Published online Jan 7, 2016. doi: 10.3748/wjg.v22.i1.300
Peer-review started: May 20, 2015
First decision: September 9, 2015
Revised: October 14, 2015
Accepted: November 24, 2015
Article in press: November 24, 2015
Published online: January 7, 2016
Processing time: 226 Days and 17.8 Hours
The multifactorial and multistage pathogenesis of hepatocellular carcinoma (HCC) has fascinated a wide spectrum of scientists for decades. While a number of major risk factors have been identified, their mechanistic roles in hepatocarcinogenesis still need to be elucidated. Many tumor suppressor genes (TSGs) have been identified as being involved in HCC. These TSGs can be classified into two groups depending on the situation with respect to allelic mutation/loss in the tumors: the recessive TSGs with two required mutated alleles and the haploinsufficient TSGs with one required mutated allele. Hepatitis B virus (HBV) is one of the most important risk factors associated with HCC. Although mice cannot be infected with HBV due to the narrow host range of HBV and the lack of a proper receptor, one advantage of mouse models for HBV/HCC research is the numerous and powerful genetic tools that help investigate the phenotypic effects of viral proteins and allow the dissection of the dose-dependent action of TSGs. Here, we mainly focus on the application of mouse models in relation to HBV-associated HCC and on TSGs that act either in a recessive or in a haploinsufficient manner. Discoveries obtained using mouse models will have a great impact on HCC translational medicine.
Core tip: Hepatitis B virus (HBV) viral products, in particular the oncogenic HBV X protein, and mutations of tumor suppressor genes (TSGs) are the driving force of hepatocellular carcinoma (HCC). Inactivation of a recessive TSG requires mutations in both alleles and fits the “two-hit” model. However, haploinsufficiency occurs when one allele is insufficient to confer the full functionality of a TSG; the gene’s effect can be partial or complete depending on tissue type, genetic modifiers/background, and environmental factors. Mouse models play a pivotal role in demonstrating the oncogenic effects of viral products and in establishing the dose-dependency and quantitative differences when analyzing a TSG involved in HCC.
- Citation: Teng YC, Shen ZQ, Kao CH, Tsai TF. Hepatocellular carcinoma mouse models: Hepatitis B virus-associated hepatocarcinogenesis and haploinsufficient tumor suppressor genes. World J Gastroenterol 2016; 22(1): 300-325
- URL: https://www.wjgnet.com/1007-9327/full/v22/i1/300.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i1.300
Liver cancer is a member of the top ten most common cancers in both men and women[1]. The most common type of primary liver cancer in adults is hepatocellular carcinoma (HCC), which is derived from hepatocytes and accounts for 70% to 80% of cases[2,3]. Liver cancer has a high mortality and poor prognosis because patients usually are diagnosed with the disease at a well-advanced stage, making liver resection or transplantation unavailable as a therapeutic option. In addition, the recurrence rate of HCC is high in patients who have received liver resection surgery, resulting in a poor cure rate and low long-term survival[1,4]. At present, liver cancer is still a worldwide health issue and remains an unsolved medical problem.
The major risk factors for HCC include: (1) hepatitis virus infection, for example hepatitis B virus (HBV) and hepatitis C virus (HCV); (2) aflatoxin B1 (AFB1) exposure; (3) alcoholic cirrhosis; and (4) metabolic factors such as obesity and diabetes[5-8]. Furthermore, in HBV-associated HCC, HBV replication and the genotype of the HBV per se are also risk factors. A higher serum level of HBV DNA is correlated with the future incidence of HCC in patients[9]. In a manner similar to the HBV infection rate, the distribution of HBV genotypes also shows significant geographical differences. For example, genotypes B and C are predominant in Asia, while genotypes A and D are predominant in Africa, Europe and India. In addition, in Asia, genotype C has been shown to be correlated with advanced liver disease, cirrhosis and HCC more than genotype B[1]. Moreover, there is a highly significant association between patient gender, HBV infection and HCC. Males have a higher HBV infection rate and a higher HCC incidence. The greater susceptibility of males in terms of HCC incidence may be attributed to the tumor-promoting effects of androgens and/or the gender-specific metabolism of carcinogens[10]. AFB1, which is produced by fungi and often contaminates maize and peanuts, is a well-recognized carcinogen of the liver. A metabolic intermediate of AFB1 is able to bind to and damage DNA, leading in HCC to a loss of function of various tumor suppressor genes such as p53[10]. Notably, areas with a high prevalence of chronic HBV infection are usually places having a high risk of AFB1 exposure. This suggests that AFB1 is a promoting factor for HBV-associated HCC and that these two risk factors may have a synergistic effect on hepatocarcinogenesis. Another risk factor related to diet is chronic alcoholism, which leads to fatty liver, fibrosis, advanced liver disease and eventually to HCC. Additionally, smoking has been recognized as a risk factor for HBV-associated HCC development. Furthermore, genetic polymorphisms present in the host are also HCC risk factors. For example, studies have revealed that a loss-of-function deletion of glutathione S-transferase seems to increase the risk of HCC[1,11]. Finally, there are synergistic effects between the different risk factors for HCC. For example, co-transfection with HCV and HBV has an additive effect on the HCC incidence, as does hepatitis virus infection plus AFB1 exposure or hepatitis virus infection plus chronic alcoholism.
Genetically modified mice, including transgenic, knock-out and knock-in mice, are able to provide animal models that help to elucidate the molecular mechanisms related to pathogenesis and carcinogenesis in the liver; furthermore, they also help us to evaluate potential chemopreventive agents and new therapeutic targets under physiological conditions. Although mice cannot be infected with HBV due to the narrow host range of HBV and the lack of a proper receptor or other factors for HBV, one of the advantages of using mouse models is the numerous and powerful resources that genetically modified mice make available. In this review, we mainly focus on the application of genetically modified mouse models to obtain a better understanding of HBV-associated carcinogenesis, the potential involvement of tumor suppressor genes in HCC development, and the potential implications of these findings in mice to translational medicine.
HBV is one of the most important risk factors for HCC[1,3,4]. Clinically, chronic infection with HBV is highly associated with the incidence of HCC and with mortality. Additionally, the geographical distribution of liver cancer seems to be highly correlated with the prevalence of HBV in a population[1]. Epidemiological studies have identified areas with a low prevalence of HBV infection, such as in North America, Western Europe and Northern Europe; areas with an intermediate prevalence of HBV infection, such as Eastern and Southern Europe; and areas with the highest prevalence of HBV infection such as East and Southeast Asia and sub-Saharan Africa[1,3]. Additionally, the epidemiological results show that in the high HBV prevalence areas there are notable differences in the mean age of diagnosis of HCC between men and women, with men showing a lower mean age of diagnosis, and in the incidence of HCC among men than women, with men showing a higher incidence[1]. The age at which infection occurs, as well as environmental and dietary factors, all seem to be related to HCC incidence[1,3]. Infection with HBV is divided into two types, acute and chronic. Most adults with an acute infection of HBV (90% of cases) spontaneously recover and develop protective immunity[12-14]. HBV chronic infection in the remaining 10% of adults can be subdivided into four phases; these are the immune tolerance phase, the immune clearance [hepatitis Be antigen (HBeAg)-positive chronic hepatitis] phase, the inactive (carrier) phase and the reactivation (HBeAg-negative chronic hepatitis) phase; all of these phases may not all be seen in a given individual patient[15]. Up to 30% of HBV carriers develop chronic hepatitis, fibrosis, cirrhosis and eventually HCC[16,17]. In an area with high HBV prevalence, chronic HBV infection often occurs during birth (mother-child transmission). Therefore, an early onset of HBV infection and long-term chronic infection-induced advance liver diseases are likely to be the main factors contributing to the high mortality and high incidence of HCC in these regions[1]. Taiwan, which is located in an HBV high-prevalence region, was a pioneer of national HBV vaccination in the 1980s. A recent study has revealed that chronic disease mortality, HCC incidence and HCC mortality have significantly declined in both males and females who have received HBV immunization[18]. Nonetheless, the life-long effects of HBV vaccination will need to be observed for decades to come. Although immunization against HBV is highly efficacious in terms of preventing HBV-related liver disease, it also results in mutations affecting the genomes of HBV in circulation[19]. In addition, there is still no effective treatment that is able to eliminate HBV from infected patients[12].
HBV is a hepatocyte-specific enveloped DNA virus that is composed of large, middle and small surface proteins. Inside the envelope of the virus, the genome of HBV is packaged within the core proteins, which is formed into nucleocapsids. The genome of HBV is a 3.2-kb partially double-stranded circular DNA that is reversely transcribed from a 3.2-kb pre-genomic RNA (pgRNA). A receptor (sodium taurocholate co-transporting peptide, NTCP), which is responsible for HBV entry into host cells, was recently identified[20,21]. After entering the host cell, the nucleocapsids transport the HBV genome DNA into the nucleus of the infected hepatocytes. The circular HBV DNA is then converted into covalently closed circular DNA (cccDNA), forming a minichromosome inside the nucleus, and these minichromosomes serve as the template for RNA transcription. In some cases, the HBV genome has been found to integrate into the host genome of a patient with chronic HBV infection; however, this is not necessary for viral replication unlike the situation with retroviruses[14,22]. The HBV polymerase, which is covalently linked to the 5’ end of the negative strand of the genome, uses the 5’ end of oligoribonucleotides on the plus strand for reverse transcription. There are four promoters and a shared polyadenylation signal in the genome that are involved in regulating the transcription of HBV proteins. The core promoter is responsible for transcripts of the pgRNA (3.5 kb) and of the pre-core protein, whose transcript is slightly longer than that of the pgRNA. The pre-core protein is able to be secreted as HBeAg after removal of residues at the N-terminus and C-terminus. In addition, in order to serve as a template for reverse transcription, the pgRNA also encodes the core and polymerase proteins. The core protein, which is also known as hepatitis B core antigen, assembles the nucleocapsids of the virus and is responsible for delivering the viral nucleocapsids into the host nucleus. There are two promoters for the surface proteins, which generate 2.4-kb and 2.1-kb transcripts. The 2.4-kb transcript encodes the large surface protein, while the 2.1kb encodes the middle and small surface proteins. These three surface proteins (HBsAg) are synthesized in the rough endoplasmic reticulum (ER) and then transported to the Golgi apparatus for glycosylation; these proteins are then able to be secreted as non-infectious subviral particles. The smallest transcript (0.9 kb) is the template for the X protein. During viral replication, the core protein packages the 3.5-kb pgRNA and polymerase into nucleocapsids. Then, the reverse transcription of pgRNA is carried out inside of the nucleocapsid; this is carried out by the viral polymerase and generates the minus-strand of the HBV DNA first. The minus strand is then used as a template for the synthesis of the plus-strand of HBV DNA. Finally, the nucleocapsids are enveloped by surface proteins during processing on the ER and Golgi apparatus, which is followed by secretion of the virus from the cell[14,23,24].
The host range of HBV is highly limited and is restricted to humans and higher primates[23]. Early studies of hepatitis virus were carried out using woodchuck hepatitis virus (WHV), duck HBV (DHBV) and ground squirrel hepatitis virus (GSHV)[23]. However, the outbred properties of the hosts of WHV and DHBV limited our understanding of the specific molecular mechanisms involved in host-virus interaction and carcinogenesis. Previous studies revealed that the transgenic mouse carrying the HBV genome is not an ideal animal model to investigate the mechanisms of HBV infection. First, the viruses produced by the transgenic liver cannot infect and re-enter their own hepatocytes in mice. Second, the cccDNA, which is the template for viral transcription in humans, was undetectable in the livers of HBV transgenic mice. Third, the transgenic mice have immune tolerance to HBV therefore cannot develop hepatitis. Currently the chimpanzee and tupaia are the only animal models that can be infected by HBV. However, their use is limited by ethics, large size, and low feasibility as genetic manipulation model systems as well as by a very high cost; this compromises their utility in research and development[25]. The problem of establishing a mouse model which can be infected by HBV may be overcome by introducing the receptor of HBV and its critical domain for host-specificity of infection into the hepatocytes of mice by transgenic technology[20,21].
Although the mouse cannot be naturally infected by HBV, transgenic mice carrying the HBV genome have helped scientists to gain insights into the molecular mechanism of viral replication and assembly, and to serve as animal models for evaluation of anti-HBV therapy. First, the viral particles generated in transgenic mouse liver are highly similar to those in human patients. In addition, viruses purified from the blood of HBV transgenic mice can infect human fetal hepatocytes in vitro[26]. This indicated that the molecular mechanism regulating the synthesis of HBV transcripts and proteins, as well as the viral package and viral secretion, probably is shared in human and mouse hepatocytes. Accordingly, these transgenic mouse models may serve as an in vivo platform of animal models to evaluate therapeutic agents against viral replication by nucleoside analog or small interfering RNA[25]. Second, the HBV particles can be generated in transgenic liver but cannot re-enter mouse hepatocytes; this suggests mechanistically that mouse hepatocytes lack a proper HBV receptor for viral internalization. Third, the replication and expression of the HBV genome is not cytopathic or toxic to the host cells; there was no sign of hepatocarcinogenesis at a late stage of mouse life[26]. Notably, the results obtained from HBV transgenic mice were consistent with those from infection experiments of HBV in chimpanzee[14,27]. Thus, it seems that transgenic mice carrying the HBV genome are capable of recapitulating several aspects of the post-infected hepatocytes in natural hosts such as the human and chimpanzee.
There have been three possible mechanisms proposed for HBV-mediated HCC. These are: (1) that a viral protein per se is oncogenic; (2) that there is an infection-promoted immune response and this triggers a long-term process of carcinogenesis in liver; and (3) that the integration of HBV DNA affects the integrity of the host genome[10]. Researchers have tried to use small animals such as the mouse, rather than primates with a longer lifespan, in order to establish platforms for the study in vivo of mechanisms related to liver carcinogenesis. In 1985, two transgenic mouse lines expressing the HBV surface proteins were independently established by two different groups using genetically modified transgenic technology[28,29]. Later, transgenic mice carrying the core gene, the X gene and even the whole genome of the HBV were created. These transgenic mouse models have helped enhance our knowledge of HBV-related biology and liver pathogenesis (Table 1).
| Transgene | Promoter | Expression | Pathology | Ref. |
| PreS/S/X | Endogenous | Surface proteins | Until 6 mo: no obvious pathology | [28] |
| PreS/S/X (two TGs) | Endogenous and mouse metallothionein I | Surface proteins | Not determined | [29] |
| PreS/S/X | Mouse metallothionein I and Albumin | Surface proteins | Ground glass | [22,30-32,34] |
| 7-9 mo: adenoma | ||||
| 12 mo: HCC | ||||
| 18 mo: HCC (100%) | ||||
| Genome (2X) | Endogenous | Surface proteins | Not determined | [241] |
| PreS/S (Knock-in) | Mouse p21 | Surface proteins | 15-24 mo: HCC [53% (♂KI/+), 72% (♂KI/KI), 0% (♀KI/+ and KI/KI)] | [37] |
| PreS/S | Endogenous | Surface proteins | HCC | [33,40] |
| X | Human α-1-antitrypsin | HBx protein | Focal necrosis, hyperplasia nodule (Authors claimed that X is not tumorigenic) | [42] |
| X | Endogenous | HBx protein | 4 mo: altered hepatocyte in multifocal area | [43,44] |
| 10 mo: tumor nodule | ||||
| 16 mo: tumor (80%) | ||||
| 24 mo: HCC (84%) | ||||
| X | Endogenous | HBx protein | 6 mo: neoplastic nodules (66.6%) | [41] |
| 15-18 mo: small neoplastic nodule (100%) and HCC (75%) | ||||
| X (Knock-in) | Mouse p21 | HBx protein | 15-24 mo: HCC [60%-64% (♂KI/+ and KI/KI), 43%-46% (♀KI/+ and KI/KI)] | [37] |
| X | Mouse albumin | HBx protein | 12 mo: dysplasia nodule (100%) | [45-47] |
| 16 mo: HCC (80%) | ||||
| 20 mo: HCC (90%-100%) | ||||
| Precore/core | Mouse metallothionein I | Precore protein | Not determined | [56] |
| Precore/core | Endogenous | Core protein | 12 mo: no hepatitis or HCC | [13,57] |
| Precore/core (two TGs) | Endogenous | Precore and core proteins | Not determined | [58] |
| Genome (2X) | Endogenous | Replicative intermediates | 10 mo: no obvious pathology | [60] |
| Genome (1.2X) | Endogenous | Replicative virus | 12 mo: no hepatitis or HCC | [59,61] |
| 24 mo: no obvious pathology | ||||
| Genome (1.3X) | Endogenous | Replicative intermediates | Until 12 mo: no obvious pathology | [62] |
In patients with HBV chronic infection, the surface protein is the dominant viral product and can be considered to be an onco-protein[28]. Aiming to mimic the human HBV carrier status, transgenic mice expressing the surface protein were the earliest HBV-related transgenic mice to be created[28,29]. One early and important observation was that the accumulation of filamentous surface protein in endoplasmic reticulum leads to hepatotoxicity and carcinogenesis[22,30-32]. The pathogenic effect of HBV surface proteins on the mouse liver are relevant to human HCC development in HBV carriers because phenomena such as ground glass cell formation are found[22,31,33,34]. The sexual dimorphism found to affect HCC incidence in humans can also be observed in transgenic mice[22,35-37]. Another research group generated HBsAg and HBV X protein (HBx) transgenic mice by the knock-in technique in order to reduce the effects of the random integration events that occur with conventional microinjection, and this model system also concluded that the HBV surface protein is oncogenic[37]. HBV vaccine is effective at decreasing HCC prevalence; however, mutated forms of the HBVs in circulation have emerged rapidly, particularly in the PreS/S region[38]. Clinically, patients harboring the PreS mutation are more susceptible to liver cirrhosis and HCC[39]. Efforts were later made to create a transgenic mouse that would help our understanding of the role of the mutated surface protein in pathology. These studies showed that the unfolded protein response (ER stress) is a major factor in HBV surface protein-induced carcinogenesis[33,40].
It has long been suggested that HBx plays a role in hepatocarcinogenesis because infection with DHBV, which lacks the X gene, does not result in the development of HCC, while infection with WHV and GSHV, which do have the X gene, does result in host HCC[41]. In addition, the X gene is highly conserved between different HBV subtypes[42]. Interestingly, the X transcript is the most abundant mRNA compared to other HBV transcripts in some clinical specimens of human HCC[41]. Importantly, several independently generated HBx transgenic mouse models have demonstrated that HBx alone is able to induce malignant transformation of hepatocytes[37,41-47] (Table 1).
The functions of the HBx protein have been extensively studied, particularly in cell culture systems, and are known to involve multiple cellular events, including transactivation of transcription factors, cell cycle progression, several signaling transduction pathways, mitochondrial homeostasis, cell death, DNA instability, glucose metabolism and lipid metabolism[4,10]. It is possible, using transgenic mice, to test in vivo whether the above HBx-mediated cellular events indeed are involved in hepatocarcinogenesis[4]. In addition, these models allow the physiological interacting partners of HBx to be evaluated. Other than mouse, various other model organisms have been used for this type of in vivo studies, for example C. elegans[48]. These studies have shown that the HBx protein binds to CED-9, which is a homolog of members of the anti-apoptotic Bcl-2 protein family; this binding triggers cell death in C. elegans and in a human hepG2 cell line[48,49]. Interestingly, these studies also discovered that when HBx is mutated, it is unable to induce CED-9-regulated cell death. Further investigation of its role in carcinogenesis using mouse models will help our understanding of the molecular mechanism of HBx-mediated HCC.
Previously, we have generated four lines of HBx transgenic mice, namely A105, A106, A110 and A112, using the C57BL/6 background[46]. All of the HBx transgenic lines generated in our laboratory spontaneously develop HCC at 13 to 16 mo of age. At the patho-histological level, the HCC developed in the HBx transgenic mice exhibits a well-differentiated morphology involving the trabecular pattern. Fibrosis, bizarre nuclei, cytoplasmic lipid droplets and hyaline globules can be observed in the HBx-induced HCC samples, which is similar to the situation observed in human HCC samples[46]. Furthermore, a gender disparity for HCC in the HBx transgenic mice has also been observed[37,43,46]. The HBx transgenic mice thus provide an animal model for mechanistic studies[45,50-53] and for the evaluation under physiological conditions of new chemopreventive agents and new therapeutic agents for HCC[47,54,55].
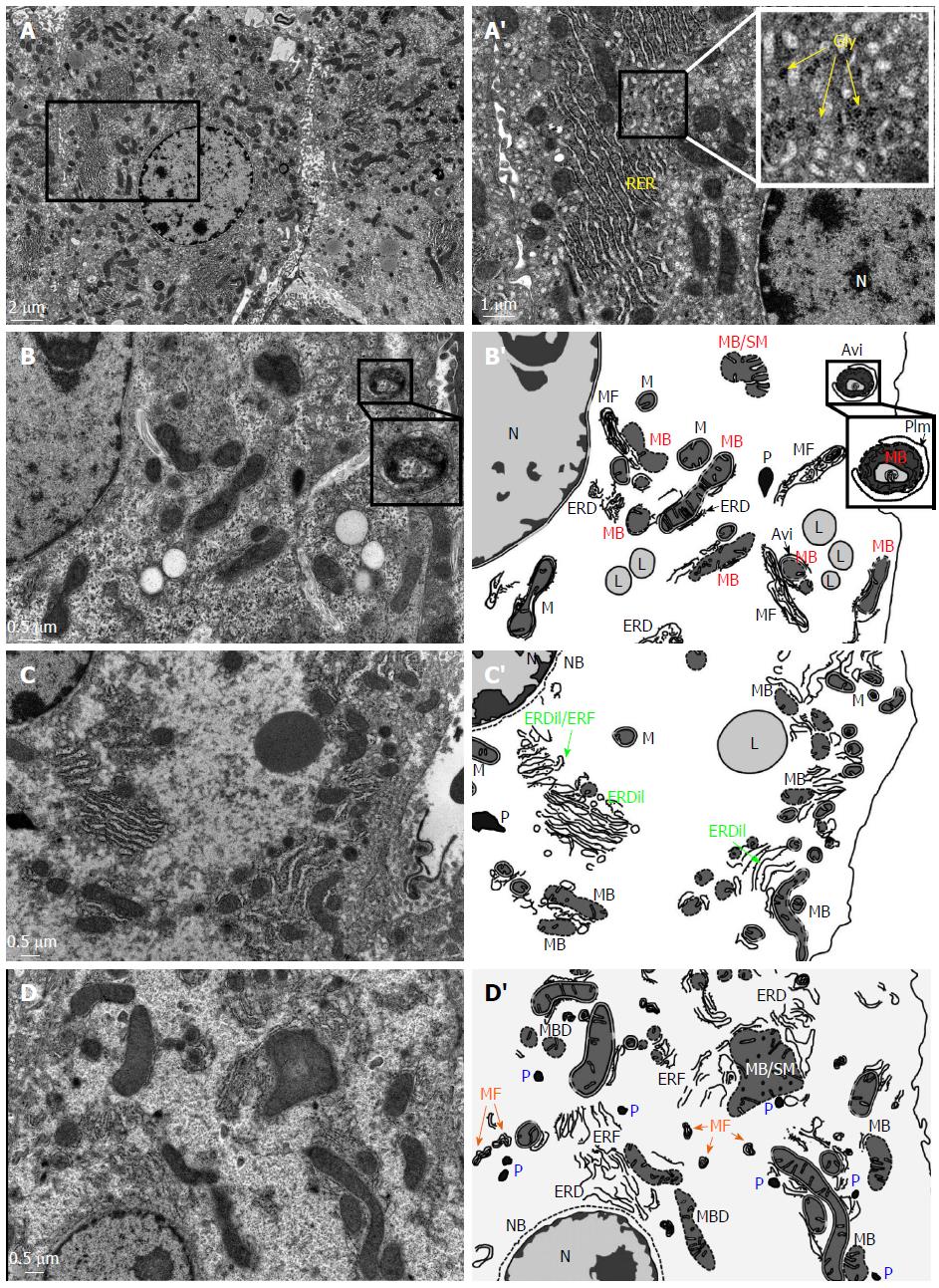
At the ultrastructural level, we have carried out transmission electron microscope (TEM) examinations and these have revealed that there are alterations in the organelles of the hepatocytes of the HBx transgenic mice (Figure 1). In wild-type mice, each normal hepatocyte typically contains a spherical nucleus with homogeneous euchromatin and heterochromatin. Long and stacked cisternae of rough endoplasmic reticulum are located parallel to the nuclear envelope and the lateral edges of the cell. Numerous mitochondria of varying lengths are present, and these range in shape from spherical mitochondria to dumbbell and rod-like shaped mitochondria. Hepatocellular glycogen is apparent in the electron micrographs as irregular non-membrane bound and rosette structures (Figure 1A and A’). In the HBx transgenic mice, TEM reveals that a dramatic decrease has occurred in the density of cytoplasmic organelles (Figure 1B-D). Many ultrastructural abnormalities of the mitochondria can be detected and these include breakdown and swelling of mitochondrial membranes, as well as an increase in the electron density of the matrix (Figure 1B and B’). In addition, the phagophore isolation membrane (PIm), which is the result of the endoplasmic reticulum engulfing degenerated mitochondria, is now present, which leads to the formation of autophagosomes (Figure 1B and B’). The endoplasmic reticulum is dilated or forms a structure with irregular fragmentation and degeneration (Figure 1C and C’). Part of the degenerated membrane debris of mitochondria and endoplasmic reticulum can be detected as irregular concentric myelin figures (Figure 1D and D’). Furthermore, some hepatocytes of HBx mice have abundant peroxisomes; this is likely to be due to the oxidative stress induced by the HBx protein (Figure 1D and D’). Moreover, the nuclear envelopes of some hepatocytes of HBx transgenic mice have undergone breakdown (Figure 1D and D’; Figure 1C and C’). All of these ultrastructural alterations and organelle degeneration events, particularly the abnormalities in the mitochondria and endoplasmic reticulum, appear at a very early stage (4-8 wk old) and contribute to the progression of carcinogenesis in the livers of HBx transgenic mice.
Previously, several groups have established transgenic mice in order to investigate HBV assembly within the hepatocyte in vivo (Table 1). Transgenic mice carrying the precore or core proteins of HBV have no overt phenotype (no sign of hepatitis and HCC formation) up to 12 mo-old[13,56-58]. Transgenic mice carrying the whole HBV genome have also been generated in order to study the HBV life cycle and its interaction with host factors[26,59-62] (Table 1). Virus particles that contain HBsAg, HBc/eAg and HBV DNA can be detected in the bloodstream of these transgenic mice. Interestingly, a gender difference in the synthesis of HBeAg and intermediates of the replicative virus can be observed in these mice[63]. However, similar to the precore/core transgenic mice, there are no obvious pathological changes detectable in the transgenic liver. Nevertheless, mice carrying the whole HBV genome do provide animal models that are amenable to the study of the HBV life cycle, and they can also help the screening of therapeutic agents against HBV replication.
No single transgenic mouse model is able to cover all aspects of HCC pathogenesis. Currently there are still some barriers when using genetically modified mice, such as their inbred genetic backgrounds versus the complications of human genetic variation, the inability to be infected by HBV, the lack of an immunopathogenesis process, the scarcity of cirrhosis, and the very rare nature of spontaneous metastasis. Accordingly, alternative strategies have been created and applied to mice in order to study HBV biology. For example, hydrodynamic injection of HBV DNA into mice[25,64] and humanized mice[65]. These models have further extended the feasibility and have helped to create an understanding of virus-host interactions as well as the pathological effects of HBV mutant proteins.
Transgenic mice carrying the HBV genome or expressing various HBV proteins provide valuable models that help to elucidate clinic observations, allowing mechanistic investigations to be performed and helping to test relevant hypotheses. These approaches also help our understanding of HBV replication and the regulation of HBV gene expression, as well as of the inflammation and innate immune response that is induced by HBV proteins under physiological conditions[13,63,66-68]. In addition, these previously published transgenic mouse models have been employed as a platform for discovery of therapeutic targets and/or for the identification of preventive chemicals[4,27,47,54,55,69]. For example, novel HBV-related microRNAs present in the hepatic and general circulation have been discovered over the past decades and are considered to be potential targets for therapy[10,70]. The possible roles of these microRNAs in carcinogenesis and their therapeutic applications can certainly be tested using the available established HBV-related mouse models. The ultimate goal, of course, is still to use transgenic mice to find a cure for patients with HBV infection and HCC. Nevertheless, there is still a great need for a good animal model for HCC and such a model would be expected to meet as many of the following requirements as possible. These are: (1) the faithful reproduction of HCC’s progression stages and incidence; (2) a reliable reproduction of the molecular and cellular events during HCC carcinogenesis; (3) the ability to mimic the tumor-host and tumor-metastases interactions; (4) a reproduction of the tumor microenvironment in human patients; and (5) the ability to manipulate in a feasible manner the genome of HBV[4].
Additionally, the HBV virus at present is also evolving in response to environmental changes such as vaccination or drug administration. For example, the emergence of mutated hepatitis B surface protein, which may potentially have vaccine-resistant properties, is a serious issue[38]. The generation of new transgenic mouse model systems is one way to dissect the effects of such mutant HBV proteins on liver carcinogenesis. Lately, a newly developed genome-editing tool, the Crips/Cas9 system, has been successfully applied and used to target the HBV DNA and reduce the production of HBV proteins[71-73]. Using a hydrodynamics-HBV persistence mouse model, Lin et al[72] have demonstrated that the Crips/Cas9 system is able to cleave the intrahepatic HBV genome containing the plasmid and facilitate its clearance in animals. These findings suggest that the genome-editing tool may have potential in the eradication of persistent HBV infection.
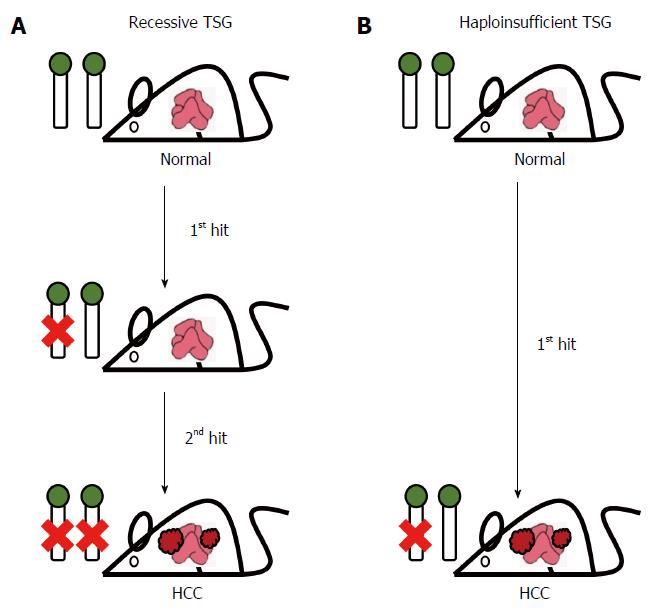
The identification of tumor suppressor genes (TSGs) is very important to the development of novel therapeutic strategies targeting HCC. Many TSGs have been identified in human HCC tissue samples, using cell culture systems and with mouse models[74-77]. These TSGs can be classified into two major groups, the recessive TSGs and the haploinsufficient TSGs (Figure 2). The recessive TSG group follows the Knudson’s two-hit hypothesis, in which a deficiency affecting both alleles of a TSG triggers HCC development. For the haploinsufficient TSG group, the loss of a single allele of a TSG is sufficient to promote tumorigenesis, with the second allele remaining intact in the tumors. These latter TSGs are functionally haploinsufficient in the heterozygous condition and it would seem that cellular level of 50% for their gene product is insufficient to suppress tumor formation[78-80]. Using mouse models, scientists have been able to delineate the molecular mechanisms underlying the roles of both recessive and haploinsufficient TSGs during the suppression of carcinogenesis in liver. These findings should contribute significantly to the development of novel chemopreventive or therapeutic strategies for HCC. Here, we will review the TSGs in HCC mainly based on the pathways in which they are involved (Table 2).
| Official gene name (common name) | Mouse genotype | Mouse genetic background | Pathology | TGS type | Functions | Human Chromosome location | Expression in human HCC tumor | Ref. |
| Autophagy regulator | ||||||||
| Becn1 (Beclin1) | Beclin1+/- | Mixed (129Sv/J; C57BL/6J) | 16-18 mo: HCC (8%) | Haplo-insufficiency | Regulation of autophagy | 17q21 | Down | [85-87] |
| 18-22 mo: HCC (26%) | ||||||||
| Promote HBV-mediated HCC | ||||||||
| Atg5 | Alb-Cre; Atg5flx/flx | C57BL/6 | 2 mo: hepatic cell death, proliferation, inflammation, fibrosis and hepatomegaly | Recessive | Control autophagosome formation | 6q21 | Down | [90,91] |
| 12 mo: hepatic adenoma (100%) | ||||||||
| Atg7 | Alb-Cre; Atg7flx/flx; | C57BL/6 | 4 mo: fatty liver | Recessive | Control autophagosome formation | 3p25.3 | Up | [92-94] |
| 12 mo: hepatic adenoma (90.9%) | ||||||||
| Cell cycle regulator | ||||||||
| Klf6 | KLF6+/- | C57BL/6 | Promote DEN-induced HCC | Haplo-insufficiency | Cell cycle regulation | 10p15 | Down | [99,101] |
| Plk4 | Plk4+/- | Mixed (129Sv;CD1) | 3 mo: mitotic failure in hepatocyte regeneration | Haplo-insufficiency | Control of cell cycle progression | 4q28 | Down | [104,105] |
| 18-24 mo: HCC (30%) | ||||||||
| Cdkn1a (p21) | p21+/- | Mixed (129Sv; C57BL/6) | Promote γ-irradiation-induced HCC | Haplo-insufficiency | Inhibits the activity of CDK2 or CDK4 | 6p21.2 | Up | [107,242] |
| p21-/- | C57BL/6 | Promote HCC in Nemo liver-specific KO | Recessive | [108] | ||||
| Trp53 (p53) | p53+/- | C57BL/6 | Promote HBV-mediated and AFB1-induced HCC | Haplo-insufficiency | DNA damage response transcription factor | 17p13.1 | Up | [111,243] |
| Aflp-Cre; p53flx/flx | C57BL/6 | 14-20 mo: HCC (90%) | Recessive | [112] | ||||
| Protein ubiquination | ||||||||
| Amfr (Gp78) | Gp78-/- | C57BL/6 | 12 mo: fatty liver, inflammation and HCC (24%) | Recessive | E3 ubiquitin ligase | 16q21 | Down | [117] |
| Park2 (Parkin) | Parkin-/- | Mixed (129/C57BL6SJL) | 11-12 mo: hepatocyte proliferation and hepatomegaly | Recessive | E3 ubiquitin ligase | 6q25.2-q27 | Down | [118,122] |
| 16-17 mo: HCC (33%) | ||||||||
| 22-23 mo: HCC (45%) | ||||||||
| Trim24 | Trim24-/- | C57BL/6 | 2 mo: hepatic cell death and inflammation | Recessive | E3 ubiquitin ligase | 7q32 | Up | [119,125] |
| 4-6 mo: fatty liver, fibrosis and liver nodule | ||||||||
| 9-21 mo: HCC (55%) | ||||||||
| Genome stability maintenance | ||||||||
| Anxa7 (Annexin 7) | Anx7+/- | C57BL/6 | 10 mo: Hepatomegaly | Haploinsufficiency | Ca2+-dependent endocrine secretion | 10q22.2 | Up | [131,132] |
| 12 mo: HCC (3.6%) | ||||||||
| Nbn (Nibrin) | Nbn+/- | Mixed (129Sv; C57BL/6) | 23 mo: HCC (8.6%) | Haploinsufficiency | DNA repair | 8q21 | Up | [134,244] |
| Pinx1 | PinX1+/- | Mixed (129Sv; C57BL/6) | 9-18 mo: HCC (17.7%) | Haploinsufficiency | Telomerase inhibitor | 8p23 | Down | [136,137] |
| Sgol1 (Shugoshin 1) | Sgo1+/- | C57BL/6 | 4 mo: hepatocyte DNA damage | Haploinsufficiency | Protector of chromosome cohesion and centrosome integrity | 3p24.3 | Up | [139,140] |
| 12 mo: HCC | ||||||||
| Promote AOM-induced HCC | ||||||||
| Metabolic function | ||||||||
| Acox1 (Acyl-CoA oxidase 1) | Aox-/- | Mixed (129/Ola; C57BL/6) | 2-4 mo: fatty liver, inflammation and hepatocyte proliferation | Recessive | Peroxisomal β-oxidation | 17q25.1 | Not determined | [144] |
| 10-15 mo: HCC (100%) | ||||||||
| Bhmt | Bhmt-/- | C57BL/6 | 1 mo: fatty liver | Recessive | Methionine metabolism | 5q14.1 | Down | [149,150] |
| 12 mo: HCC (50%) | ||||||||
| Nr1h4 (Farnesoid X receptor) | Fxr-/- | C57BL/6N | 3 mo: hepatic cell death and proliferation | Recessive | Transcriptional regulation of synthesis and transport of bile acids | 12q23.1 | Down | [155,157] |
| 6-9 mo: fatty liver and inflammation | ||||||||
| 12 mo: fibrosis and HCC (16.1%) | ||||||||
| Gnmt (Glycine N-methyltransferase) | Gnmt-/- | Mixed (129Sv; C57BL/6) | 3 mo: fatty liver and fibrosis | Recessive | Methionine metabolism | 6p12 | Down | [148,151] |
| 8 mo: HCC (100%) at 8-mo | ||||||||
| Abcb4 (Mdr2) | Mdr2-/- | 129/OlaHsd | 3 mo: inflammation and fibrosis | Recessive | Phosphatidylcholine translocase | 7q21.1 | Down | [156,158] |
| 6-mo: HCC | ||||||||
| Hippo signaling pathway | ||||||||
| Stk4 (Mst1) | Alb-Cre; Mst1/2flx/flx | Mixed (129Sv; C57BL/6;CD1) | 1 mo: hepatocyte proliferation and hepatomegaly | Recessive | Kinase (negative regulator) | 20q11.2 | Not determined | [165,166] |
| 6 mo: HCC (100%) | ||||||||
| Stk3 (Mst2) | Alb-Cre; Mst1/2flx/flx | Mixed (129Sv; C57BL/6;CD1) | 1 mo: hepatocyte proliferation and hepatomegaly | Recessive | Kinase (negative regulator) | 8q22.2 | Not determined | [165,166] |
| 6 mo: HCC (100%) | ||||||||
| Nf2 (Neurofibromin 2) | Alb-Cre; Nf2flx/flx | Mixed (FVB/N; C57BL/6) | 2 mo: oval cell over-proliferation and hepatomegaly | Recessive | Negative regulator | 22q12.2 | Not determined | [168] |
| 7 mo: HCC (100%) | ||||||||
| Jak/Stat signaling pathway | ||||||||
| Socs1 | Socs1+/- | C57BL/6 | Promote DEN-induced HCC | Haploinsufficiency | Negative regulator | 16p13.13 | Down | [174] |
| Socs3 | Socs3+/- | Mixed (129Sv; C57BL/6) | Promote DEN-induced HCC | Haploinsufficiency | Negative regulator | 17q25.3 | Down | [176] |
| Ptpn11 (Shp2) | Alb-Cre; Shp2flx/flx | C57BL/6 | 2-3 mo: inflammation and hepatic cell death | Recessive | Protein tyrosine phosphatase (negative regulator) | 12q24 | Down | [179] |
| 8-mo: hyperplasia nodule | ||||||||
| 12-18 mo: hepatic adenoma (68%) | ||||||||
| Promote DEN-induced HCC | ||||||||
| Ptpro | Ptpro-/- | C57BL/6 | Promote DEN-induced HCC | Recessive | Receptor-like protein tyrosine phosphatase (negative regulator) | 12p13.3 | Down | [181,182] |
| NF-κB signaling pathway | ||||||||
| Cyld (Cylindromatosis) | Alfp-Cre; Cyldflx/flx | C57BL/6 congenic | 1-2 mo: hepatic cell death, inflammation and fibrosis | Recessive | Deubiquitinase (negative regulator) | 16q12.1 | Down | [188,189] |
| 12 mo: HCC | ||||||||
| Lgals3 (Galectin-3) | Gal3-/- | CD1 | 6 mo: fatty liver and inflammation | Recessive | Regulation of inflammatory responses | 14q22.3 | Up | [192-194] |
| 15 mo: fibrosis and liver nodule | ||||||||
| 25 mo: HCC (100%) | ||||||||
| Ikbkg (Nemo) | Alfp-Cre; Nemoflx/flx | C57BL/6 | 2 mo: fatty liver, inflammation, hepatic cell death and proliferation | Recessive | Activation of NF-κB | Xq28 | Down | [195,196] |
| 6 mo: dysplastic nodule | ||||||||
| 12 mo: HCC (100%) | ||||||||
| Map3k7 (Tak1) | Alfp-Cre; Tak1flx/flx | Mixed (129/Ola; C57BL/6) | 1-2 mo: hepatic cell death, proliferation and fibrosis | Recessive | Serine/threonine protein kinase (Activation of the NF-κB) | 6q15 | Not determined | [197] |
| 4-8 mo: HCC (88%) | ||||||||
| PI3K/Akt/mTOR signaling pathway | ||||||||
| Stk11 (Lkb1) | Lkb1+/- | Mixed (129Sv; C57BL/6) | 10-12 mo: Hepatic hyperplasia, HCC (29%) | Recessive | Serine/threonine kinase (activator of AMPK) | 19p13.3 | Down | [201,202] |
| 14 mo: HCC (75%) | ||||||||
| Pten | Alb-Cre; Ptenflx/flx | C57BL/6 | 2-3 mo: fatty liver and hepatomegaly | Recessive | Phospholipid phosphatase (negative regulator) | 10q23.3 | Down | [204,208] |
| 9-10 mo: inflammation and fibrosis | ||||||||
| 17-18 mo: HCC (66%) | ||||||||
| Raptor (Regulatory associated protein of mTOR, complex 1) | Alb-Cre; Raptorflx/flx | C57BL/6 | 2 mo: hepatic cell death, inflammation and fibrosis | Recessive | Activation of mTOR activity | 17q25.3 | Up | [209,210] |
| Promote DEN-induced HCC | ||||||||
| TGF-β signaling pathway | ||||||||
| Sptbn1 (β-spectrin) | Elf+/- | Mixed (129SvEv; Black Swiss) | 15 mo: Fatty liver, HCC (40%) | Haploinsufficiency | Propagation of TGF-β signal | 2p21 | Down | [214] |
| Tgfb1 (TGF-β1) | Tgf-β1+/- | C57BL/6NCr | Promote DEN-induced HCC | Haploinsufficiency | Growth factor | 19q13.1 | Up | [216,245] |
| Tgfbr2 (TGF-β type II receptor) | TβR-II+/- | Mixed (129Sv; C57BL/6) | Promote DEN-induced HCC | Recessive | TGF-β receptor | 3p22 | Down | [217,218] |
| Wnt signaling pathway | ||||||||
| Apc | Adenovirus-Cre; Apcflx/flx | C57BL/6N | 9 mo: HCC (67%) | Recessive | Antagonist | 5q21 | Down | [222,223] |
| MicroRNA | ||||||||
| Mir122 | Mir122a-/- | C57BL/6 | 3 mo: fatty liver, inflammation and fibrosis | Recessive | Post-transcriptional regulation of gene expression | 18q21.31 | Down | [227,228] |
| 11 mo: HCC (75%) | ||||||||
| Mir140 | Mir140-/- | C57BL/6 | Promote DEN-induced HCC | Recessive | Post-transcriptional regulation of gene expression | 16q22.1 | Not determined | [229] |
| Miscellaneous | ||||||||
| Ncoa5 (Nuclear receptor coactivator 5) | Ncoa5+/- | Mixed (129Sv; C57BL/6) | 6 mo: fatty liver | Haploinsufficiency | Estrogen receptor coactivator | 20q13.12 | Down | [231] |
| 10 mo: inflammation and fibrosis | ||||||||
| 10-18 mo: HCC (94%) | ||||||||
| Prkar1a | Prkar1a+/- | Mixed (129Sv/J; C57BL/6) | 9-19 mo: HCC (29.4%) | Haploinsufficiency | Regulation of the serine/ threonine kinase activity | 17q24.2 | Up | [233,246] |
| Ncoa2 (Nuclear receptor coactivator 2) | Ncoa2-/- | Mixed (129Sv/J; C57BL/6) | Promote DEN-induced HCC | Recessive | Transcriptional coactivator | 8q13.3 | Down | [235] |
| Nfe2l1 (Nuclear factor, erythroid 2-like 1) | Alb-Cre; Nrf1flx/flx | Mixed (129Sv; C57BL/6) | 1-2 mo: fatty liver, necrosis and inflammation | Recessive | Transcription factor | 17q21.3 | Not determined | [238] |
| 6 mo: fibrosis | ||||||||
| 12 mo: HCC (100%) | ||||||||
Autophagy is a highly regulated process that degrades damaged cellular organelles and macromolecules allowing recycling of bioenergetic molecules. Many proteins are involved in the autophagy pathway, including the Beclin1 and autophagy-related gene (Atg) proteins. Autophagy plays dual roles in hepatocarcinogenesis; firstly, it acts as a tumor suppressor during the initiation stages of HCC, while exerting a tumor supportive function during the promotion and progression stages. In normal cells, autophagy has a tumor suppressor function that aims to maintain normal metabolism, preserve genetic stability and inhibit inflammation. On the other hand, autophagy also plays a role in supporting tumor progression by helping cancer cells survive stress-induced cell death[81,82]. The tumor suppressor function of autophagy has been demonstrated using mouse models.
Beclin1: Beclin1 is one of the major mediators of autophagy; this gene is located on human chromosome 17q21. Mono-allelic deletion has been detected in various human cancers including ovarian, breast and prostate cancers[83]. A decrease in the levels of autophagy has been observed in a highly malignant HCC cell line compared with immortalized normal hepatic cells. Furthermore, the expression level of Beclin1 has been found to be decreased in human HCC tissues compared with the adjacent non-tumor tissues[84,85]. Homozygous knockout of Beclin1 leads to embryonic lethality; this finding demonstrates that Beclin1 is essential for early embryonic development in mice. Importantly, heterozygous deficiency of Beclin1 leads to the development of spontaneous HCC and accelerates HBV-induced hepatocarcinogenesis in mice. Moreover, the wild-type allele of Beclin1 has been found not to be mutated/lost in the HCC tissue of the Beclin1+/- mice[86,87]. These studies demonstrated that Beclin1 is a haploinsufficient TSG in relation to HCC.
ATG5 and ATG7:Atg5 and Atg7 are autophagy-related genes (Atg) that promote the elongation of the autophagosome membrane. Homozygous knockout of either Atg5 or Atg7 leads to impairment of autophagosome formation and postnatal lethality in mice[88,89]. Regarding ATG5, mutation and/or loss of ATG5 expression has been observed in human HCC tissues[90]. In mice, hepatocyte-specific knockout of Atg5 was found to result in hepatomegaly, hepatic cell death, compensatory proliferation of hepatocytes, inflammation and fibrosis, as well as the development of hepatocellular adenomas in mice[91]. Regarding ATG7, the expression of the ATG7 protein is increased in human HCC tissue samples compared with their adjacent non-tumor tissue samples, but the mechanism is currently unknown[92]. However, mice with a hepatocyte-specific knockout of Atg7 develop fatty liver and hepatocellular adenoma, indicating that autophagy is involved in lipid metabolism and plays a role in suppressing tumor formation in the liver[93,94].
Cell cycle progression is regulated by cyclin-dependent kinases (CDKs) together with several activators (cyclins) and CDKs inhibitors (p21 and p27). In normal tissue, the cell cycle is carefully controlled and regulated in order to maintain cell number homeostasis[95]. Abnormal cell cycle progression and sustained cell proliferation is one of the hallmarks of cancer cells[96]. Many genes involved in cell cycle control have potential tumor suppressive roles in HCC.
Kruppel-like factor 6: Kruppel-like factor 6 (KLF6) is a ubiquitously expressed zinc finger transcription factor that regulates the cell cycle and signal transduction. KLF6 is frequently inactivated by loss of heterozygosity, somatic mutation or promoter methylation in various cancers including prostate, colon and liver cancer[97,98]. In mice, heterozygous deficiency of Klf6 promotes diethylnitrosamine (DEN)-induced hepatocarcinogenesis in Klf6+/- mice; however, whether the wild-type allele of Klf6 remained intact in the HCC was not determined during this study[99]. Liver-specific knockout of the two alleles of Klf6 enhanced DEN-induced HCC in mice[100]. In humans, expression of KLF6 is decreased in HCV-related HCC tissue samples[99]. Down-regulation of KLF6 has also been observed in HBV-related human HCC[101]. These studies in humans and mice suggest that KLF6 may function as a haploinsufficient TSG in HCC.
Polo-like kinase 4: Polo-like kinase 4 (PLK4) is a member of the polo-like kinase protein family that plays a critical role in cell cycle progression[102]. Homozygous knockout of Plk4 resulted in embryonic lethality due to a mitotic defect in mice[103]. Importantly, heterozygous deficiency of Plk4 leads to mitotic failure of regenerated hepatocytes and spontaneous HCC development in the heterozygous knockout mice. Furthermore, the wild-type allele of Plk4 has been shown to remain intact in these HCC tissue samples[104]. In humans, the expression level of PLK4 is down-regulated in HCC tissue samples[105]. These studies indicate that PLK4 functions as a haploinsufficient TSG in HCC.
p21: The p21 protein is an inhibitor of CDK and is involved in cell cycle control, cell senescence and cell death[106]. Heterozygous and homozygous knockout of p21 promotes γ-irradiation-induced tumor formation in many types of cancers including HCC. The p21 protein has been found to be expressed in malignant tumors from irradiated p21+/- mice, suggesting that p21 functions as a haploinsufficient TSG in this situation[107]. Moreover, homozygous knockout of p21 in hepatocytes has been shown to enhance NEMO-mediated hepatocarcinogenesis in mice[108]. These results suggested that p21 is able to function as a recessive or haploinsufficient TSG across various different tissues under a range of different physiological/environmental settings in mice[107].
p53: The p53 protein is a transcription factor that regulates DNA damage and stress responses. This protein functions as a tumor suppressor in many types of cancers[109,110]. Heterozygous knockout of p53 has been shown to promote HBV-mediated and AFB1-induced HCC development in mice; furthermore, the wild-type allele of p53 has been found to remain intact in the HCC tissues of these p53+/- mice, suggesting that p53 functions as a haploinsufficient TSG in these circumstances[111]. Liver-specific knockout of the two alleles of p53 leads to spontaneous HCC development in mice. These findings suggest that p53, like p21, is able to function as either a recessive TSG or a haploinsufficient TSG in different tissues under a range of different physiological/environmental settings in mice[112].
Intracellular protein degradation occurs via two major pathways; these are the ubiquitin-proteasome system and the lysosomal-mediated proteolysis pathway. The ubiquitin-proteasome system is a multi-step process involving protein labeling by polyubiquitination, which is followed by protein degradation via the proteasome. Protein ubiquitination is a tightly regulated process that involves three major enzymes, namely E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase[113,114]. The E3 ubiquitin ligases act by targeting specific proteins, including gene products from oncogenes and tumor suppressor genes. Abnormal regulation of the E3 ubiquitin ligase is involved in the development of many types of cancer, including HCC[115,116]. Many E3 ubiquitin ligases have been demonstrated to function as tumor suppressors of HCC in mice, including Gp78, Parkin and Trim24[117-119]. These mouse models have been able to uncover the role of E3 ubiquitin ligases in hepatocarcinogenesis.
Glycoprotein 78: Glycoprotein 78 (GP78) is an endoplasmic reticulum (ER) membrane-anchored E3 ubiquitin ligase, which is involved in the ER-associated degradation (ERAD) pathway. The ERAD pathway is a protective mechanism that helps to maintain ER protein homeostasis through degradation of misfolded proteins[120]. In mice, homozygous knockout of Gp78 results in the development of non-alcoholic steatohepatitis (NASH), liver fibrosis and HCC. In humans, GP78 has been shown to be down-regulated in HCC tissue samples compared to adjacent non-tumor liver tissue samples[117]. These studies suggest that Gp78 is a regulator of ER homeostasis in the normal liver, and functions as a recessive TSG in HCC.
Parkin: Parkin is an E3 ubiquitin ligase involved in protein turnover, the stress response, mitochondrial homeostasis, metabolism and cell growth. Inactivation of Parkin has been frequently detected in many types of human cancers including HCC[121]. In humans, the protein levels of Parkin have been shown to be decreased in HCC tissue samples and HCC cell lines[122]. In mice, Parkin deficiency leads to hepatomegaly and HCC via an increase in the hepatocyte proliferation and a reduction in hepatic cell death. Additionally, disruption of Parkin results in impaired lipid uptake by hepatocytes when the mice are fed a high fat diet (HFD)[118,123]. These studies reveal that Parkin plays an important role in maintaining cell numbers and lipid homeostasis in the liver and functions as a recessive TSG in HCC.
Tripartite motif 24: Tripartite motif 24 (TRIM24) is located in the HCC critical region on human chromosome 7q32, suggesting that TRIM24 may function as a tumor suppressor in HCC. TRIM24 is an E3 ubiquitin ligase, which negatively regulates the level of p53[124]. Upregulation of the TRIM24 protein has been observed in human HCC tissue samples compared with their adjacent non-tumor tissue samples[125]. Trim24 deficiency has been found to result in the development of fatty liver, hepatic injury and spontaneous HCC in homozygous knockout mice, which suggests that Trim24 functions as a recessive TSG in HCC[119]. In addition, these studies have also revealed that the expression level of Trim24 is an important factor in the progression of HCC.
Genomic stability is very important because it allows cells to avoid neoplastic transformation and tumorigenesis. The major mechanisms for maintaining genomic stability are high-fidelity DNA replication, accurate chromosome segregation, faithful DNA repair and cell cycle checkpoint control[126]. Genomic instability is a hallmark of most cancers and significantly contributes to cancer initiation and progression[127,128]. Many haploinsufficient TSGs in HCC are functionally involved in the maintenance of genomic stability.
Annexin 7: Annexin 7 (ANX7) is a member of the annexin family and is a Ca2+-dependent phospholipid-binding protein[129]. Homozygous knockout of Anx7 in mice leads to postnatal lethality at embryonic day 10 due to cerebral hemorrhage. Interestingly, heterozygous knockout of Anx7 results in a Ca2+-dependent endocrine secretory defect in mice, indicating that Anx7 plays an important role in the regulation of endocrine secretion[130,131]. In addition, heterozygous deficiency of Anx7 exhibits a phenotype of hepatomegaly and HCC develops spontaneously in these mice; the wild-type allele of Anx7 has been found to remain intact in the HCC tissue from these Anx7+/- mice[132]. Genomic instability and downregulation of tumor suppressor genes have been observed in the HCC of Anx7+/- mice. These findings suggest that Anx7 helps to maintain genomic stability and functions as a haploinsufficient TSG in HCC.
Nibrin: Nibrin (NBN) is involved in the repair of DNA double stranded breaks and has the potential to function as a tumor suppressor gene[133]. Homozygous knockout of Nbn causes embryonic lethality between embryonic day E3.5 and E7.5 in mice. Importantly, heterozygous deficiency of Nbn results in many types of spontaneous tumors including prostate cancer, mammary gland tumors, lymphoma and HCC in mice. Various chromosome aberrations, including fragmentation and end-to-end fusion, have been observed in the mouse embryonic fibroblasts (MEFs) of Nbn+/- mice. Loss of heterozygosity was found not to occur in the HCC tissues from the Nbn+/- mice[134]. These findings indicate that Nbn plays an important role in the maintenance of chromosome stability and would seem to function as a haploinsufficient TSG in HCC.
PIN2/TRF1-interacting telomerase inhibitor 1: PIN2/TRF1-interacting telomerase inhibitor 1 (PinX1) is a potent telomerase inhibitor involved in maintaining telomeres at optimal length and its expression level is frequently down-regulated in the HBV-related HCC[135,136]. Heterozygous deficiency of PinX1 has been found to result in telomere elongation and chromosome instability in MEF. Spontaneous HCC development has been observed in the PinX1+/- mice. The expression levels of PinX1 are similar when HCC and its surrounding non-tumor tissues in the PinX1+/- mice are compared[137]. These findings indicate that PinX1 may function as a haploinsufficient TSG and is an essential factor for chromosome stability.
Shugoshin 1: Shugoshin 1 (SGO1) is a guardian of chromosome cohesion that is associated with the fidelity of chromosome segregation during mitosis[138]. In mice, heterozygous deficiency of Sgo1 has been found to lead to persistent hepatocyte DNA damage, to promote azoxymethane (AOM)-induced HCC and to result in the development of spontaneous HCC in Sgo+/- mice. Interestingly, the SGO1 protein level has been shown to be higher in HCC tissue samples compared with the adjacent non-tumor tissues from the Sgo1+/- mice, suggesting that the wild-type allele of Sgo1 is still present and is likely to be upregulated in HCC tissue[139]. In humans, upregulation of SGO1 expression has been observed in the HCC tissue samples compared with their adjacent non-tumor liver tissue samples[140]. These findings reveal that Sgo1 helps maintain the accuracy of chromosome segregation and may function as a haploinsufficient TSG in HCC.
The liver is an important metabolic organ and plays key roles in glucose, lipid, protein, bile acid and methionine metabolism. Dysregulation of metabolism may cause liver disease, including HCC[141]. Abnormal lipid metabolism, including decreased fatty acid oxidation, very low-density lipoprotein (VLDL) secretion or enhanced lipid uptake and lipogenesis, leads to hepatic steatosis, which is one of the risk factors for HCC. Lipid accumulation in hepatocytes results in lipotoxicity, oxidative stress, chronic liver damage and liver regeneration, which all contribute to hepatocarcinogenesis[142,143]. The effect of abnormal metabolic functioning, including lipid metabolism, methionine metabolism and bile acid metabolism, on hepatocarcinogenesis had been clearly demonstrated using mouse models.
The fatty acyl-CoA oxidase: The fatty acyl-CoA oxidase (AOX) protein, which is involved in lipid metabolism, is the first step enzyme of peroxisomal β-oxidation. AOX deficiency leads to steatohepatitis, oxidative stress, chronic liver damage and liver regeneration, as well as the development of spontaneous HCC in mice. Homozygous knockout of AOX results in the development of HCC due to sustained activation of peroxisome proliferator-activated receptor alpha (PPARα), indicating that abnormal lipid metabolism may contribute to HCC development[144].
BHMT and GNMT: Methionine metabolism is very important in maintaining the metabolic homeostasis of liver. One of the key metabolites is S-adenosylmethionine (AdoMet), which is the major methyl donor for methylation of several substrates, including DNA, RNA, histones, and various other small molecules. Abnormal methionine metabolism can cause fatty liver and HCC[145-147]. Reduced expression of the main enzymes involved in methionine metabolism, including methionine adenosyltransferase (MAT), glycine methyltransferase (GNMT) and betaine homocysteine (BHMT), have been observed in human HCC[148,149]. These findings suggest that impairment of methionine homeostasis may contribute to hepatocarcinogenesis.
BHMT is an enzyme that catalyzes the conversion of homocysteine (Hcy) to methionine. High levels of activity of BHMT have been detected in the livers of humans and mice. Disruption of Bhmt perturbs methionine metabolism and results in fatty liver and HCC development in mice. The fatty liver in Bhmt-deficient mice has been found to be due to a decrease in hepatic VLDL secretion[150]. These studies indicate that BHMT is important for methionine metabolism, the maintenance of liver homeostasis and the suppression of liver cancer.
GNMT is a methyltransferase that contributes to the maintenance of AdoMet homeostasis, which helps to avoid aberrant methylation. Loss of Gnmt causes fatty liver and HCC in mice. Aberrant methylation of DNA, which contributed to activation of the Janus kinase (JAK)/signal transducer of activators of transcription (STAT) pathway, has been observed in the livers of Gnmt-deficient mice[151]. These findings indicate that GNMT suppresses HCC development by maintaining AdoMet levels and allowing normal DNA methylation to occur.
Farnesoid X receptor and multidrug resistance 2: Bile acids are cholesterol metabolites and largely synthesized by hepatocytes. Hepatocellular bile acid synthesis and transport are highly regulated by canalicular transporter multidrug resistance 2 (MDR2) and nuclear transcriptional regulator farnesoid X receptor (FXR). Bile acids play key roles in the liver, including lipid metabolism, glucose metabolism and liver regeneration. Abnormal bile acid homeostasis may contribute to the development of fatty liver and HCC[152-154]. In mice, deficiency in the Fxr or Mdr2 results in impairment of bile acids metabolism, an increase in inflammation, and the development of spontaneous HCC[155,156]. In humans, reduced expression levels of FXR and MDR2 have been observed in HCC tissue samples[157,158]. These findings suggest that deregulation of bile acid homeostasis may be involved in hepatocarcinogenesis.
Aberrant cellular signaling pathways are critical events in the process of hepatocarcinogenesis. Previous studies have identified many critical signaling pathways involved in HCC development[159-161]. Previous studies in mice have revealed that dysregulation of many important signaling pathways contribute to the development of HCC; these included the Hippo pathway, the JAK/STAT pathway, the NF-κB pathway, the PI3K/AKT/mTOR pathway, the TGFβ pathway and the WNT/β-catenin pathway.
The Hippo signaling pathway is a highly conserved kinase cascade that regulates cell proliferation, cell survival and organ size control. The Hippo pathway can be divided into three major parts, namely the upstream regulators, the Hippo core kinase components, and the downstream target genes. The Hippo pathway is critical for the maintenance of hepatocyte quiescence and the regulation of liver size. Dysregulation of the Hippo pathway has been observed in many types of human cancers, including HCC[162-164]. In mouse models, the tumor suppressive role of various Hippo signaling components has been identified, including the core kinases MST1/MST2 and the upstream regulator NF1.
Mammalian sterile 20-like kinase 1 and 2: Mammalian sterile 20-like kinase 1 (Mst1) and Mst2 are the components of the Hippo core kinase cascade. The single knockout of Mst1 or Mst2 results in no obvious phenotypes in mice, which suggests the functional redundancy of Mst1 and Mst2 during embryonic development and hepatocarcinogenesis. However, hepatocyte-specific knockout of both the Mst1 and Mst2 genes causes an increase in hepatocyte proliferation, hepatomegaly and spontaneous HCC development in mice[165,166]. These studies demonstrate that Mst1 and Mst2 are required for organ size control and tumor suppression.
Neurofibromatosis type 2: Neurofibromatosis type 2 (NF2) is the positive regulator of the Hippo signaling pathway[167]. Nf2 deficiency leads to hepatic progenitor cell proliferation, hepatomegaly and hepatocarcinogenesis in mice. The tumor promoting phenotypes in the Nf2-deficient liver have been shown to be disrupted by loss of the Yes-associated protein (YAP), which is inhibited by the Hippo pathway[168,169]. These studies indicate that the tumor suppressor function of Nf2 in HCC occurs via inhibiting YAP activation.
JAK/STAT signaling pathway: JAK/STAT pathway is a plasma membrane to nucleus signaling pathway that regulates cell proliferation, differentiation and cell death. Activation of the JAK/STAT pathway is regulated by various positive (cytokine and tyrosine kinase) and negative regulators [protein tyrosine phosphatase (PTP) and suppressor of cytokine signaling (SOCS) proteins]. Constitutive activation of the JAK/STAT pathway is associated with tumorigenesis. The SOCS protein is one of the negative regulators of JAK/STAT pathway and its expression is decreased in many types of cancers including HCC[170-172]. The role of the negative regulators of the JAK/STAT pathway in hepatocarcinogenesis has been studied and evaluated using mouse models.
Suppressor of the cytokine signaling-1: Suppressor of the cytokine signaling-1 (SOCS1) is a negative regulator of JAK-mediated cytokine signaling via the protein’s direct binding to JAK[173]. In humans, a lower level of SOCS1 expression has been observed in HCV-associated HCC samples compared with adjacent non-tumor liver tissue samples[174]. In mice, heterozygous deficiency of Socs1 has been found to increase dimethylnitrosamine (DMN)-induced and diet-induced liver fibrosis; furthermore, heterozygous deficiency has also been found to promote DEN-induced hepatocarcinogenesis in the Socs1+/- mice. These results suggested that SOCS1 is a haploinsufficient TSG in HCC.
Suppressor of the cytokine signaling-3: Suppressor of the cytokine signaling-3 (SOCS3) is a negative regulator for the JAK and interleukin (IL)-6-related signaling pathways[175]. In humans, expression of SOCS3 has been shown to be reduced in HCV-mediated HCC samples and this has been found to be accompanied by enhanced activation of STAT3. In mice, heterozygous deficiency of Socs3 has been found to promote DEN-induced HCC in Socs3+/- mice[176]. These findings indicate that SOCS3 may function as a haploinsufficient TSG in HCC.
Protein tyrosine phosphatase non-receptor type 11: Protein tyrosine phosphatase non-receptor type 11 (PTPN11) is a tyrosine phosphatase that dephosphorylates activated STAT3 and attenuates proinflammatory IL-6 signaling[177,178]. In humans, a decrease in PTPN11 protein expression has been observed in HCC tissue samples compared with the adjacent non-tumor tissue samples[179]. In mice, Ptpn11 deficiency in hepatocytes has been shown to result in hepatic inflammation, cell death and hepatocellular adenoma formation. In addition, Ptpn11 deficiency also promotes DEN-induced HCC development in mice. Furthermore, Stat3 is required for the tumor promoting effect of Ptpn11 deficiency to occur. Together, these studies indicate that Ptpn11 functions as a tumor suppressor in HCC by controlling the oncogenic activity of Stat3.
Protein tyrosine phosphatase receptor type O: Protein tyrosine phosphatase receptor type O (PTPRO) is a receptor type of protein tyrosine phosphatase that is frequently down-regulated in human HCC tissues through promoter hypermethylation[180,181]. Studies have revealed that Ptpro deficiency promotes DEN-induced HCC and enhances Stat3 activity in mice[181,182]. These findings indicate that PTPRO suppresses HCC development via control of the activity of Stat3.
Nuclear factor-κB (NF-κB) is an important transcriptional regulator of inflammation and is required for survival of hepatocytes in the liver. Upon tumor necrosis factor (TNF) stimulation, the IκB kinase (IKK) complex is recruited and activated by the kinase TAK1, and this then stimulates NF-κB activity by phosphorylating the negative inhibitor IκB protein[183]. Various studies have revealed that NF-κB is constitutively activated in many cancers including HCC. This suggests a tumor promoting role for NF-κB signaling[184]. However, evidence from different groups has indicated that NF-κB signaling may also have a tumor suppressor role in HCC. It is likely that NF-κB signaling has a dual role and that this role changes at different stages of carcinogenesis[185,186].
Cylindromatosis: Cylindromatosis (CYLD) is a deubiquitinase that inhibits the NF-κB pathway via deubiquitination of upstream regulatory factors[187]. In humans, reduced expression levels of CYLD have been observed in HCC tissue samples compared with the adjacent non-tumor tissue samples[188]. In mice, liver-specific disruption of CYLD triggers hepatic cell death, inflammation, fibrosis and spontaneous HCC development. Constitutive hyperactivation of TAK1, a NF-κB upstream regulatory factor, is required for liver pathogenesis in liver-specific CYLD knockout mice[189]. These findings show that CYLD suppresses hepatocarcinogenesis via control of the NF-κB signaling pathway.
Galectin-3: The galectin-3 is a down-stream target gene of NF-κB and plays an important role in inflammation response. Under normal conditions, galectin-3 is expressed in the bile duct epithelial and Kupffer cells, but not in the hepatocytes of livers[190,191]. However, expression of galectin-3 has been detected in human HCC cells, indicating that galectin-3 may be involved in HCC development[192]. In mice, homozygous knockout of galectin-3 results in fatty liver, inflammation, fibrosis and spontaneous HCC formation[193,194]. These studies indicate that galectin-3 plays an important role in hepatocarcinogenesis.
NF-κB-essential-modulator: NF-κB-essential-modulator (NEMO) is a regulatory subunit of the IKK complex and is essential for NF-κB activation. Ablation of NEMO blocks NF-κB activation in hepatocytes. In humans, loss of NEMO protein expression has been found in a substantial proportion of HCC tissue samples compared with their adjacent non-tumor tissue samples[195]. In mice, disruption of NEMO in hepatocytes leads to nonalcoholic steatohepatitis (NASH) and the spontaneous development of HCC[196]. These findings show that NEMO acts as a tumor suppressor in the liver.
TGF-β-activated kinase 1: TGF-β-activated kinase 1 (TAK1) is a kinase that activates the IKK complex upon TNF stimulation. Ablation of TAK1 in the hepatocytes of mice results in liver damage and the early onset of spontaneous HCC development, which suggests a tumor suppressor function for TAK1 in liver cancer[197].
The PI3K/AKT/mTOR pathway has a critical role in the regulation of cell growth, proliferation and metabolism. The PI3K/AKT/mTOR pathway is frequently activated in diverse cancers including HCC. The pathway is negatively regulated by phosphatase and tensin homolog deleted from chromosome 10 (PTEN) and AMP-activated protein kinase (AMPK). Alterations in PTEN or the AMPK activator LKB1 have been observed in many types of human cancers[198,199]. Knock-out mice having loss of PTEN and LKB1 have been generated and used to evaluate the roles of the PI3K/AKT/mTOR signaling pathway in HCC development.
Liver kinase B1: Liver kinase B1 (LKB1) is a serine/threonine kinase. LKB1 positively regulates downstream kinases including AMPK, which inhibits mTOR pathway. LKB1 has identified as the disease gene of Peutz-Jegher polyposis and cancer syndrome[200]. In addition, down-regulation of LBK1 has been observed in human HCC tissue samples compared with the adjacent non-tumor tissue samples[201]. In mice, heterozygous knockout of Lkb1 leads to increased proliferation of hepatocytes and the development of spontaneous HCC. Loss of heterozygosity of the wild-type allele of Lkb1 has been detected in the HCC tissues from the heterozygous knockout of Lkb1 mice, indicating that Lkb1 acts as a recessive TSG in HCC[202].
PTEN: PTEN is a phospholipid phosphatase that negatively regulates the highly oncogenic pro-survival PI3K/AKT signaling pathway[203]. In humans, expression levels of PTEN are decreased in HCC tissue samples compared with the adjacent non-tumor tissue samples[204]. In mice, homozygous knockout of Pten results in embryonic lethality. Furthermore, Pten heterozygous mice develop tumors in multiple organs including the liver. Loss of the wild-type Pten allele has been observed in liver tumors from Pten+/- mice[205-207]. Moreover, Pten deficiency in hepatocytes leads to steatohepatitis and HCC formation in mice[208]. These studies indicate that Pten functions as a recessive TSG in HCC.
Regulatory-associated protein of mTOR: Regulatory-associated protein of mTOR (RAPTOR) is one of the associated proteins of mTOR and is required for mTOR activity. RAPTOR expression has been found to be up-regulated in advanced human HCC tissue samples compared with the adjacent non-tumor tissue samples, suggesting that the RAPTOR may contribute to hepatocarcinogenesis[209]. Hepatocyte-specific disruption of RAPTOR has been shown to result in hepatic damage, inflammation, fibrosis and the acceleration of DEN-induced hepatocarcinogenesis in mice. The HCC promoting effect of RAPTOR deficiency may be attributed partly to the hyper-activation of AKT in such livers[210]. These loss-of-function studies indicated that a persistent inhibition of mTOR may not be a suitable way to treat HCC.
TGF-β signaling is involved in many important pathways related to cell growth, including proliferation, motility and cell death. TGF-β inhibits hepatocyte proliferation in the liver, suggesting that it has potential tumor suppressive effects in HCC[211]. Although TGF-β has tumor suppressive functions in benign and early-stage tumors, it can also act as a tumor promoter in advanced-stage cancers. TGF-β has dual functions of tumor prevention and tumor promotion at difference cancer stages[212,213]. Scientists have applied genetically modified mouse models to evaluate the complex roles of the TGF-β signaling pathway in HCC development.
Embryonic liver fodrin: Embryonic liver fodrin (ELF) is an adaptor protein that mediates access to the receptor and SMAD3 activation in the TGF-β signaling pathway. In humans, ELF has been found to be significantly decreased in HCC tissue samples compared with the adjacent non-tumor tissue samples[214]. In mice, heterozygous deficiency of ELF leads to the development of fatty liver and spontaneous HCC, which is accompanied by enhanced activation of cyclin D1. These findings indicate that ELF functions as a haploinsufficient TSG in HCC.
Transforming growth factorβ1: Transforming growth factor β1 (TGF-β1) is a negative factor that inhibits hepatocyte proliferation[215]. In mice, heterozygous deficiency of TGF-β1 increases hepatocyte proliferation and promotes DEN-induced HCC in TGF-β1+/- mice. Tumors from TGF-β1+/- mice do not show loss of the wild-type allele of TGF-β1[216]. These findings indicate that the TGF-β1 functions as a haploinsufficient TSG in HCC.
TGF-β type II receptor: TGF-β type II receptor (TβR-II) is a receptor of TGF-β growth factor. In humans, down-regulation of TβR-II has been frequently observed in HCC tissue[217]. In mice, heterozygous deficiency of TβR-II results in increased hepatocyte proliferation and promotes DEN-induced HCC carcinogenesis; in addition, a decrease of TβR-II expression has been observed in the DEN-induced HCC tissue samples compared to the non-tumor liver in TβR-II+/- mice suggesting loss of the wild-type allele of TβR-II in HCC[218]. These studies show that TβR-II functions as a recessive TSG in HCC.
The WNT/β-catenin signaling pathway plays critical roles in liver development, growth and metabolism. Moreover, WNT/β-catenin signaling is important for cancer development, including tumor initiation, growth and metastasis. Aberrant activation of WNT/β-catenin signaling has been observed in human HCC, suggesting that this pathway is involved in hepatocarcinogenesis[219-221].
Adenomatous polyposis coli: Adenomatous polyposis coli (APC) is the antagonist of WNT signaling and acts by enhancing β-catenin degradation. In humans, the expression level of the APC protein has been found to be decreased in HCC tissue samples compared to the adjacent non-tumor tissue samples[222]. In mice, liver-specific disruption of Apc causes activation of β-catenin signaling and spontaneous HCC formation[223]. These findings demonstrate that APC functions as a tumor suppressor in HCC via regulation of the β-catenin signaling.
MicroRNAs (miRNAs) are short noncoding RNAs and act as post-transcriptional regulators of gene expression. miRNAs may function as oncogenes or tumor suppressor genes through their targeting of various different genes involved in tumorigenesis. Deregulation of miRNA expression has been observed in human HCC indicating that miRNAs are involved in hepatocarcinogenesis. Importantly, the direct role of miRNA in hepatocarcinogenesis has been demonstrated using mouse models; examples of such miRNAs include miRNA-122 and miRNA-140[224-226].
miR-122 and miR-140: miR-122 is a major form of miRNA found in the liver. Downregulation of miR-122 has been observed in human HCC[227]. Disruption of miR-122 leads to hepatic steatosis, inflammation, fibrosis and spontaneous HCC in mice. The fatty liver in miR-122-deficient mice is caused by impaired VLDL secretion in the liver[228]. These studies demonstrate the metabolic and tumor suppressor function of miR-122 in livers. Another miRNA, miR-140, has also been suggested to act as a tumor suppressor in HCC. miR-140 deficiency results in enhanced NF-κB activity and promotes DEN-induced HCC development, indicating that the miR-140 functions as a tumor suppressor via regulation of the NF-κB activity in mice[229].
Nuclear receptor coactivator 5: Nuclear receptor coactivator 5 (NCOA5) is an estrogen receptor coactivator that is able to enhance estrogen receptor α activity in the presence of estradiol[230]. In humans, expression of NCOA5 mRNA has been found to be decreased in about 40% of HCC tissue samples compared with their adjacent non-tumor tissue samples[231]. In mice, heterozygous deficiency of Ncoa5 leads to fatty liver, inflammation, fibrosis and spontaneous HCC development. In addition, expression of the NCOA5 protein has been detected in HCC samples from Ncoa5+/- mice. These findings indicate that Ncoa5 functions as a haploinsufficient TSG in HCC.
Regulatory subunit 1a of protein kinase A: Regulatory subunit 1a of protein kinase A (PRKAR1A) is the major component of type I protein kinase A and regulates protein kinase activity[232]. Heterozygous deficiency of Prkar1a leads to multiple tumor formation events including HCC. Loss of heterozygosity of the Prkar1a allele was not observed in the tumors of Prkar1a+/- mice, suggesting that Prkar1a functions as a haploinsufficient tumor suppressor[233].
Nuclear receptor coactivator 2: Nuclear receptor coactivator 2 (NCOA2) is a transcriptional coactivator that regulates fasting hepatic glucose release by controlling the expression of glucose-6-phosphatase, which is the key enzyme of gluconeogenesis[234]. Homozygous deficiency of Ncoa2 promotes DEN-induced hepatocarcinogenesis in mice revealing a tumor suppressor role for Ncoa2 in liver cancer[235].
Nuclear factor erythroid 2-like 1: Nuclear factor erythroid 2-like 1 (NFE2L1; also known as NRF1, LCR-F1 and TCF11) is a transcription factor that controls the redox balance and lipid metabolism[236,237]. Hepatocyte-specific disruption of the Nrf1 gene causes hepatic steatosis, cell death, inflammation, oxidative stress, fibrosis and spontaneous hepatic cancer in mice[238]. These findings suggested that Nrf1 may function as a tumor suppressor in HCC by reducing oxidative stress in liver.
Spontaneous tumor formation largely occurs due to an accumulation of multiple somatic mutations. Discoveries from genetically modified mice are very informative and allow the identification of potential TSG based on clinical findings and characterization of carcinogenic mechanisms. HCC is tremendously heterogeneous at the pathological, clinical, genomic and molecular levels; this may be due to the natural function of liver, which serves as a major organ responsible for metabolism and detoxification of the whole body[239]. This situation also indicates the complexity of the signaling pathways active in the liver with multiple signaling pathways and cellular processes intercrossing in hepatocytes in order to regulate and control the cell cycle, metabolism and detoxification.
Inactivation of TSGs might involve a range of different modulations in addition to mutation, deletion and loss-of-heterozygosity; for example promoter silencing by DNA methylation. One of the therapeutic strategies is to take a TSG as a molecular target, for example p53. Not only the TSG itself can be a target; its interacting proteins and/or the downstream targets of the TSG are also candidates for suppressing tumor formation. In this regard, HCC mouse models, which recapitulate the pathology and progression of hepatocarcinogenesis in human HCC, provide a useful in vivo animal model for therapeutic testing and mechanistic studies under physiological conditions. Several TSGs are currently under evaluation in order to test whether they can serve as a target for treating lung or ovarian cancer[240]. Taking into consideration tissue specificity and the possibility of potential off-target problems, it may be useful to identify liver-specific TSGs for the development of precision medicine when tackling HCC.
P- Reviewer: Tomizawa M S- Editor: Gong ZM L- Editor: Logan S E- Editor: Wang CH
| 1. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2507] [Article Influence: 192.8] [Reference Citation Analysis (2)] |
| 2. | Bakiri L, Wagner EF. Mouse models for liver cancer. Mol Oncol. 2013;7:206-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 127] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 3. | Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28:753-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 400] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 4. | Li Y, Tang ZY, Hou JX. Hepatocellular carcinoma: insight from animal models. Nat Rev Gastroenterol Hepatol. 2012;9:32-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 5. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 6. | Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34 Suppl 1:S75-S78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 133] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Kremsdorf D, Soussan P, Paterlini-Brechot P, Brechot C. Hepatitis B virus-related hepatocellular carcinoma: paradigms for viral-related human carcinogenesis. Oncogene. 2006;25:3823-3833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 210] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 8. | Chen CL, Yang HI, Yang WS, Liu CJ, Chen PJ, You SL, Wang LY, Sun CA, Lu SN, Chen DS. Metabolic factors and risk of hepatocellular carcinoma by chronic hepatitis B/C infection: a follow-up study in Taiwan. Gastroenterology. 2008;135:111-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 423] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 9. | Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN, Huang GT, Iloeje UH. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2309] [Cited by in RCA: 2364] [Article Influence: 124.4] [Reference Citation Analysis (0)] |
| 10. | Tan A, Yeh SH, Liu CJ, Cheung C, Chen PJ. Viral hepatocarcinogenesis: from infection to cancer. Liver Int. 2008;28:175-188. [RCA] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 11. | Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593-2604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 246] [Cited by in RCA: 243] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 12. | Halegoua-De Marzio D, Hann HW. Then and now: the progress in hepatitis B treatment over the past 20 years. World J Gastroenterol. 2014;20:401-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 47] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (1)] |
| 13. | Milich DR, Jones JE, Hughes JL, Maruyama T, Price J, Melhado I, Jirik F. Extrathymic expression of the intracellular hepatitis B core antigen results in T cell tolerance in transgenic mice. J Immunol. 1994;152:455-466. [PubMed] |
| 14. | Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1212] [Article Influence: 60.6] [Reference Citation Analysis (0)] |
| 15. | Kwon H, Lok AS. Hepatitis B therapy. Nat Rev Gastroenterol Hepatol. 2011;8:275-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 211] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 16. | Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 633] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 17. | Ilan E, Burakova T, Dagan S, Nussbaum O, Lubin I, Eren R, Ben-Moshe O, Arazi J, Berr S, Neville L. The hepatitis B virus-trimera mouse: a model for human HBV infection and evaluation of anti-HBV therapeutic agents. Hepatology. 1999;29:553-562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Chiang CJ, Yang YW, You SL, Lai MS, Chen CJ. Thirty-year outcomes of the national hepatitis B immunization program in Taiwan. JAMA. 2013;310:974-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 179] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 19. | Hudu SA, Malik YA, Niazlin MT, Harmal NS, Sekawi Z. An Overview of Hepatitis B Virus Surface Antigen Mutant in the Asia Pacific. Curr Issues Mol Biol. 2014;16:69-78. [PubMed] |
| 20. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1599] [Article Influence: 123.0] [Reference Citation Analysis (1)] |
| 21. | Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070-1083. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 622] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 22. | Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 518] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 23. | Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 827] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 24. | Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J Viral Hepat. 2002;9:323-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 165] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Inuzuka T, Takahashi K, Chiba T, Marusawa H. Mouse models of hepatitis B virus infection comprising host-virus immunologic interactions. Pathogens. 2014;3:377-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Araki K, Nishimura S, Ochiya T, Okubo K, Miyazaki J, Matsubara K, Yamamura K. Production and effect of infectious Dane particles in transgenic mice. Jpn J Cancer Res. 1991;82:235-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 27. | Cavanaugh VJ, Guidotti LG, Chisari FV. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. J Virol. 1997;71:3236-3243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 146] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 28. | Babinet C, Farza H, Morello D, Hadchouel M, Pourcel C. Specific expression of hepatitis B surface antigen (HBsAg) in transgenic mice. Science. 1985;230:1160-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 112] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Chisari FV, Pinkert CA, Milich DR, Filippi P, McLachlan A, Palmiter RD, Brinster RL. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 202] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 30. | Chisari FV, Filippi P, McLachlan A, Milich DR, Riggs M, Lee S, Palmiter RD, Pinkert CA, Brinster RL. Expression of hepatitis B virus large envelope polypeptide inhibits hepatitis B surface antigen secretion in transgenic mice. J Virol. 1986;60:880-887. [PubMed] |
| 31. | Chisari FV, Filippi P, Buras J, McLachlan A, Popper H, Pinkert CA, Palmiter RD, Brinster RL. Structural and pathological effects of synthesis of hepatitis B virus large envelope polypeptide in transgenic mice. Proc Natl Acad Sci USA. 1987;84:6909-6913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 270] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 32. | Toshkov I, Chisari FV, Bannasch P. Hepatic preneoplasia in hepatitis B virus transgenic mice. Hepatology. 1994;20:1162-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Wang HC, Chang WT, Chang WW, Wu HC, Huang W, Lei HY, Lai MD, Fausto N, Su IJ. Hepatitis B virus pre-S2 mutant upregulates cyclin A expression and induces nodular proliferation of hepatocytes. Hepatology. 2005;41:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 106] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 34. | Dunsford HA, Sell S, Chisari FV. Hepatocarcinogenesis due to chronic liver cell injury in hepatitis B virus transgenic mice. Cancer Res. 1990;50:3400-3407. [PubMed] |
| 35. | DeLoia JA, Burk RD, Gearhart JD. Developmental regulation of hepatitis B surface antigen expression in two lines of hepatitis B virus transgenic mice. J Virol. 1989;63:4069-4073. [PubMed] |
| 36. | Farza H, Salmon AM, Hadchouel M, Moreau JL, Babinet C, Tiollais P, Pourcel C. Hepatitis B surface antigen gene expression is regulated by sex steroids and glucocorticoids in transgenic mice. Proc Natl Acad Sci USA. 1987;84:1187-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 81] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 37. | Wang Y, Cui F, Lv Y, Li C, Xu X, Deng C, Wang D, Sun Y, Hu G, Lang Z. HBsAg and HBx knocked into the p21 locus causes hepatocellular carcinoma in mice. Hepatology. 2004;39:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 112] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 38. | Su IJ, Wang LH, Hsieh WC, Wu HC, Teng CF, Tsai HW, Huang W. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 39. | Abe K, Thung SN, Wu HC, Tran TT, Le Hoang P, Truong KD, Inui A, Jang JJ, Su IJ. Pre-S2 deletion mutants of hepatitis B virus could have an important role in hepatocarcinogenesis in Asian children. Cancer Sci. 2009;100:2249-2254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 40. | Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 41. | Yu DY, Moon HB, Son JK, Jeong S, Yu SL, Yoon H, Han YM, Lee CS, Park JS, Lee CH. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 214] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 42. | Lee TH, Finegold MJ, Shen RF, DeMayo JL, Woo SL, Butel JS. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64:5939-5947. [PubMed] |
| 43. | Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 880] [Cited by in RCA: 867] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 44. | Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T, Kurokawa K. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19:810-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 182] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 45. | Lu JW, Hsia Y, Yang WY, Lin YI, Li CC, Tsai TF, Chang KW, Shieh GS, Tsai SF, Wang HD. Identification of the common regulators for hepatocellular carcinoma induced by hepatitis B virus X antigen in a mouse model. Carcinogenesis. 2012;33:209-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 46. | Wu BK, Li CC, Chen HJ, Chang JL, Jeng KS, Chou CK, Hsu MT, Tsai TF. Blocking of G1/S transition and cell death in the regenerating liver of Hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun. 2006;340:916-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 47. | Wu YF, Fu SL, Kao CH, Yang CW, Lin CH, Hsu MT, Tsai TF. Chemopreventive effect of silymarin on liver pathology in HBV X protein transgenic mice. Cancer Res. 2008;68:2033-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 48. | Geng X, Harry BL, Zhou Q, Skeen-Gaar RR, Ge X, Lee ES, Mitani S, Xue D. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:18465-18470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Geng X, Huang C, Qin Y, McCombs JE, Yuan Q, Harry BL, Palmer AE, Xia NS, Xue D. Hepatitis B virus X protein targets Bcl-2 proteins to increase intracellular calcium, required for virus replication and cell death induction. Proc Natl Acad Sci USA. 2012;109:18471-18476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 50. | Yen CJ, Lin YJ, Yen CS, Tsai HW, Tsai TF, Chang KY, Huang WC, Lin PW, Chiang CW, Chang TT. Hepatitis B virus X protein upregulates mTOR signaling through IKKβ to increase cell proliferation and VEGF production in hepatocellular carcinoma. PLoS One. 2012;7:e41931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 51. | Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy. 2014;10:1687-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 52. | Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 53. | Teng CF, Hsieh WC, Yang CW, Su HM, Tsai TF, Sung WC, Huang W, Su IJ. A biphasic response pattern of lipid metabolomics in the stage progression of hepatitis B virus X tumorigenesis. Mol Carcinog. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 54. | Lin HC, Chen YF, Hsu WH, Yang CW, Kao CH, Tsai TF. Resveratrol helps recovery from fatty liver and protects against hepatocellular carcinoma induced by hepatitis B virus X protein in a mouse model. Cancer Prev Res (Phila). 2012;5:952-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 55. | Hsieh WC, Yang CW, Haung YS, Chao TW, Tsai TF, Su IJ. Chemoprevention of HBV-related hepatocellular carcinoma by the combined product of resveratrol and silymarin in transgenic mice. Funct Foods Heal Dis. 2013;3:341-352. |
| 56. | Milich DR, Jones JE, Hughes JL, Price J, Raney AK, McLachlan A. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci USA. 1990;87:6599-6603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 415] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 57. | Takashima H, Araki K, Miyazaki J, Yamamura K, Kimoto M. Characterization of T-cell tolerance to hepatitis B virus (HBV) antigen in transgenic mice. Immunology. 1992;75:398-405. [PubMed] |
| 58. | Guidotti LG, Martinez V, Loh YT, Rogler CE, Chisari FV. Hepatitis B virus nucleocapsid particles do not cross the hepatocyte nuclear membrane in transgenic mice. J Virol. 1994;68:5469-5475. [PubMed] |
| 59. | Yamamura K, Araki K, Hino O, Tomita N, Miyazaki J, Matsubara K. HBV production in transgenic mice. Gastroenterol Jpn. 1990;25 Suppl 2:49-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 60. | Farza H, Hadchouel M, Scotto J, Tiollais P, Babinet C, Pourcel C. Replication and gene expression of hepatitis B virus in a transgenic mouse that contains the complete viral genome. J Virol. 1988;62:4144-4152. [PubMed] |
| 61. | Araki K, Miyazaki J, Hino O, Tomita N, Chisaka O, Matsubara K, Yamamura K. Expression and replication of hepatitis B virus genome in transgenic mice. Proc Natl Acad Sci USA. 1989;86:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 148] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 62. | Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158-6169. [PubMed] |
| 63. | Guidotti LG, Eggers CM, Raney AK, Chi SY, Peters JM, Gonzalez FJ, McLachlan A. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J Virol. 1999;73:10377-10386. [PubMed] |
| 64. | Chen SH, Wu HL, Kao JH, Hwang LH. Persistent hepatitis B viral replication in a FVB/N mouse model: impact of host and viral factors. PLoS One. 2012;7:e36984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 65. | Grompe M, Strom S. Mice with human livers. Gastroenterology. 2013;145:1209-1214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 66. | Moriyama T, Guilhot S, Klopchin K, Moss B, Pinkert CA, Palmiter RD, Brinster RL, Kanagawa O, Chisari FV. Immunobiology and pathogenesis of hepatocellular injury in hepatitis B virus transgenic mice. Science. 1990;248:361-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 266] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 67. | Nakamoto Y, Guidotti LG, Kuhlen CV, Fowler P, Chisari FV. Immune pathogenesis of hepatocellular carcinoma. J Exp Med. 1998;188:341-350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 258] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 68. | Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol. 2001;75:2900-2911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 96] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 69. | Julander JG, Sidwell RW, Morrey JD. Characterizing antiviral activity of adefovir dipivoxil in transgenic mice expressing hepatitis B virus. Antiviral Res. 2002;55:27-40. [PubMed] |
| 70. | Fan HX, Tang H. Complex interactions between microRNAs and hepatitis B/C viruses. World J Gastroenterol. 2014;20:13477-13492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 28] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 71. | Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 185] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 72. | Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Mol Ther Nucleic Acids. 2014;3:e186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 73. | Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids. 2014;3:e216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 220] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 74. | Martin J, Dufour JF. Tumor suppressor and hepatocellular carcinoma. World J Gastroenterol. 2008;14:1720-1733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 75. | Imbeaud S, Ladeiro Y, Zucman-Rossi J. Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis. 2010;30:75-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 76. | Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 962] [Cited by in RCA: 1149] [Article Influence: 88.4] [Reference Citation Analysis (0)] |
| 77. | Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422-1433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 412] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 78. | Payne SR, Kemp CJ. Tumor suppressor genetics. Carcinogenesis. 2005;26:2031-2045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 79. | Smilenov LB. Tumor development: haploinsufficiency and local network assembly. Cancer Lett. 2006;240:17-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 80. | Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J Pathol. 2011;223:137-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 81. | Cui J, Gong Z, Shen HM. The role of autophagy in liver cancer: molecular mechanisms and potential therapeutic targets. Biochim Biophys Acta. 2013;1836:15-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 82. | Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34:856-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 905] [Cited by in RCA: 952] [Article Influence: 95.2] [Reference Citation Analysis (0)] |
| 83. | Rautou PE, Mansouri A, Lebrec D, Durand F, Valla D, Moreau R. Autophagy in liver diseases. J Hepatol. 2010;53:1123-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 323] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 84. | Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008;68:9167-9175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 218] [Cited by in RCA: 227] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 85. | Kotsafti A, Farinati F, Cardin R, Cillo U, Nitti D, Bortolami M. Autophagy and apoptosis-related genes in chronic liver disease and hepatocellular carcinoma. BMC Gastroenterol. 2012;12:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 86. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077-15082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1534] [Cited by in RCA: 1721] [Article Influence: 78.2] [Reference Citation Analysis (0)] |
| 87. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 83] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 88. | Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2154] [Cited by in RCA: 2294] [Article Influence: 109.2] [Reference Citation Analysis (0)] |
| 89. | Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1755] [Cited by in RCA: 1926] [Article Influence: 96.3] [Reference Citation Analysis (0)] |
| 90. | An CH, Kim MS, Yoo NJ, Park SW, Lee SH. Mutational and expressional analyses of ATG5, an autophagy-related gene, in gastrointestinal cancers. Pathol Res Pract. 2011;207:433-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 91. | Ni HM, Woolbright BL, Williams J, Copple B, Cui W, Luyendyk JP, Jaeschke H, Ding WX. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J Hepatol. 2014;61:617-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 224] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 92. | Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177-187.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 93. | Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 923] [Cited by in RCA: 1059] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 94. | Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-1135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3239] [Cited by in RCA: 3105] [Article Influence: 194.1] [Reference Citation Analysis (0)] |
| 95. | Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226:352-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 495] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 96. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47087] [Article Influence: 3363.4] [Reference Citation Analysis (5)] |
| 97. | Andreoli V, Gehrau RC, Bocco JL. Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death. IUBMB Life. 2010;62:896-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 98. | Tetreault MP, Yang Y, Katz JP. Krüppel-like factors in cancer. Nat Rev Cancer. 2013;13:701-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 303] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 99. | Tarocchi M, Hannivoort R, Hoshida Y, Lee UE, Vetter D, Narla G, Villanueva A, Oren M, Llovet JM, Friedman SL. Carcinogen-induced hepatic tumors in KLF6+/- mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology. 2011;54:522-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 100. | Vetter D, Cohen-Naftaly M, Villanueva A, Lee YA, Kocabayoglu P, Hannivoort R, Narla G, M Llovet J, Thung SN, Friedman SL. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology. 2012;56:1361-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 101. | Kremer-Tal S, Narla G, Chen Y, Hod E, DiFeo A, Yea S, Lee JS, Schwartz M, Thung SN, Fiel IM. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol. 2007;46:645-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 102. | Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW. Sak/Plk4 and mitotic fidelity. Oncogene. 2005;24:306-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 103. | Hudson JW, Kozarova A, Cheung P, Macmillan JC, Swallow CJ, Cross JC, Dennis JW. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr Biol. 2001;11:441-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 104. | Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 164] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 105. | Pellegrino R, Calvisi DF, Ladu S, Ehemann V, Staniscia T, Evert M, Dombrowski F, Schirmacher P, Longerich T. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51:857-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 106. | Warfel NA, El-Deiry WS. p21WAF1 and tumourigenesis: 20 years after. Curr Opin Oncol. 2013;25:52-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 107. | Jackson RJ, Engelman RW, Coppola D, Cantor AB, Wharton W, Pledger WJ. p21Cip1 nullizygosity increases tumor metastasis in irradiated mice. Cancer Res. 2003;63:3021-3025. [PubMed] |
| 108. | Ehedego H, Boekschoten MV, Hu W, Doler C, Haybaeck J, Gaβler N, Müller M, Liedtke C, Trautwein C. p21 ablation in liver enhances DNA damage, cholestasis, and carcinogenesis. Cancer Res. 2015;75:1144-1155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 109. | Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5482] [Cited by in RCA: 5462] [Article Influence: 195.1] [Reference Citation Analysis (0)] |
| 110. | Gurpinar E, Vousden KH. Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 2015;25:486-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 111. | Ghebranious N, Sell S. Hepatitis B injury, male gender, aflatoxin, and p53 expression each contribute to hepatocarcinogenesis in transgenic mice. Hepatology. 1998;27:383-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 112. | Katz SF, Lechel A, Obenauf AC, Begus-Nahrmann Y, Kraus JM, Hoffmann EM, Duda J, Eshraghi P, Hartmann D, Liss B. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology. 2012;142:1229-1239.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 113. | Liu J, Shaik S, Dai X, Wu Q, Zhou X, Wang Z, Wei W. Targeting the ubiquitin pathway for cancer treatment. Biochim Biophys Acta. 2015;1855:50-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 114. | Johnson DE. The ubiquitin-proteasome system: opportunities for therapeutic intervention in solid tumors. Endocr Relat Cancer. 2015;22:T1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 115. | Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 318] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 116. | Yin J, Zhu JM, Shen XZ. The role and therapeutic implications of RING-finger E3 ubiquitin ligases in hepatocellular carcinoma. Int J Cancer. 2015;136:249-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 117. | Zhang T, Kho DH, Wang Y, Harazono Y, Nakajima K, Xie Y, Raz A. Gp78, an E3 ubiquitin ligase acts as a gatekeeper suppressing nonalcoholic steatohepatitis (NASH) and liver cancer. PLoS One. 2015;10:e0118448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 118. | Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R, Chiba T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002-6011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 184] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 119. | Jiang S, Minter LC, Stratton SA, Yang P, Abbas HA, Akdemir ZC, Pant V, Post S, Gagea M, Lee RG. TRIM24 suppresses development of spontaneous hepatic lipid accumulation and hepatocellular carcinoma in mice. J Hepatol. 2015;62:371-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 120. | Chen Z, Du S, Fang S. gp78: a multifaceted ubiquitin ligase that integrates a unique protein degradation pathway from the endoplasmic reticulum. Curr Protein Pept Sci. 2012;13:414-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 121. | Xu L, Lin DC, Yin D, Koeffler HP. An emerging role of PARK2 in cancer. J Mol Med (Berl). 2014;92:31-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 122. | Wang F, Denison S, Lai JP, Philips LA, Montoya D, Kock N, Schüle B, Klein C, Shridhar V, Roberts LR. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosomes Cancer. 2004;40:85-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 123. | Kim KY, Stevens MV, Akter MH, Rusk SE, Huang RJ, Cohen A, Noguchi A, Springer D, Bocharov AV, Eggerman TL. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest. 2011;121:3701-3712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 176] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 124. | Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 629] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 125. | Liu X, Huang Y, Yang D, Li X, Liang J, Lin L, Zhang M, Zhong K, Liang B, Li J. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PLoS One. 2014;9:e85462. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 126. | Shen Z. Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011;3:1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 127. | Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1407] [Cited by in RCA: 1651] [Article Influence: 110.1] [Reference Citation Analysis (0)] |
| 128. | Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest. 2012;122:1138-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 203] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 129. | Guo C, Liu S, Greenaway F, Sun MZ. Potential role of annexin A7 in cancers. Clin Chim Acta. 2013;423:83-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 130. | Srivastava M, Atwater I, Glasman M, Leighton X, Goping G, Caohuy H, Miller G, Pichel J, Westphal H, Mears D. Defects in inositol 1,4,5-trisphosphate receptor expression, Ca(2+) signaling, and insulin secretion in the anx7(+/-) knockout mouse. Proc Natl Acad Sci USA. 1999;96:13783-13788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 95] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 131. | Srivastava M, Torosyan Y, Raffeld M, Eidelman O, Pollard HB, Bubendorf L. ANXA7 expression represents hormone-relevant tumor suppression in different cancers. Int J Cancer. 2007;121:2628-2636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 132. | Srivastava M, Montagna C, Leighton X, Glasman M, Naga S, Eidelman O, Ried T, Pollard HB. Haploinsufficiency of Anx7 tumor suppressor gene and consequent genomic instability promotes tumorigenesis in the Anx7(+/-) mouse. Proc Natl Acad Sci USA. 2003;100:14287-14292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 133. | Tauchi H, Matsuura S, Kobayashi J, Sakamoto S, Komatsu K. Nijmegen breakage syndrome gene, NBS1, and molecular links to factors for genome stability. Oncogene. 2002;21:8967-8980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 138] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 134. | Dumon-Jones V, Frappart PO, Tong WM, Sajithlal G, Hulla W, Schmid G, Herceg Z, Digweed M, Wang ZQ. Nbn heterozygosity renders mice susceptible to tumor formation and ionizing radiation-induced tumorigenesis. Cancer Res. 2003;63:7263-7269. [PubMed] |
| 135. | Soohoo CY, Shi R, Lee TH, Huang P, Lu KP, Zhou XZ. Telomerase inhibitor PinX1 provides a link between TRF1 and telomerase to prevent telomere elongation. J Biol Chem. 2011;286:3894-3906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 136. | Liao C, Zhao M, Song H, Uchida K, Yokoyama KK, Li T. Identification of the gene for a novel liver-related putative tumor suppressor at a high-frequency loss of heterozygosity region of chromosome 8p23 in human hepatocellular carcinoma. Hepatology. 2000;32:721-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 71] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 137. | Zhou XZ, Huang P, Shi R, Lee TH, Lu G, Zhang Z, Bronson R, Lu KP. The telomerase inhibitor PinX1 is a major haploinsufficient tumor suppressor essential for chromosome stability in mice. J Clin Invest. 2011;121:1266-1282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 138. | Marston AL. Shugoshins: tension-sensitive pericentromeric adaptors safeguarding chromosome segregation. Mol Cell Biol. 2015;35:634-648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 139. | Yamada HY, Zhang Y, Reddy A, Mohammed A, Lightfoot S, Dai W, Rao CV. Tumor-promoting/progressing role of additional chromosome instability in hepatic carcinogenesis in Sgo1 (Shugoshin 1) haploinsufficient mice. Carcinogenesis. 2015;36:429-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 140. | Wang LH, Yen CJ, Li TN, Elowe S, Wang WC, Wang LH. Sgo1 is a potential therapeutic target for hepatocellular carcinoma. Oncotarget. 2015;6:2023-2033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 141. | Lade A, Noon LA, Friedman SL. Contributions of metabolic dysregulation and inflammation to nonalcoholic steatohepatitis, hepatic fibrosis, and cancer. Curr Opin Oncol. 2014;26:100-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 142. | Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol. 2012;56:952-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 593] [Cited by in RCA: 706] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 143. | Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10:656-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 772] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 144. | Fan CY, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem. 1998;273:15639-15645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 261] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 145. | Anstee QM, Day CP. S-adenosylmethionine (SAMe) therapy in liver disease: a review of current evidence and clinical utility. J Hepatol. 2012;57:1097-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 167] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 146. | Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiol Rev. 2012;92:1515-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 415] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 147. | Mato JM, Martínez-Chantar ML, Lu SC. S-adenosylmethionine metabolism and liver disease. Ann Hepatol. 2013;12:183-189. [PubMed] |
| 148. | Chen YM, Shiu JY, Tzeng SJ, Shih LS, Chen YJ, Lui WY, Chen PH. Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. Int J Cancer. 1998;75:787-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 149. | Avila MA, Berasain C, Torres L, Martín-Duce A, Corrales FJ, Yang H, Prieto J, Lu SC, Caballería J, Rodés J. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907-914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 150. | Teng YW, Mehedint MG, Garrow TA, Zeisel SH. Deletion of betaine-homocysteine S-methyltransferase in mice perturbs choline and 1-carbon metabolism, resulting in fatty liver and hepatocellular carcinomas. J Biol Chem. 2011;286:36258-36267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 165] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 151. | Martínez-Chantar ML, Vázquez-Chantada M, Ariz U, Martínez N, Varela M, Luka Z, Capdevila A, Rodríguez J, Aransay AM, Matthiesen R. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191-1199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 252] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 152. | Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1046] [Cited by in RCA: 1234] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 153. | Baptissart M, Vega A, Maqdasy S, Caira F, Baron S, Lobaccaro JM, Volle DH. Bile acids: from digestion to cancers. Biochimie. 2013;95:504-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 154. | Halilbasic E, Claudel T, Trauner M. Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J Hepatol. 2013;58:155-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 299] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 155. | Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 289] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 156. | Mauad TH, van Nieuwkerk CM, Dingemans KP, Smit JJ, Schinkel AH, Notenboom RG, van den Bergh Weerman MA, Verkruisen RP, Groen AK, Oude Elferink RP. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am J Pathol. 1994;145:1237-1245. [PubMed] |
| 157. | Liu N, Meng Z, Lou G, Zhou W, Wang X, Zhang Y, Zhang L, Liu X, Yen Y, Lai L. Hepatocarcinogenesis in FXR-/- mice mimics human HCC progression that operates through HNF1α regulation of FXR expression. Mol Endocrinol. 2012;26:775-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 158. | Zollner G, Wagner M, Fickert P, Silbert D, Fuchsbichler A, Zatloukal K, Denk H, Trauner M. Hepatobiliary transporter expression in human hepatocellular carcinoma. Liver Int. 2005;25:367-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 159. | Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989-5005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 680] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 160. | Nault JC, Zucman-Rossi J. Genetics of hepatobiliary carcinogenesis. Semin Liver Dis. 2011;31:173-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 161. | Marquardt JU, Galle PR, Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J Hepatol. 2012;56:267-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 139] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 162. | Ma Y, Yang Y, Wang F, Wei Q, Qin H. Hippo-YAP signaling pathway: A new paradigm for cancer therapy. Int J Cancer. 2015;137:2275-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 80] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 163. | Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 515] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 164. | Yu FX, Meng Z, Plouffe SW, Guan KL. Hippo pathway regulation of gastrointestinal tissues. Annu Rev Physiol. 2015;77:201-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 165. | Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 471] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 166. | Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437-1442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 562] [Cited by in RCA: 604] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 167. | Cooper J, Giancotti FG. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett. 2014;588:2743-2752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 140] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 168. | Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718-1730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 229] [Cited by in RCA: 222] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 169. | Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 677] [Cited by in RCA: 644] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 170. | Quintás-Cardama A, Verstovsek S. Molecular pathways: Jak/STAT pathway: mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19:1933-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 234] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 171. | Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30:1005-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 412] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 172. | O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1042] [Cited by in RCA: 1104] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 173. | Inagaki-Ohara K, Kondo T, Ito M, Yoshimura A. SOCS, inflammation, and cancer. JAKSTAT. 2013;2:e24053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 174. | Yoshida T, Ogata H, Kamio M, Joo A, Shiraishi H, Tokunaga Y, Sata M, Nagai H, Yoshimura A. SOCS1 is a suppressor of liver fibrosis and hepatitis-induced carcinogenesis. J Exp Med. 2004;199:1701-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 148] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 175. | Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4:551-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 597] [Cited by in RCA: 623] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 176. | Ogata H, Kobayashi T, Chinen T, Takaki H, Sanada T, Minoda Y, Koga K, Takaesu G, Maehara Y, Iida M. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology. 2006;131:179-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 145] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 177. | Lehmann U, Schmitz J, Weissenbach M, Sobota RM, Hortner M, Friederichs K, Behrmann I, Tsiaris W, Sasaki A, Schneider-Mergener J. SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130. J Biol Chem. 2003;278:661-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 178. | Nevzorova YA, Liedtke C. Sh(i)pping signals protect against Stat3-driven liver cancer. Hepatology. 2012;55:322-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 179. | Bard-Chapeau EA, Li S, Ding J, Zhang SS, Zhu HH, Princen F, Fang DD, Han T, Bailly-Maitre B, Poli V. Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell. 2011;19:629-639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 277] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 180. | Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713-2722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 320] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 181. | Hou J, Xu J, Jiang R, Wang Y, Chen C, Deng L, Huang X, Wang X, Sun B. Estrogen-sensitive PTPRO expression represses hepatocellular carcinoma progression by control of STAT3. Hepatology. 2013;57:678-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 182. | Zhang W, Hou J, Wang X, Jiang R, Yin Y, Ji J, Deng L, Huang X, Wang K, Sun B. PTPRO-mediated autophagy prevents hepatosteatosis and tumorigenesis. Oncotarget. 2015;6:9420-9433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 183. | Luedde T, Schwabe RF. NF-κB in the liver--linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1094] [Cited by in RCA: 1087] [Article Influence: 77.6] [Reference Citation Analysis (0)] |
| 184. | Qiao L, Zhang H, Yu J, Francisco R, Dent P, Ebert MP, Röcken C, Farrell G. Constitutive activation of NF-kappaB in human hepatocellular carcinoma: evidence of a cytoprotective role. Hum Gene Ther. 2006;17:280-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 185. | Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF-κB addiction and its role in cancer: ‘one size does not fit all’. Oncogene. 2011;30:1615-1630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 394] [Cited by in RCA: 384] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 186. | He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 879] [Cited by in RCA: 950] [Article Influence: 67.9] [Reference Citation Analysis (0)] |
| 187. | Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 337] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 188. | Hellerbrand C, Bumes E, Bataille F, Dietmaier W, Massoumi R, Bosserhoff AK. Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis. 2007;28:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 189. | Nikolaou K, Tsagaratou A, Eftychi C, Kollias G, Mosialos G, Talianidis I. Inactivation of the deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation, fibrosis, and cancer. Cancer Cell. 2012;21:738-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 190. | Liu L, Sakai T, Sano N, Fukui K. Nucling mediates apoptosis by inhibiting expression of galectin-3 through interference with nuclear factor kappaB signalling. Biochem J. 2004;380:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 191. | Henderson NC, Sethi T. The regulation of inflammation by galectin-3. Immunol Rev. 2009;230:160-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 407] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 192. | Matsuda Y, Yamagiwa Y, Fukushima K, Ueno Y, Shimosegawa T. Expression of galectin-3 involved in prognosis of patients with hepatocellular carcinoma. Hepatol Res. 2008;38:1098-1111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 193. | Nomoto K, Tsuneyama K, Abdel Aziz HO, Takahashi H, Murai Y, Cui ZG, Fujimoto M, Kato I, Hiraga K, Hsu DK. Disrupted galectin-3 causes non-alcoholic fatty liver disease in male mice. J Pathol. 2006;210:469-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 194. | Nakanishi Y, Tsuneyama K, Nomoto K, Fujimoto M, Salunga TL, Nakajima T, Miwa S, Murai Y, Hayashi S, Kato I. Nonalcoholic steatohepatitis and hepatocellular carcinoma in galectin-3 knockout mice. Hepatol Res. 2008;38:1241-1251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 195. | Aigelsreiter A, Haybaeck J, Schauer S, Kiesslich T, Bettermann K, Griessbacher A, Stojakovic T, Bauernhofer T, Samonigg H, Kornprat P. NEMO expression in human hepatocellular carcinoma and its association with clinical outcome. Hum Pathol. 2012;43:1012-1019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 196. | Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 468] [Cited by in RCA: 492] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 197. | Bettermann K, Vucur M, Haybaeck J, Koppe C, Janssen J, Heymann F, Weber A, Weiskirchen R, Liedtke C, Gassler N. TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell. 2010;17:481-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 198. | Bhat M, Sonenberg N, Gores GJ. The mTOR pathway in hepatic malignancies. Hepatology. 2013;58:810-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 199. | Polivka J, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142:164-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 485] [Cited by in RCA: 613] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 200. | Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27:6908-6919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 178] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 201. | Miyoshi H, Deguchi A, Nakau M, Kojima Y, Mori A, Oshima M, Aoki M, Taketo MM. Hepatocellular carcinoma development induced by conditional beta-catenin activation in Lkb1+/- mice. Cancer Sci. 2009;100:2046-2053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 202. | Nakau M, Miyoshi H, Seldin MF, Imamura M, Oshima M, Taketo MM. Hepatocellular carcinoma caused by loss of heterozygosity in Lkb1 gene knockout mice. Cancer Res. 2002;62:4549-4553. [PubMed] |
| 203. | Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 786] [Cited by in RCA: 847] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 204. | Hu TH, Huang CC, Lin PR, Chang HW, Ger LP, Lin YW, Changchien CS, Lee CM, Tai MH. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer. 2003;97:1929-1940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 224] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 205. | Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1158] [Cited by in RCA: 1170] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 206. | Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169-1178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 610] [Cited by in RCA: 627] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 207. | Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563-1568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 741] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 208. | Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, Mizuno K, Hasegawa G, Kishimoto H, Iizuka M. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113:1774-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 30] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 209. | Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972-1983, 1983.e1-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 591] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 210. | Umemura A, Park EJ, Taniguchi K, Lee JH, Shalapour S, Valasek MA, Aghajan M, Nakagawa H, Seki E, Hall MN. Liver damage, inflammation, and enhanced tumorigenesis after persistent mTORC1 inhibition. Cell Metab. 2014;20:133-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 211. | Majumdar A, Curley SA, Wu X, Brown P, Hwang JP, Shetty K, Yao ZX, He AR, Li S, Katz L. Hepatic stem cells and transforming growth factor β in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2012;9:530-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 212. | Inman GJ. Switching TGFβ from a tumor suppressor to a tumor promoter. Curr Opin Genet Dev. 2011;21:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 167] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 213. | Principe DR, Doll JA, Bauer J, Jung B, Munshi HG, Bartholin L, Pasche B, Lee C, Grippo PJ. TGF-β: duality of function between tumor prevention and carcinogenesis. J Natl Cancer Inst. 2014;106:djt369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 426] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 214. | Kitisin K, Ganesan N, Tang Y, Jogunoori W, Volpe EA, Kim SS, Katuri V, Kallakury B, Pishvaian M, Albanese C. Disruption of transforming growth factor-beta signaling through beta-spectrin ELF leads to hepatocellular cancer through cyclin D1 activation. Oncogene. 2007;26:7103-7110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 215. | Russell WE, Coffey RJ, Ouellette AJ, Moses HL. Type beta transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Natl Acad Sci USA. 1988;85:5126-5130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 283] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 216. | Tang B, Böttinger EP, Jakowlew SB, Bagnall KM, Mariano J, Anver MR, Letterio JJ, Wakefield LM. Transforming growth factor-beta1 is a new form of tumor suppressor with true haploid insufficiency. Nat Med. 1998;4:802-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 231] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 217. | Kiss A, Wang NJ, Xie JP, Thorgeirsson SS. Analysis of transforming growth factor (TGF)-alpha/epidermal growth factor receptor, hepatocyte growth Factor/c-met,TGF-beta receptor type II, and p53 expression in human hepatocellular carcinomas. Clin Cancer Res. 1997;3:1059-1066. [PubMed] |
| 218. | Im YH, Kim HT, Kim IY, Factor VM, Hahm KB, Anzano M, Jang JJ, Flanders K, Haines DC, Thorgeirsson SS. Heterozygous mice for the transforming growth factor-beta type II receptor gene have increased susceptibility to hepatocellular carcinogenesis. Cancer Res. 2001;61:6665-6668. [PubMed] |
| 219. | Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 639] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 220. | Monga SP. Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int J Biochem Cell Biol. 2011;43:1021-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 221. | Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1384] [Cited by in RCA: 1527] [Article Influence: 127.3] [Reference Citation Analysis (0)] |
| 222. | Csepregi A, Röcken C, Hoffmann J, Gu P, Saliger S, Müller O, Schneider-Stock R, Kutzner N, Roessner A, Malfertheiner P. APC promoter methylation and protein expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:579-589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 223. | Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, Giovannini M, Perret C. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc Natl Acad Sci USA. 2004;101:17216-17221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 253] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 224. | Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 225. | George J, Patel T. Noncoding RNA as therapeutic targets for hepatocellular carcinoma. Semin Liver Dis. 2015;35:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 59] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 226. | Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev. 2015;81:62-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 185] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 227. | Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 488] [Cited by in RCA: 467] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 228. | Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884-2897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 669] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 229. | Takata A, Otsuka M, Yoshikawa T, Kishikawa T, Hikiba Y, Obi S, Goto T, Kang YJ, Maeda S, Yoshida H. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology. 2013;57:162-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 230. | Sauvé F, McBroom LD, Gallant J, Moraitis AN, Labrie F, Giguère V. CIA, a novel estrogen receptor coactivator with a bifunctional nuclear receptor interacting determinant. Mol Cell Biol. 2001;21:343-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 231. | Gao S, Li A, Liu F, Chen F, Williams M, Zhang C, Kelley Z, Wu CL, Luo R, Xiao H. NCOA5 haploinsufficiency results in glucose intolerance and subsequent hepatocellular carcinoma. Cancer Cell. 2013;24:725-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 232. | Bossis I, Stratakis CA. Minireview: PRKAR1A: normal and abnormal functions. Endocrinology. 2004;145:5452-5458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 143] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 233. | Veugelers M, Wilkes D, Burton K, McDermott DA, Song Y, Goldstein MM, La Perle K, Vaughan CJ, O’Hagan A, Bennett KR. Comparative PRKAR1A genotype-phenotype analyses in humans with Carney complex and prkar1a haploinsufficient mice. Proc Natl Acad Sci USA. 2004;101:14222-14227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 111] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 234. | Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, York B, Karpen S, Finegold M, Moore D. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322:1395-1399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 133] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 235. | O’Donnell KA, Keng VW, York B, Reineke EL, Seo D, Fan D, Silverstein KA, Schrum CT, Xie WR, Mularoni L. A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer. Proc Natl Acad Sci USA. 2012;109:E1377-E1386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 236. | Chen L, Kwong M, Lu R, Ginzinger D, Lee C, Leung L, Chan JY. Nrf1 is critical for redox balance and survival of liver cells during development. Mol Cell Biol. 2003;23:4673-4686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 121] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 237. | Hirotsu Y, Hataya N, Katsuoka F, Yamamoto M. NF-E2-related factor 1 (Nrf1) serves as a novel regulator of hepatic lipid metabolism through regulation of the Lipin1 and PGC-1β genes. Mol Cell Biol. 2012;32:2760-2770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 238. | Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci USA. 2005;102:4120-4125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 232] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 239. | Vilarinho S, Calvisi DF. New advances in precision medicine for hepatocellular carcinoma recurrence prediction and treatment. Hepatology. 2014;60:1812-1814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 240. | Shanker M, Jin J, Branch CD, Miyamoto S, Grimm EA, Roth JA, Ramesh R. Tumor suppressor gene-based nanotherapy: from test tube to the clinic. J Drug Deliv. 2011;2011:465845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 241. | Burk RD, DeLoia JA, elAwady MK, Gearhart JD. Tissue preferential expression of the hepatitis B virus (HBV) surface antigen gene in two lines of HBV transgenic mice. J Virol. 1988;62:649-654. [PubMed] |
| 242. | Qin LF, Ng IO. Expression of p27(KIP1) and p21(WAF1/CIP1) in primary hepatocellular carcinoma: clinicopathologic correlation and survival analysis. Hum Pathol. 2001;32:778-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 243. | Zhang MF, Zhang ZY, Fu J, Yang YF, Yun JP. Correlation between expression of p53, p21/WAF1, and MDM2 proteins and their prognostic significance in primary hepatocellular carcinoma. J Transl Med. 2009;7:110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 244. | Wang Y, Li M, Long J, Shi XY, Li Q, Chen J, Tong WM, Jia JD, Huang J. Clinical significance of increased expression of Nijmegen breakage syndrome gene (NBS1) in human primary liver cancer. Hepatol Int. 2014;8:250-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 245. | Abou-Shady M, Baer HU, Friess H, Berberat P, Zimmermann A, Graber H, Gold LI, Korc M, Büchler MW. Transforming growth factor betas and their signaling receptors in human hepatocellular carcinoma. Am J Surg. 1999;177:209-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 99] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 246. | Chaerkady R, Harsha HC, Nalli A, Gucek M, Vivekanandan P, Akhtar J, Cole RN, Simmers J, Schulick RD, Singh S. A quantitative proteomic approach for identification of potential biomarkers in hepatocellular carcinoma. J Proteome Res. 2008;7:4289-4298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |