Published online Nov 28, 2015. doi: 10.3748/wjg.v21.i44.12667
Peer-review started: June 3, 2015
First decision: July 10, 2015
Revised: August 3, 2015
Accepted: September 28, 2015
Article in press: September 30, 2015
Published online: November 28, 2015
Processing time: 178 Days and 21.7 Hours
AIM: To evaluate a multiplex PCR assay for the detection of bacterial and viral enteropathogens in stool samples from patients with ulcerative colitis (UC).
METHODS: We prospectively analyzed 300 individuals, including immunocompetent patients, immunocompromised patients, and patients with UC. Stool samples were collected from the recto-sigmoid region of the colon by endoscopy. The samples were qualitatively analyzed for bacterial and viral enteropathogens with a multiplex PCR assay using a Seeplex® Kit. Additional clinical and laboratory data were collected from the medical records.
RESULTS: A multiplex PCR assay detected 397 pathogens (191 bacteria and 206 viruses) in 215 samples (71.7%). The most frequently detected bacteria were Escherichia coli H7, 85 (28.3%); followed by Aeromonas spp., 43 (14.3%); and Clostridium perfringens, 36 (12.0%) samples. The most prevalent viruses were Epstein-Barr virus (EBV), 90 (30.0%); followed by human herpes virus-6 (HHV-6), 53 (17.7%); and cytomegalovirus (CMV), 37 (12.3%) samples. The prevalence rate of CMV infection was significantly higher in the immunocompromised group than in the immunocompetent group (P < 0.01). CMV infection was more common in patients with UC (26/71; 36.6%) than in the immunocompetent patients excluding UC (6/188; 3.2%) (P < 0.01). CMV infection was more prevalent in UC active patients (25/58; 43.1%) than in UC inactive patients (1/13; 7.7%) (P < 0.05). Among 4 groups which defined by the UC activity and immunosuppressive drugs, the prevalence rate of CMV infection was highest in the UC active patients with immunosuppressive drugs (19/34; 55.8%). Epstein-Barr virus (EBV) infection was more common in the immunocompromised patients excluding UC (18/41; 43.9%) than in the immunocompetent patients excluding UC (47/188; 25.0%) (P < 0.05). The simultaneous presence of CMV and EBV and/or HHV6 in UC active patients (14/58; 24.1%) was greater than in immunocompromised patients excluding UC (5/41; 12.2%) (P < 0.05).
CONCLUSION: The multiplex PCR assay that was used to analyze the stool samples in this study may serve as a non-invasive approach that can be used to exclude the possibility of CMV infection in patients with active UC who are treated with immunosuppressive therapy.
Core tip: Infection with cytomegalovirus (CMV) can cause exacerbation of ulcerative colitis (UC). Thus, early diagnosis of CMV infection is important. Although endoscopic biopsy is the best approach for the diagnosis of CMV infection, this procedure may be invasive for patients and damaging to the inflamed intestine. Our prospective study on the use of the qualitative multiplex PCR assay in stool samples revealed that CMV infection is significantly more prevalent in UC active patients with immunosuppressive drugs. The multiplex PCR assay for stool samples may prove useful, non-invasive method to exclude the presence of CMV infection in patients with active UC who are treated with immunosuppressive drugs.
- Citation: Nahar S, Iraha A, Hokama A, Uehara A, Parrott G, Ohira T, Kaida M, Kinjo T, Kinjo T, Hirata T, Kinjo N, Fujita J. Evaluation of a multiplex PCR assay for detection of cytomegalovirus in stool samples from patients with ulcerative colitis. World J Gastroenterol 2015; 21(44): 12667-12675
- URL: https://www.wjgnet.com/1007-9327/full/v21/i44/12667.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i44.12667
Cytomegalovirus (CMV), a double-stranded enveloped DNA virus and a member of β-herpesviridae family, commonly infects 40%-100% of adult populations[1] and 15.8%-34% of patients with inflammatory bowel disease (IBD) who are treated with steroids and/or other immunosuppressive drugs[2]. The eyes, lungs, central nervous system, liver and intestine are the primary target organs for CMV infection. Typically, individuals who are infected with CMV remain asymptomatic, but the infection may manifest with mild mononucleosis-like symptoms. CMV, like other herpes viruses, persists in a life-long latency coupled with a risk of intermittent reactivation in some situations, such as the following: recipients of solid organ transplants, patients undergoing hemodialysis, patients with HIV, and patients who are treated with steroids and other immunosuppressive drugs.
Typically, enteric infections are self-limiting and acute, but serious illness can occur in immunocompromised patients. The number of immunocompromised patients has been increasing dramatically in recent years due to the increased number of organ transplants, increased numbers of patients on hemodialysis, or infected with HIV, and widespread use of immunosuppressive drugs and steroids. Due to defective or altered cellular and humoral immunity, immunocompromised patients are more susceptible to infections. Any infection has the possibility to cause an overwhelming disease in these populations[3,4].
Patients with IBD such as ulcerative colitis (UC) and Crohn’s disease (CD) are often immunosuppressed. Patients with severe, steroid-refractory or steroid-dependent states, as well as those treated with other biologic therapies undergo even more intensive immunosuppression. Together with the disease activity, these factors may contribute to the increased risk of colonic reactivation of latent CMV or CMV reinfection in patients with UC[5]. CMV can cause the exacerbation of UC, particularly in those with steroid-dependent/steroid- refractory diseases[2,6-8]. Histopathology and the identification of CMV DNA in colonic tissue by PCR or immunohistochemistry were recommended as the gold standard diagnostic tool for the diagnosis of CMV infection in immunosuppressed groups by the European Crohn’s and Colitis Organization in 2009[9]. Although histopathology may be the most specific diagnostic approach, a biopsy is an invasive procedure that requires an endoscopic examination. In patients with UC, the inflamed colonic tissue is friable, which leads to an increased risk of hemorrhage or perforation during invasive procedures[10]. In addition, an extended length of time is required to obtain results of a histopathological analysis, and during this time, a given patient’s condition may deteriorate clinically[11].
Molecular diagnostic tools based on the PCR method, have been developed to improve the detection of enteropathogens[12-14]. Of late, additional advances have been made to simultaneously identify enteropathogens using multiplex real-time PCR[15-18], but these methods can be low-throughput and expensive. Therefore, the implementation of a rapid, sensitive, powerful, and non-invasive molecular tool is necessary to determine the etiology of diarrhea in UC patients. Only then will it be possible to provide early and specific interventions for the prevention and control of infections in both individuals and the community.
The aim of this prospective study was to evaluate the feasibility of a qualitative multiplex PCR assay of stool for the simultaneous detection of bacterial and viral enteropathogens, focusing on CMV infection for adult patients, including patients with UC, immunocompetent patients, and immunocompromised patients.
We prospectively analyzed 300 patients who underwent colonoscopy at the Department of Endoscopy at the University of the Ryukyus Hospital from August 2014 to January 2015. Stool samples were collected endoscopically from the recto-sigmoid region of the colon and were transported to the laboratory. Stool samples were immediately processed for the multiplex PCR. Additional clinical and laboratory data were collected from medical records. UC activity was assessed using the Mayo scoring system[19].
DNA was extracted using Ribospin™ vRD kit (GeneAll Biotechnology, Seoul, South Korea) according to manufacturer’s instructions. Briefly, 300 μL of endoscopically collected stool was transferred into a 1.5-mL microcentrifuge tube followed by the addition of 500 μL of buffer to lyse the fecal matter. After an incubation of 10-15 min at room temperature, 700 μL of nucleic binding buffer was added; the solution was then vortexed and mixed. The resultant solution was transferred to a mini spin column and was centrifuged at 10000 g for 30-60 s. Total DNA was bound to the glass fiber membrane while the remaining impurities on the membrane were removed by two successive wash buffers. Pure DNA was eluted to a final volume of 50 μL of nuclease-free water. All procedures were performed at room temperature. The extracted DNA was stored at 4 °C for immediate use or at -20 °C for long-term use.
The multiplex PCR was performed according to the manufacturer’s instructions using a Seeplex® Meningitis V1, Diarrhea B1 and Diarrhea B2 ACE detection kits (Seegene, Seoul, South Korea). In regards to the Meningitis V1 ACE Detection assay, multiplex PCR was performed in a 20-μL total volume, which included 2 μL of each 10 × MV1 PM and 8-MOP solution, 1 μL of the Meningitis ACE internal control, 10 μL of the 2 × multiplex master mix and 5 μL of nucleic acid template. Negative and positive control samples were included in every PCR procedure and contained 5 μL of the Meningitis ACE NC and PC, respectively. DNA amplification was performed in an Eppendorf Mastercycler under the following conditions: an initial denaturation at 94 °C for 15 min, 94 °C for 30 s followed by 40 cycles at 63 °C for 1.5 min, 72 °C for 1.5 min with a final cycle at 72 °C for 10 min and then a hold at 4 °C.
In regards to the Diarrhea B1/B2 ACE Detection assay, the multiplex PCR was performed in a 20-μL volume that contained 4 μL of 5 × DB1 PM, 3 μL of 8-MOP solution, 10 μL of 2 × master mix and 3 μL of nucleic acid template. Negative and positive controls samples were included in each PCR reaction and contained 3 μL of DB1 NC and DB1 PC, respectively, instead of nucleic acid.
All multiplex PCR mixtures underwent the same amplification conditions shown above. The Seeplex® Meningitis V1 kit is able to detect five pathogens in a single reaction tube including CMV, human herpes virus-6 (HHV-6), Epstein-Barr virus (EBV), herpes simplex virus type 1 (HSV-1), HSV-2, and varicella zoster virus (VZV). The Seeplex® Diarrhea B1 & B2 ACE detection assay permits the simultaneous amplification of the target DNA of the following: Salmonella spp. (S. bongori and S. enterica), Shigella spp. (S. flexneri, S boydii, S. sonnei, and S. dysenteriae), Vibrio spp. (V. cholerae, V. parahaemolyticus, and V. vulnificus), Clostridium difficile toxin B, Campylobacter spp. (C. jejuni and C. coli), Clostridium perfringens toxin, Yersinia enterocolitica, Aeromonas spp. (A. salmonicida, A. sobria, A. bivalvium, and A. hydrophila), Escherichia coli O157: H7, and Verotoxin-producing E. coli (VTEC). In total, we were able to test for 25 pathogens simultaneously.
PCR products were analyzed by microchip electrophoresis system using the DNA-1000 Reagent kit in the MCE-202 MultiNA machine (Shimadzu, Japan). The data was analyzed using MultiNA viewer software (Shimadzu, Japan).
This prospective study was approved by the institutional review board of the University of the Ryukyus, and written informed consent was obtained from all patients prior to their inclusion in the study.
Statistical comparisons were conducted by two-tailed χ2 test with Yates’ correction using SPSS (version 21.0, SPSS Inc., Chicago, IL, United States). A P value < 0.05 was considered statistically significant.
The clinical characteristics of the included patients are summarized in Table 1. Three hundred patients were enrolled, including 41 immunocompromised patients and 71 patients with UC. The average age of the patients was 59.04 years (range 16-86 years), and there were more males (59%) than females. All of the UC active patients had diarrhea. A total of 300 samples collected by colonoscopy were examined. Among the 300 samples, multiplex PCR showed a positive reaction in 215 samples (71.7%) (Table 1). Among the 300 samples, one or more bacteria and viruses were identified in 146 (48.7%) and 148 (49.3%) samples, respectively. One bacterium was detected in 108 (36.0%) samples. Two and three other types of bacteria were detected in 33 (11.0%) and 5 (1.67%) samples, respectively. Ninety six (32.0%) had a single viral infection while 45 (15.0%) and 7 (2.3%) had 2 and 3 viruses, respectively. The most frequently detected bacteria were E. coli H7, 85 (28.3%); followed by Aeromonas spp., 43 (14.3%); C. perfringens, 36 (12.0%); and C. difficile Toxin B, 15 (5.0%). Shigella spp. and Y. enterocolitica were not detected. The most prevalent viruses were EBV, 91 (30.3%); followed by HHV-6, 53 (17.7%); and CMV, 37 (12.3%). The least prevalent viruses were HSV2, 6 (2.0%) and VZV, 6 (2.0%). Internal control was positive for all samples.
| Characteristics | Number of patients 300 (100) |
| Gender | |
| Male | 177 (59) |
| Female | 123 (41) |
| Age in years | 59.04 (16-86 ) |
| Underlying disease | |
| Immunocompromised | 41 (13.7) |
| Ulcerative colitis | 71 (23.7) |
| One or more bacteria/viruses detected | 215 (71.7) |
| One or more bacteria detected | 146 (48.7) |
| Number of bacteria detected | |
| 1 | 108 (36.0) |
| 2 | 33 (11.0) |
| 3 | 5 (1.67) |
| One or more viruses detected | 148 (49.3) |
| Number of viruses detected | |
| 1 | 96 (32.0) |
| 2 | 45 (15.0) |
| 3 | 7 (2.3) |
| Bacteria detected | |
| Clostridium difficile toxin B | 15 (5.0) |
| Salmonella spp. | 4 (1.3) |
| Campylobacter spp. | 3 (1.0) |
| Vibrio spp. | 1 (0.3) |
| Clostridium perfringens | 36 (12.0) |
| E. coli H7 | 85 (28.3) |
| E. coli O157 | 1 (0.3) |
| VTEC | 1 (0.3) |
| Aeromonas spp. | 43 (14.3) |
| Virus detected | |
| CMV | 37 (12.3) |
| HHV6 | 53 (17.7) |
| EBV | 91 (30.3) |
| HSV1 | 14 (4.7) |
| HSV2 | 6 (2.0) |
| VZV | 6 (2.0) |
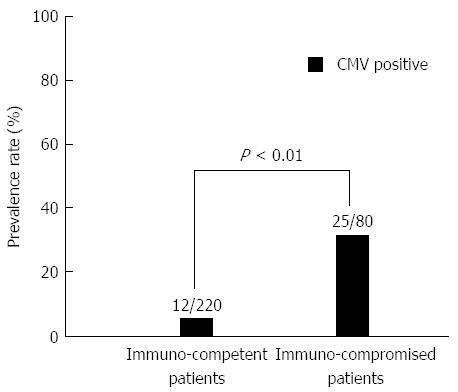
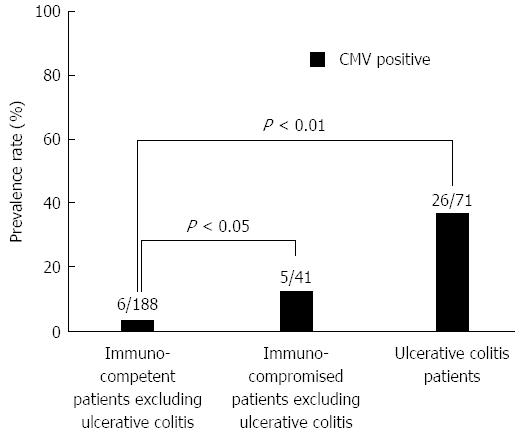
Patients who were treated with immunosuppressive drugs (e.g., anticancer drugs, T-cell inhibitors, steroids), HIV patients, and patients on hemodialysis for chronic kidney disease were included in the immunocompromised group (80; 26.7%). Patients who were not treated with immunosuppressive drugs were incorporated into the immunocompetent group (220; 73.3%). The prevalence rate of CMV infection was significantly higher in the immunocompromised group than in the immunocompetent group (P < 0.01) (Figure 1). CMV infection was more common in patients with UC (26/71; 36.6%) than in the immunocompetent patients excluding UC (6/188; 3.2%) (P < 0.01) (Figure 2).
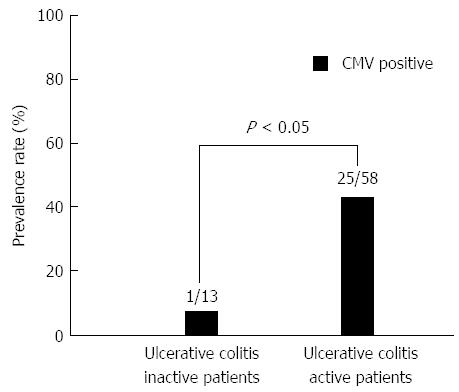
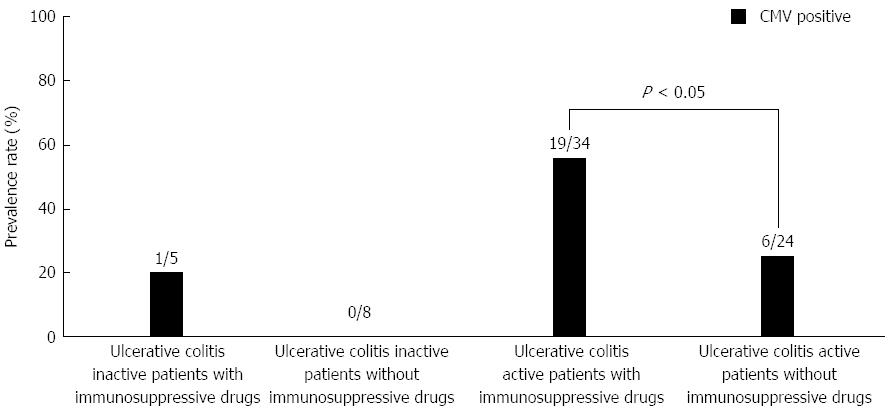
In UC patients, CMV infection was more prevalent in UC active patients (25/58; 43.1%) than in UC inactive patients (1/13; 7.7%) (P < 0.05) (Figure 3). UC patients were further categorized into 4 groups based on the UC activity and the administration of immunosuppressive drugs. The prevalence rate of CMV infection was highest among individuals in the active UC group who were treated with immunosuppressive drugs compared with individuals in the other 3 groups (Figure 4). Among the 58 patients with active UC, immunosuppressive drugs were prescribed for 34 patients. The CMV infection rate was significantly higher in those who were treated with immunosuppressive drugs (19/34; 55.8%) compared with those who were not treated with immunosuppressive drugs (6/24; 25.0%) (P < 0.05).
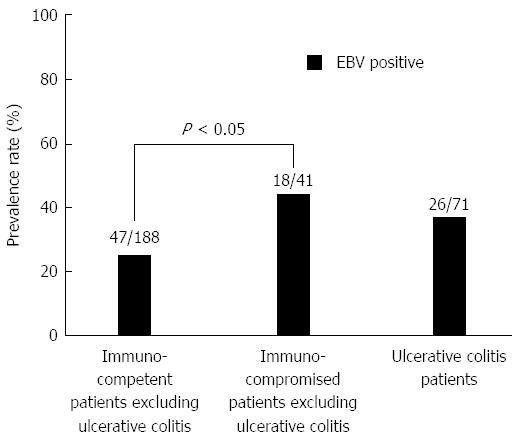
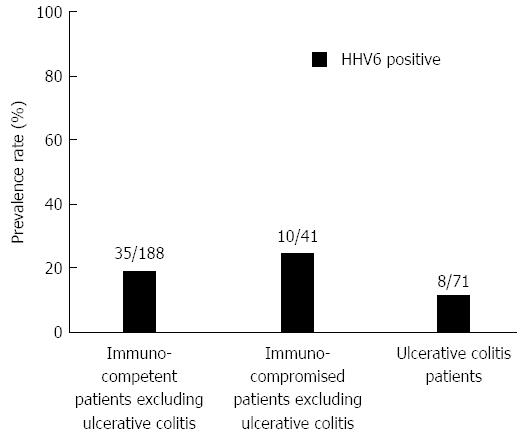
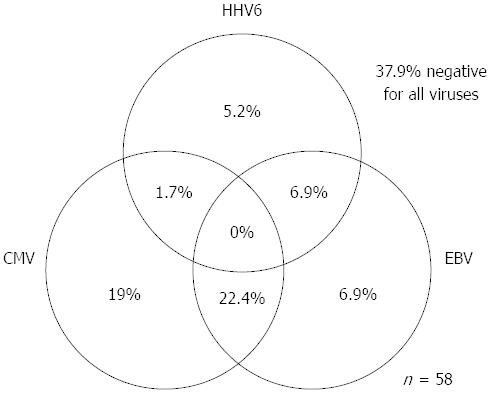
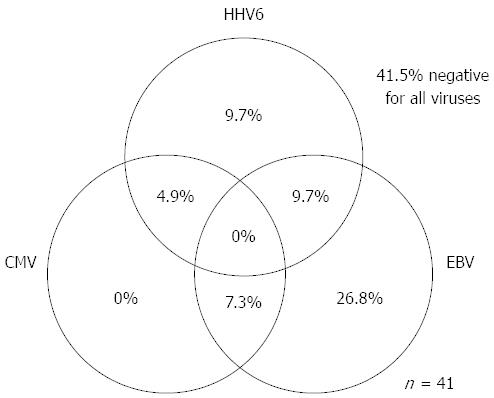
As for EBV infection, EBV infection was more common in the immunocompromised patients excluding UC (18/41; 43.9%) than in the immunocompetent patients excluding UC (47/188; 25.0%) (P < 0.05) (Figure 5). Figure 6 shows no significant differences of the prevalence rate of HHV6 infection among the groups. The overlapping prevalence patterns for CMV, EBV, and HHV6 are shown for UC active patients (Figure 7) and the immunocompromised patients excluding UC (Figure 8). The simultaneous presence of CMV and EBV and/or HHV6 in UC active patients (14/58; 24.1%) was greater than in immunocompromised patients excluding UC (5/41; 12.2%) (P < 0.05). No patients in our cohort were simultaneously infected with all three viruses.
Multiplex PCR has been widely applied to the diagnosis of gastrointestinal infectious diseases[18]. Notably, the Seeplex® Diarrhea ACE Detection assay has shown itself to be a rapid, sensitive, specific, and reliable diagnostic tool for the direct detection of the most common enteropathogens in stool samples[20]. Other multiplex PCR assays (e.g., the FilmArray gastrointestinal panel and Luminex xTag gastrointestinal pathogen panel) have also yielded an increased percent positive rate compared with routine testing[21]. Based on these recent developments, we have conducted this prospective assay with the Seeplex® Diarrhea and Meningitis Detection method. We have analyzed (1) the results of a multiplex PCR assay as a rapid and non-invasive molecular diagnostic tool for the early diagnosis of CMV infection in patients with UC; (2) the prevalence rate of CMV infection in UC in comparison with control populations; and (3) the relationship of CMV infection with immunosuppressive drugs and disease activity. We have found that CMV infection is significantly more prevalent in the active UC group compared with the immunocompetent group and the immunocompromised group (P < 0.05), which further indicates the strong association of CMV infection in patients with active UC.
In addition, CMV infection is significantly correlated with the administration of immunosuppressive drugs among the patients with active UC (P < 0.05). UC patients who were treated with immunosuppressive drugs such as corticosteroids, tacrolimus, azathioprine, 6-mercaptopurine, or cyclosporine A, and those with existing inflammation are considered to be at an increased risk for the development of CMV infection[22-24]. Although some studies have suggested that CMV infection may be an occasional finding or that CMV may be an inactive participant in UC[22,25,26], CMV itself can be a major cause of exacerbation of UC. As a result, CMV subsequently leads to the worst clinical conditions seen among patients with UC[27]. Therefore, an early diagnosis of CMV infection in UC is crucial, and several modalities have been developed for the diagnosis of CMV infection.
Currently, the guidelines of the European Colitis and Crohn’s Organization recommend the combined use of hematoxylin-eosin (HE) staining, immunohistochemical (IHC) staining with a CMV-specific monoclonal antibody and PCR for CMV in colonic tissue for the detection of colonic CMV infection in patients with UC[9]. IHC staining has a higher sensitivity than conventional histology (78%-93%) for the detection of CMV in colonic tissue[28].
Several studies have reported that the quantitative real-time PCR technique is a highly sensitive method for the diagnosis of CMV infection in inflamed colonic tissue compared with non-inflamed colonic tissue in the setting of UC[29]. The detection of CMV DNA in mucosal biopsies by PCR analysis has been regarded as the most sensitive assay for the diagnosis of CMV infection of the intestinal tract[30]. However, this technique requires invasive procedures such as sigmoidoscopy or colonoscopy to collect biopsy material and in some instances a physician may not be able to perform a colonoscopy. Furthermore, inflamed colonic tissue in cases of UC is mostly friable, edematous, and eroded and bleeds when touched. As a result, there is a high risk of bleeding during the collection of tissues. Additionally, endoscopic biopsy may lead to false-negative results due to sampling bias. To reduce sampling error, tissues in different locations along the colon are needed. Therefore, serology, a CMV antigenemia assay and PCR of blood samples for CMV are more convenient than invasive procedures[11,31]. Serological tests, such as the detection of CMV-specific IgM antibodies, have been shown to have a 100% sensitivity and a 99% specificity[32]. However, the presence of IgM antibodies can only reveal acute infection, and as such, cases of reactivation are often undiagnosed. Therefore, serology also has limited value in the diagnosis of CMV infection in patients with UC.
A recent study[11] evaluated the diagnostic performance of a CMV antigenemia assay and PCR of blood for the detection of CMV in patients with UC and showed lower sensitivities for the diagnosis of CMV infection compared with a previous study[10]. The presence of a CMV infection in the intestine cannot be ruled out in the case of a negative CMV antigenemia assay. In many cases, additional testing will be required to reach the diagnosis of CMV infection. The CMV antigenemia assay also has limited clinical value in the prediction of the reactivation of CMV infection in the gastrointestinal tract[33] and is therefore ineffective at preventing the development of CMV colitis. The CMV antigenemia assay is also relatively time consuming and demands expert pathologists to reduce subjective bias during interpretation of the slides. As the pp65 antigen for the CMV antigenemia assay is examined in blood leukocytes, it may also reveal false-negative results in leucopenic patients[27]. The diagnostic accuracy of CMV antigenemia may depend on site of organ/tissue involvement[34]. Under these circumstances, the testing of stool samples by PCR was chosen as an alternative to these techniques.
We have found advantages in the detection of CMV DNA in stool samples by the multiplex PCR technique. Although this prospective study was designed so that fresh samples would be prepared from stool samples obtained by endoscopy, stool analysis is traditionally a non-invasive procedure. The procurement of tissues by biopsy may have potential risks when the intestine is inflamed. In addition, the stool may better reflect the CMV enterocolitis that is located in the proximal colon and small intestine. We are planning to analyse stool samples which are collected by the patients at home.
Several studies have evaluated CMV DNA in stool samples[26,35,36]. Ganzenmueller et al[37] have retrospectively evaluated quantitative real-time PCR in 66 fecal samples and intestinal biopsies for the diagnosis of CMV intestinal disease. Their study also compared CMV DNA in stool and colonic biopsies with the results of histopathology and the CMV antigenemia assay. In their study, the sensitivity and specificity of stool CMV DNA for the diagnosis of CMV intestinal disease are 67% and 96%, respectively. Therefore, CMV DNA in the stool is more sensitive and specific than the gold standard method (histopathology and IHC) for the diagnosis of CMV in inflamed colonic tissue.
Additional studies have evaluated the sensitivity, specificity and reliability of the multiplex PCR assay as a rapid molecular diagnostic tool for the investigation of the etiology of enteric infections in pediatric patients[20]. However, the diagnostic significance of the multiplex PCR assay in stool samples for the diagnosis of CMV infection in patients with UC has not yet been evaluated. In our prospective study, we evaluated the ability of a commercially available multiplex PCR assay to diagnose CMV infection in UC using stool samples. To our knowledge, this is the first large-scale prospective study that assesses the multiplex PCR assay as a diagnostic tool for the diagnosis of CMV in patients with active UC, and in both immunocompetent and immunocompromised patients. As for EBV and HHV6, this study has showed a possible synergistic role for these viruses in the pathogenesis in active UC activation, as suggested by the prior reports[38,39].
Our study has some limitations. First, we did not quantify the CMV DNA. However, in patients with UC, any positive result with respect to CMV is clinically significant without quantification of the viral load. Quantification of CMV in stool specimens is not feasible during specimen processing (e.g., diluted versus non-diluted)[40], and positive results should be carefully considered, especially in patients with UC who are treated with immunosuppressive drugs[37]. Furthermore, this prospective study was not designed to compare the different modalities for the detection of CMV, including histopathology, CMV antigenemia assay, serology, and tissue CMV PCR. Herfarth et al[36] have shown that the sensitivity and specificity of the stool PCR analysis compared with PCR in mucosal biopsies were 83% and 93%, respectively. They have noted that it is not clear whether CMV DNA detected in the stool samples was due to leakage from the blood compartment into the intestinal tract, or if it was derived from intestinal CMV infection.
In conclusion, this prospective study proposes multiplex PCR as a successful, non-invasive diagnostic technique for rapid detection of CMV infection among UC patients in clinical in-patient and out-patient settings. Additionally, we present a new protocol for the broad analysis of the enteropathogens in stool samples in adult populations. This method will also help predict CMV infection prior to the development of intestinal symptoms, which is important for the prevention of exacerbation of UC by CMV reactivation. Positive PCR results may help to rapidly diagnose patients at a high risk for CMV infection. Further studies are needed to confirm these results. Future studies that involve colonic biopsies will be needed to confirm the reliability of the multiplex PCR test results.
The authors are grateful to clinical staff in the Department of Infectious, Respiratory, and Digestive Medicine and the Department of Endoscopy at the University of the Ryukyus.
Infection with cytomegalovirus (CMV) can cause exacerbation of ulcerative colitis (UC). Thus, early diagnosis of CMV infection is important. Although endoscopic biopsy is the best approach for the diagnosis of CMV infection, this procedure may be invasive for patients and damaging to the inflamed intestine.
Molecular diagnostic advances have been made to simultaneously identify enteropathogens using multiplex polymerase chain reaction (PCR). The aim of this prospective study was to evaluate the feasibility of a qualitative multiplex PCR assay of stool for the simultaneous detection of bacterial and viral enteropathogens, focusing on CMV infection for adult patients, including patients with UC, immunocompetent patients, and immunocompromised patients.
This is the first large-scale prospective study that assesses the multiplex PCR assay in stool samples as a diagnostic tool for the diagnosis of CMV in patients with UC. This assay may prove useful, non-invasive method to exclude the presence of CMV infection in patients with active UC who are treated with immunosuppressive drugs.
This multiplex PCR as a successful, non-invasive diagnostic technique will help predict CMV infection prior to the development of intestinal symptoms, which is important for the prevention of exacerbation of UC by CMV reactivation.
Multiplex PCR assay: A rapid, sensitive, specific, and reliable diagnostic tool for the simultaneous detection of bacterial and viral enteropathogens by PCR.
In this study, the authors detected CMV and other enteric pathogens using multiplex PCR in stool samples collected from 300 patients who underwent colonoscopy including patients with UC.
P- Reviewer: Annese V, Zhang L S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
| 1. | Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis. 2006;43:1143-1151. [PubMed] |
| 2. | Papadakis KA, Tung JK, Binder SW, Kam LY, Abreu MT, Targan SR, Vasiliauskas EA. Outcome of cytomegalovirus infections in patients with inflammatory bowel disease. Am J Gastroenterol. 2001;96:2137-2142. [PubMed] |
| 3. | Rozenblyum EV, Allen UD, Silverman ED, Levy DM. Cytomegalovirus infection in childhood-onset systemic lupus erythematosus. Int J Clin Rheumtol. 2013;8:137-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 4. | Taylor GH. Cytomegalovirus. Am Fam Physician. 2003;67:519-524. [PubMed] |
| 5. | McCurdy JD, Jones A, Enders FT, Killian JM, Loftus EV, Smyrk TC, Bruining DH. A model for identifying cytomegalovirus in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol. 2015;13:131-137; quiz e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 6. | Cottone M, Pietrosi G, Martorana G, Casà A, Pecoraro G, Oliva L, Orlando A, Rosselli M, Rizzo A, Pagliaro L. Prevalence of cytomegalovirus infection in severe refractory ulcerative and Crohn’s colitis. Am J Gastroenterol. 2001;96:773-775. [PubMed] |
| 7. | Domènech E, Vega R, Ojanguren I, Hernández A, Garcia-Planella E, Bernal I, Rosinach M, Boix J, Cabré E, Gassull MA. Cytomegalovirus infection in ulcerative colitis: a prospective, comparative study on prevalence and diagnostic strategy. Inflamm Bowel Dis. 2008;14:1373-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 170] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 8. | Maher MM, Nassar MI. Acute cytomegalovirus infection is a risk factor in refractory and complicated inflammatory bowel disease. Dig Dis Sci. 2009;54:2456-2462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 9. | Rahier JF, Ben-Horin S, Chowers Y, Conlon C, De Munter P, D’Haens G, Domènech E, Eliakim R, Eser A, Frater J. European evidence-based Consensus on the prevention, diagnosis and management of opportunistic infections in inflammatory bowel disease. J Crohns Colitis. 2009;3:47-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 400] [Cited by in RCA: 372] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 10. | Nagata N, Kobayakawa M, Shimbo T, Hoshimoto K, Yada T, Gotoda T, Akiyama J, Oka S, Uemura N. Diagnostic value of antigenemia assay for cytomegalovirus gastrointestinal disease in immunocompromised patients. World J Gastroenterol. 2011;17:1185-1191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Kim JW, Boo SJ, Ye BD, Kim CL, Yang SK, Kim J, Kim SA, Park SH, Park SK, Yang DH. Clinical utility of cytomegalovirus antigenemia assay and blood cytomegalovirus DNA PCR for cytomegaloviral colitis patients with moderate to severe ulcerative colitis. J Crohns Colitis. 2014;8:693-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 12. | de Boer RF, Ott A, Kesztyüs B, Kooistra-Smid AM. Improved detection of five major gastrointestinal pathogens by use of a molecular screening approach. J Clin Microbiol. 2010;48:4140-4146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 13. | Wolffs PF, Bruggeman CA, van Well GT, van Loo IH. Replacing traditional diagnostics of fecal viral pathogens by a comprehensive panel of real-time PCRs. J Clin Microbiol. 2011;49:1926-1931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 14. | Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J Clin Microbiol. 2010;48:2929-2933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 15. | Barletta F, Mercado EH, Lluque A, Ruiz J, Cleary TG, Ochoa TJ. Multiplex real-time PCR for detection of Campylobacter, Salmonella, and Shigella. J Clin Microbiol. 2013;51:2822-2829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Barbut F, Monot M, Rousseau A, Cavelot S, Simon T, Burghoffer B, Lalande V, Tankovic J, Petit JC, Dupuy B. Rapid diagnosis of Clostridium difficile infection by multiplex real-time PCR. Eur J Clin Microbiol Infect Dis. 2011;30:1279-1285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Enserink R, Scholts R, Bruijning-Verhagen P, Duizer E, Vennema H, de Boer R, Kortbeek T, Roelfsema J, Smit H, Kooistra-Smid M. High detection rates of enteropathogens in asymptomatic children attending day care. PLoS One. 2014;9:e89496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 18. | McAuliffe GN, Anderson TP, Stevens M, Adams J, Coleman R, Mahagamasekera P, Young S, Henderson T, Hofmann M, Jennings LC. Systematic application of multiplex PCR enhances the detection of bacteria, parasites, and viruses in stool samples. J Infect. 2013;67:122-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625-1629. [PubMed] |
| 20. | Onori M, Coltella L, Mancinelli L, Argentieri M, Menichella D, Villani A, Grandin A, Valentini D, Raponi M, Russo C. Evaluation of a multiplex PCR assay for simultaneous detection of bacterial and viral enteropathogens in stool samples of paediatric patients. Diagn Microbiol Infect Dis. 2014;79:149-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 21. | Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol. 2014;52:3667-3673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 227] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 22. | Lawlor G, Moss AC. Cytomegalovirus in inflammatory bowel disease: pathogen or innocent bystander? Inflamm Bowel Dis. 2010;16:1620-1627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 197] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 23. | Nakase H, Honzawa Y, Toyonaga T, Yamada S, Minami N, Yoshino T, Matsuura M. Diagnosis and treatment of ulcerative colitis with cytomegalovirus infection: importance of controlling mucosal inflammation to prevent cytomegalovirus reactivation. Intest Res. 2014;12:5-11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Goodman AL, Murray CD, Watkins J, Griffiths PD, Webster DP. CMV in the gut: a critical review of CMV detection in the immunocompetent host with colitis. Eur J Clin Microbiol Infect Dis. 2015;34:13-18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 25. | Matsuoka K, Iwao Y, Mori T, Sakuraba A, Yajima T, Hisamatsu T, Okamoto S, Morohoshi Y, Izumiya M, Ichikawa H. Cytomegalovirus is frequently reactivated and disappears without antiviral agents in ulcerative colitis patients. Am J Gastroenterol. 2007;102:331-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 166] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 26. | do Carmo AM, Santos FM, Ortiz-Agostinho CL, Nishitokukado I, Frota CS, Gomes FU, Leite AZ, Pannuti CS, Boas LS, Teixeira MG. Cytomegalovirus infection in inflammatory bowel disease is not associated with worsening of intestinal inflammatory activity. PLoS One. 2014;9:e111574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Nakase H, Matsumura K, Yoshino T, Chiba T. Systematic review: cytomegalovirus infection in inflammatory bowel disease. J Gastroenterol. 2008;43:735-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 28. | Garrido E, Carrera E, Manzano R, Lopez-Sanroman A. Clinical significance of cytomegalovirus infection in patients with inflammatory bowel disease. World J Gastroenterol. 2013;19:17-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 51] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 29. | Yoshino T, Nakase H, Ueno S, Uza N, Inoue S, Mikami S, Matsuura M, Ohmori K, Sakurai T, Nagayama S. Usefulness of quantitative real-time PCR assay for early detection of cytomegalovirus infection in patients with ulcerative colitis refractory to immunosuppressive therapies. Inflamm Bowel Dis. 2007;13:1516-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 126] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 30. | Kandiel A, Lashner B. Cytomegalovirus colitis complicating inflammatory bowel disease. Am J Gastroenterol. 2006;101:2857-2865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 217] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 31. | Chun J, Lee C, Kwon JE, Hwang SW, Kim SG, Kim JS, Jung HC, Im JP. Usefulness of the cytomegalovirus antigenemia assay in patients with ulcerative colitis. Intest Res. 2015;13:50-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 32. | Revello MG, Gerna G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin Microbiol Rev. 2002;15:680-715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 33. | Ganzenmueller T, Henke-Gendo C, Schlué J, Wedemeyer J, Huebner S, Heim A. Quantification of cytomegalovirus DNA levels in intestinal biopsies as a diagnostic tool for CMV intestinal disease. J Clin Virol. 2009;46:254-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 34. | Hamada Y, Nagata N, Shimbo T, Igari T, Nakashima R, Asayama N, Nishimura S, Yazaki H, Teruya K, Gatanaga H. Assessment of antigenemia assay for the diagnosis of cytomegalovirus gastrointestinal diseases in HIV-infected patients. AIDS Patient Care STDS. 2013;27:387-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 35. | Chan KS, Yang CC, Chen CM, Yang HH, Lee CC, Chuang YC, Yu WL. Cytomegalovirus colitis in intensive care unit patients: difficulties in clinical diagnosis. J Crit Care. 2014;29:474.e1-474.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 36. | Herfarth HH, Long MD, Rubinas TC, Sandridge M, Miller MB. Evaluation of a non-invasive method to detect cytomegalovirus (CMV)-DNA in stool samples of patients with inflammatory bowel disease (IBD): a pilot study. Dig Dis Sci. 2010;55:1053-1058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 37. | Ganzenmueller T, Kluba J, Becker JU, Bachmann O, Heim A. Detection of cytomegalovirus (CMV) by real-time PCR in fecal samples for the non-invasive diagnosis of CMV intestinal disease. J Clin Virol. 2014;61:517-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Wakefield AJ, Fox JD, Sawyerr AM, Taylor JE, Sweenie CH, Smith M, Emery VC, Hudson M, Tedder RS, Pounder RE. Detection of herpesvirus DNA in the large intestine of patients with ulcerative colitis and Crohn’s disease using the nested polymerase chain reaction. J Med Virol. 1992;38:183-190. [PubMed] |
| 39. | Bertalot G, Villanacci V, Gramegna M, Orvieto E, Negrini R, Saleri A, Terraroli C, Ravelli P, Cestari R, Viale G. Evidence of Epstein-Barr virus infection in ulcerative colitis. Dig Liver Dis. 2001;33:551-558. [PubMed] |
| 40. | Binnicker MJ, Espy ME. Comparison of six real-time PCR assays for qualitative detection of cytomegalovirus in clinical specimens. J Clin Microbiol. 2013;51:3749-3752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |