, Janisch L, Wen M, Ramírez J, Karrison T, Fleming GF, Vokes EE, Schilsky RL, Ratain MJ. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43-47. [PubMed] |
| 27. | Stewart CF, Panetta JC, O’Shaughnessy MA, Throm SL, Fraga CH, Owens T, Liu T, Billups C, Rodriguez-Galindo C, Gajjar A. UGT1A1 promoter genotype correlates with SN-38 pharmacokinetics, but not severe toxicity in patients receiving low-dose irinotecan. J Clin Oncol. 2007;25:2594-2600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Dias MM, Pignon JP, Karapetis CS, Boige V, Glimelius B, Kweekel DM, Lara PN, Laurent-Puig P, Martinez-Balibrea E, Páez D. The effect of the UGT1A1*28 allele on survival after irinotecan-based chemotherapy: a collaborative meta-analysis. Pharmacogenomics J. 2014;14:424-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | McLeod HL, Sargent DJ, Marsh S, Green EM, King CR, Fuchs CS, Ramanathan RK, Williamson SK, Findlay BP, Thibodeau SN. Pharmacogenetic predictors of adverse events and response to chemotherapy in metastatic colorectal cancer: results from North American Gastrointestinal Intergroup Trial N9741. J Clin Oncol. 2010;28:3227-3233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 30. | Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D’Andrea M, Pasetto LM, Pessa S, Errante D, De Pangher V. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061-3068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 259] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 31. | Freyer G, Rougier P, Bugat R, Droz JP, Marty M, Bleiberg H, Mignard D, Awad L, Herait P, Culine S. Prognostic factors for tumour response, progression-free survival and toxicity in metastatic colorectal cancer patients given irinotecan (CPT-11) as second-line chemotherapy after 5FU failure. CPT-11 F205, F220, F221 and V222 study groups. Br J Cancer. 2000;83:431-437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 32. | Meyerhardt JA, Kwok A, Ratain MJ, McGovren JP, Fuchs CS. Relationship of baseline serum bilirubin to efficacy and toxicity of single-agent irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2004;22:1439-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Dréanic J, Maillet M, Dhooge M, Mir O, Brezault C, Goldwasser F, Chaussade S, Coriat R. Prognostic value of the Glasgow Prognostic Score in metastatic colorectal cancer in the era of anti-EGFR therapies. Med Oncol. 2013;30:656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |