Published online Oct 28, 2015. doi: 10.3748/wjg.v21.i40.11387
Peer-review started: June 3, 2015
First decision: June 23, 2015
Revised: July 15, 2015
Accepted: September 30, 2015
Article in press: September 30, 2015
Published online: October 28, 2015
Processing time: 147 Days and 4.2 Hours
Pancreatic cancer is usually diagnosed at an advanced stage and curative resection is feasible in only a small minority of patients at the time of diagnosis. Diagnosis at an early stage is unequivocally associated with better long-term survival. Several candidate molecular markers for early detection are currently under investigation in different phases of discovery and validation. Recent advances in the technology for whole genome, methylome, ribonucleome, and proteome interrogation has enabled rapid advancements in the field of biomarker discovery. In this review we discuss the current status of molecular markers for detection of pancreatic cancer in blood, pancreatic cyst fluid, pancreatic juice and stool and briefly highlight some promising preliminary results of new approaches that have the potential of advancing this field in the near future.
Core tip: Pancreatic cancer is a leading cause of cancer mortality worldwide. Early detection at a resectable stage is associated with the best long-term prognosis. There are ongoing efforts globally to discover, validate and optimize molecular markers for early diagnosis. The challenge is to develop highly sensitive markers not only for earliest stage cancer but also to accurately detect premalignant lesions with high grade dysplasia that would maximally benefit from resection. In this review, we summarize some of the most promising biomarkers for molecular detection of pancreatic cancer and discuss evolving molecular approaches.
- Citation: Majumder S, Chari ST, Ahlquist DA. Molecular detection of pancreatic neoplasia: Current status and future promise. World J Gastroenterol 2015; 21(40): 11387-11395
- URL: https://www.wjgnet.com/1007-9327/full/v21/i40/11387.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i40.11387
In 2015, an estimated 48960 people will be diagnosed with pancreas cancer (PC) in the United States[1]. Based on current SEER (Surveillance, Epidemiology, and End Results Program) statistics factsheets published by the National Cancer Institute (NCI), only a small minority of them (7.2%) will survive for 5 years or more[1]. In 2012, 337872 new cases of PC were reported worldwide along with 330372 PC-related deaths[2]. PC is currently the seventh leading cause of cancer mortality worldwide and is projected to become the second leading cause of cancer mortality in the United States by 2030[3,4].
Over the past two decades the 5-year survival rate of PC has improved from 3.6% to 7.2%[1]. This has been attributed to improvements in peri-operative care and decreased surgical mortality[2,5]. However, definitive surgical intervention is feasible in only a small minority of patients diagnosed with PC. The disease is localized to the pancreas in less than 10% cases at the time of initial diagnosis and patients with localized disease have a significantly better outcome with 5-year survival rate of 27%[1]. In instances of incidentally discovered asymptomatic stage I disease, 5-year survival may exceed 70%[6]. It is obvious that diagnosing PC in the early stages will significantly improve survival.
There is an urgent need to develop early detection methods to improve these outcomes. New molecular approaches offer the promise of accuracy, ready distribution, and affordability that will be needed to deliver practical screening tools. For a diagnostic biomarker to be clinically useful in early detection of PC, the most important characteristics are high sensitivity, high specificity, and ability to discriminate low-grade dysplasia from high-grade dysplasia and early cancer. Such performance features would help select patients for early endoscopic or surgical intervention with curative intent. Highly sensitive markers that lack specificity can result in a large number of false positive tests and lead to unnecessary and expensive tests and procedures.
Contrary to common belief that pancreatic ductal adenocarcinoma (PDAC) is a rapidly-growing malignancy, its progression through the stages of precancer to metastatic disease may take an average of two decades[7]. This is a critically important observation suggesting that there may be an ample period during which to screen the key target lesions-earliest stage cancer and those precancers at greatest risk of progression[8].
Although the earliest precancer lesions in PDAC, also known as pancreatic intra-epithelial neoplasia (PanIN), are well-defined from a histologic standpoint, currently available imaging techniques and biomarkers lack sensitivity for their detection. The cystic premalignant lesions which include intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs) are more readily identifiable by imaging. However, as the large majority of these cystic lesions do not progress to cancer, their incidental detection often results in a management dilemma. Current risk prediction models are imperfect and the proportion of pancreatic resections for non-neoplastic lesions appears to be much higher than the proportion of “missed” cancer[9].
Thus, the natural history of PDAC appears to present a window of opportunity for detection of early stage and potentially-curable neoplasms. The challenge is to develop highly sensitive molecular markers not only for earliest stage PDAC but also with the ability to accurately detect pancreatic premalignant lesions with high grade dysplasia that would maximally benefit from surgical resection.
The past decade has witnessed explosive progress in whole genome, methylome, ribonucleome, and proteome interrogation that has accelerated the identification of candidate biomarkers for early detection of PDAC. While candidate markers generated from these discovery engines have potential to yield discriminant tests when applied to distant media, clinical validation and test development are largely at early stages. The use of a variety of assay platforms has been explored on diverse media such as plasma, serum, cyst fluid, pancreatic juice and stool. Molecular markers have been evaluated to detect both cancer and premalignant pancreatic lesions, assess prognosis, and predict tumor response to specific pharmacotherapy. In the future, such molecular tests may be applied to guide individualized cancer therapy.
Over the past decade, several molecular tests have been studied in pancreatic neoplasms (Table 1). In this review, we describe the molecular markers that are currently in the pipeline and also discuss evolving molecular approaches which may potentially alter the diagnostic paradigm for pancreatic cancer in the future. The focus is on PDAC and cystic neoplasms of the exocrine pancreas since these comprise the majority of malignant pancreatic tumors.
| Bio specimen | Type of biomarker | Examples of molecular markers |
| Blood | Conventional protein markers | Carbohydrate antigen 19-9 (CA 19-9) |
| Carcinoembryonic antigen (CEA) | ||
| Novel proteins | Intercellular adhesion molecule-1 (ICAM-1) | |
| Osteoprotegerin (OPG) | ||
| Macrophage inhibitory cytokine-1 (MIC-1) | ||
| Tissue inhibitor of metalloproteinases-1 (TIMP-1) | ||
| S100 calcium-binding protein P (S100P) | ||
| Mutated genes | KRAS, TP53, SMAD4, CDKN2A, KDM6A, PREX2 | |
| Aberrantly methylated genes | p16, ppENK, cyclin D2, SPARC/osteonectin SOCS-1, TSLC1 | |
| Micro-RNAs | miR-1290, miR-145, miR-150, miR-223, miR-636, miR-26b, miR-34a, miR-122, miR-126, miR-145, miR-150, miR-223, miR-505, miR-636, miR-885.5p. | |
| Circulating tumor cells | (molecular markers not yet interrogated) | |
| Cyst fluid | Mutated genes | KRAS, GNAS |
| Aberrantly methylated genes | BNIP3, PTCHD2, SOX17, NXPH1 and EBF3 | |
| Micro-RNAs | miR-138, miR-195, miR-204, miR-216a, miR-217, miR-218, miR-802, miR-155, miR-214, miR-26a, miR-30b, miR-31, and miR-125 | |
| Tumor tissue | Novel proteins | Gelsolin, Lumican, Galectin-1 and Laminin |
| Pancreatic juice | Mutated genes | KRAS, TP53 |
| Aberrantly methylated genes | ADCY1, CD1D, BMP3 | |
| Stool | Mutated genes | KRAS, BMP3 |
Conventional approach: Conventional biomarkers for the detection of PDAC have been largely protein-based and have several limitations. Carbohydrate antigen 19-9 (CA 19-9) is the most widely studied conventional biomarker for PDAC. Importantly, levels are often normal in early disease and falsely elevated in patients with various conditions, such as biliary obstruction[10]. Furthermore, it is well known that up to 15% of individuals with a high tumor burden have normal or undetectable CA 19-9 levels. About 5% of Caucasians lack the Lewis blood group antigen and have undetectable levels of CA 19-9 since they lack the characteristic fucosylation pattern required for CA 19-9 detection by commercially available assays[10]. The combination of these factors makes it an unreliable screening tool. Its scope in current practice is largely restricted to detection of tumor recurrence after surgical resection[11,12]. Carcinoembryonic antigen (CEA) has also been studied as a diagnostic test for PDAC and found to have poor performance characteristics. In isolation, it has low diagnostic accuracy for aggregate stages of PDAC with a sensitivity of 54% and specificity of 79%; these respective metrics change to 86% and 72%, when CEA is used in combination with CA 19-9[13,14].
Novel proteins: A variety of discovery approaches have been pursued to profile the pancreatic cancer proteome and identify biomarkers for diagnosis. Several sample types, including whole tumor tissue, isolated neoplastic cells and isolated tumor stromal cells have been used in these discovery and validation efforts[15]. In addition to tissue profiling, the proteome has also been studied in pancreatic juice and serum using liquid chromatography tandem mass spectrometry as well as high-throughput immunologic proteomic strategies[16]. In the past, antibodies to osteoprotegerin, intercellular adhesion molecule-1 (ICAM-1) and tissue inhibitor of metalloproteinases-1 (TIMP-1) have been proposed as potential diagnostic biomarkers for PDAC. In a case-control study of 333 patients with PDAC, a combination of CA 19-9, ICAM-1 and OPG antibodies was found to have a sensitivity and specificity of 88% and 90%, respectively, for distinguishing PDAC from healthy controls[17]. However, a more recent case-control study evaluating ICAM-1 and TIMP-1 levels argued against their utility in the early diagnosis of PDAC. In this study investigators tested the levels of these proteins in pre-diagnosis samples of patients 0-12 mo before a diagnosis of PDAC and a quantitative comparison with non-cancer control samples revealed no significant difference[18]. Interestingly, patients with jaundice secondary to both benign and malignant etiology had elevated circulating levels of these proteins leading the investigators to believe that failure to account for biliary obstruction may have contributed false positive results in prior studies. Other circulating proteins that have been identified as candidate biomarkers include macrophage inhibitory cytokine-1 (MIC-1) and S100 calcium-binding protein P (S100P). Serum MIC-1 may have a higher sensitivity (62.5% vs 25.0%) and similar specificity compared to CA19-9 for detection of early stage PDAC[19]. A meta-analysis evaluating the diagnostic performance of S100P reported pooled sensitivity and specificity of 87% and 88%, respectively, for aggregate stages of PDAC with an AUC of 0.93[20].
Further investigation is clearly indicated to corroborate early observations with these novel differentially-expressed candidate biomarkers and to better define their biological significance and clinical utility.
DNA: Mutations - The genomic landscape of PDAC is complex. In recent years, in-depth analysis of the coding region of the genome using high-throughput studies of gene expression has revealed a large number of gene expression abnormalities associated with PDAC. In one of the early studies, Jones et al[21] performed SAGE (serial analysis of gene expression) analysis of 24 PDACs with tumor cells passaged in vitro as cell lines or in nude mice as xenografts. This study identified 10-fold overexpression of 541 genes in more than 90% of the tumors when compared to normal pancreatic duct cells[21].
Commonly mutated genes that characterize PDAC include KRAS, TP53, SMAD4 and CDKN2A. A recently published study describing deep whole-genome sequencing and copy number variation (CNV) analysis of 100 patients with PDAC has reaffirmed that chromosomal rearrangement in these four genes is an important mechanism of DNA damage in pancreatic carcinogenesis[22]. Activating mutations in KRAS were identified in nearly all patients in this study and the prevalence of inactivation events for the other genes was 74% for TP53, 35% for CDKN2A and 31% for SMAD4. This study also identified inactivating mutations in two new genes KDM6A and PREX2, occurring in 18% and 10% of patients[22].
The reported mutation rate of KRAS in PDAC ranges from 75%-95%, making it the most commonly mutated gene in PDAC[23,24]. Although KRAS mutation testing appears to be an attractive diagnostic target, commercially available plasma assays lack specificity and are relatively insensitive in the detection of early PDAC[25]. KRAS mutations are known to be present with greater frequency in smokers and in patients with chronic pancreatitis both of which are risk factors for PDAC and further confound the diagnostic utility of this marker[24]. A recent meta-analysis of 8 prospective studies assessing the accuracy of KRAS gene mutation analysis for diagnosing of PDAC reported a pooled sensitivity and specificity of 88.7% and 92% for KRAS mutation analysis combined with cytology of an endoscopic ultrasound (EUS)-guided fine needle aspirate (FNA) compared to 80.6% and 97% for EUS-FNA alone. The authors concluded that there may be a role of using KRAS as a diagnostic marker in cases where the cytology is inconclusive[26]. Recent interest has focused on quantification of KRAS mutants in blood of patients with PDAC as a marker of early diagnosis[27,28]. Kinugasa et al[28] recently demonstrated that the ratio of mutant to wild type KRAS could be used as a biomarker in early PDAC.
Although currently available data clearly demonstrate that genetic mutations and alterations in gene expression unequivocally contribute to pancreatic carcinogenesis, none of these genomic markers have demonstrated favorable performance characteristics as a diagnostic biomarker in clinical application. Furthermore, the numerous mutational sites across involved genes render impractical their use as clinical biomarkers with current assay platforms. The evolution of high-speed sequencing platforms could overcome this limitation in the future.
Another challenge of detecting tumor DNA in blood pertains to their very low circulating concentrations at the earliest stages of cancer. Also, unlike RNA and various protein markers, there is typically only one copy number of a given DNA marker per tumor cell. Accordingly, assay platforms with exquisitely high analytical sensitivity are required to render this approach feasible. The availability of digital PCR, next-generation sequencing, and other innovative platforms in recent years may provide the requisite performance to detect very low levels of circulating mutant DNA[29]. These results are preliminary and need to be validated in larger studies.
Methylation - Several studies have focused on the methylation status of PDAC and cystic pancreatic neoplasms. Genes that have been identified to undergo aberrant methylation during pancreatic carcinogenesis include p16, ppENK, cyclin D2, SPARC/osteonectin SOCS-1 and TSLC1[30-35]. Genome-wide study of DNA methylation patterns in 167 resected untreated PDACs demonstrated enrichment of various aberrantly methylated genes, some of which appear to be highly discriminant for PDAC as single markers[36]. Our group has also identified novel methylated DNA marker candidates via whole methylome interrogation and has demonstrated the potential utility of methylation markers in site-prediction of gastrointestinal malignancies[37]. The use of these epigenetic alterations as early detection markers for PDAC is encouraging, and rigorous assessment of their application to distant media is now needed.
RNA: Micro-RNAs are non-coding RNAs that regulate posttranscriptional gene expression. The increasingly recognized role of micro-RNAs in oncogenesis and tumor metastasis has been described[38,39]. MicroRNA profiles specific for PDAC have been found in serum, pancreatic tissue, cyst fluid and more recently in whole blood[40-43].
Some of the earlier studies focused on serum and plasma microRNA profiles to distinguish patients with PDAC from healthy volunteers. Li et al[40] reported that serum miR-1290 levels appeared to have an excellent discriminatory ability (AUC of 0.96) to distinguish patients with early pancreatic cancer cases from healthy controls. Corroboration and extension of these early observations are needed. Several other investigators proposed the role of other serum microRNAs but most of these early studies lacked independent validation. The Danish BIOPAC (Biomarkers in Patients with Pancreatic Cancer) study analyzed microRNA expression in whole blood in 409 patients with pancreatic cancer. This prospective case-control study identified 2 miRNA panels consisting of sets of 4 (miR-145, miR-150, miR-223, miR-636) and 10 mi-RNAs (miR-26b, miR-34a, miR-122, miR-126*, miR-145, miR-150, miR-223, miR-505, miR-636, miR-885.5p) that distinguished patients with PDAC from healthy controls[41]. The larger panel had a comparatively higher AUC for detection of early pancreatic cancer (AUC of 0.91 vs 0.80). Although currently available studies have focused on identifying free miRNA in plasma or serum, there is growing interest in exploring circulating exosomes as a concentrated and potentially more discriminant source of miRNA in patients with PDAC[44,45].
As is evident from the above data, a large number of mi-RNAs have been implicated as potential biomarkers. The widespread availability of next generation sequencing techniques has facilitated rapid advancement in this field of research. Next steps must involve validation of some of the more promising markers in larger cohorts, especially focusing on discriminating between low-grade and high-grade dysplasia.
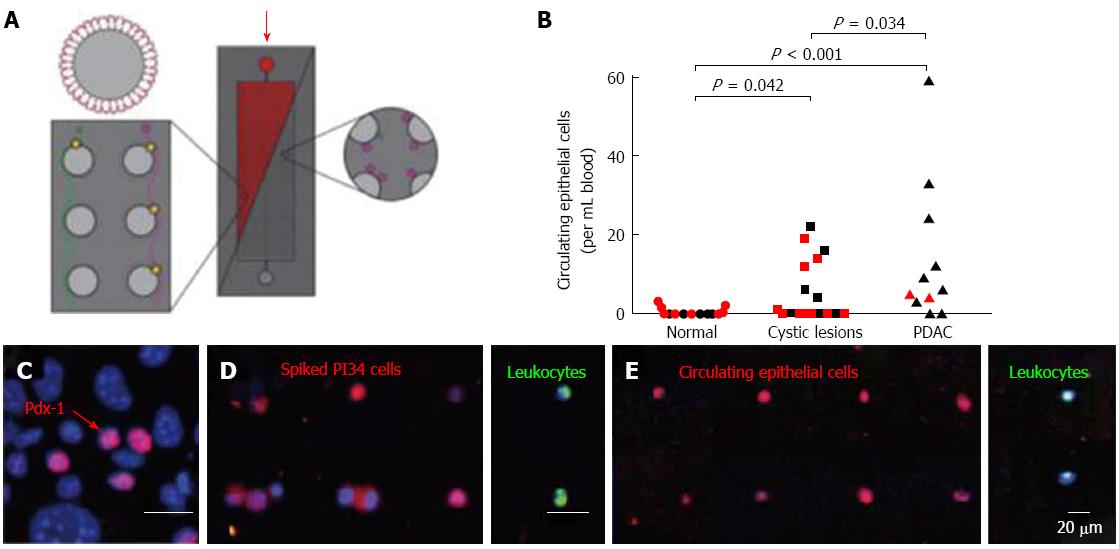
Circulating tumor cells: Recent data suggest that hematogenous spread of pancreatic tumor cells may be an early event during pancreatic oncogenesis and circulating tumor cells may be detectable before the primary tumor can be visualized on any imaging test. In mouse models, Rhim and colleagues demonstrated that an epithelial to mesenchymal transition of PDAC tumor cells occurs as an early phenomenon, even before histologic emergence of cancer[46]. Additionally, they used cell lineage labelling techniques in transgenic mice with PDAC for a proof of concept study to show that pancreatic epithelial cells may enter the circulation prior to tumor formation, and followed that by observing the same phenomenon in a subset of patients with cystic pancreatic lesions (Figure 1)[29,46]. Circulating epithelial cells were also detected in 8 of 11 (73%) patients with PDAC, but in 0 of 19 control patients without cysts or cancer. These results are preliminary and need to be validated in larger studies. Other studies have explored the possibility of using circulating tumor cell detection as a marker of predicting prognosis after curative surgical resection of the primary tumor. Based on preliminary data circulating tumor cell testing appears to be more sensitive (75% vs 69%) and specific (95% vs 81%) than CA 19-9 for diagnosing recurrent disease[47].
Advances in endoscopic ultrasound have provided a relatively safe modality for tissue acquisition from malignant and premalignant pancreatic lesions. Several investigators have studied fine-needle aspirate and cyst fluid specimens for biomarker detection.
The multicenter PANDA (Pancreatic Cyst DNA Analysis) study found the combined presence of a KRAS mutation and allelic loss in cyst fluid to have a sensitivity of only 37% at a specificity of 96% for discriminating malignant from benign pancreatic cysts[48]. In another study by Wu et al[49], cyst fluid GNAS mutations were detected exclusively in IPMNs and not in any other mucinous, non-mucinous or malignant pancreatic cysts. Hong and associates identified hypermethylation of several genes including BNIP3, PTCHD2, SOX17, NXPH1 and EBF3 in cyst fluid from IPMNs with high-grade dysplasia[50]. In a study comparing the miRNome of low-grade and high-grade pancreatic cystic neoplasms, thirteen miRNAs (miR-138, miR-195, miR-204, miR-216a, miR-217, miR-218, miR-802, miR-155, miR-214, miR-26a, miR-30b, miR-31, and miR-125) were differentially enriched in cyst fluid samples from patients with PDAC[42]. Matthaei and colleagues identified a panel of 9 mi-RNAs in cyst fluid and microdissected formalin-fixed, paraffin-embedded (FFPE) tissue from surgically resected IPMN specimens, which distinguished potentially malignant cysts requiring surgical intervention from benign cysts that could be managed conservatively with a sensitivity of 89%, a specificity of 100%, and AUC of 1[43]. Such high discrimination in this early report clearly warrants further exploration.
Mass spectrometric analysis of whole tumor tissue has identified several proteins associated with pancreatic oncogenesis. Gelsolin and lumican are two such proteins initially identified in cancer tissue that were found to have 80% sensitivity and 95% specificity as a plasma biomarker in discriminating early stage pancreatic cancer patients and healthy controls[51]. A quantitative proteomic study of PanIN 3 lesions identified more than 200 dysregulated proteins, the majority being cytoskeleton proteins involved in cell motility. Galectin-1 and laminin were overexpressed both in these advanced PanIN lesions and the adjacent pancreatic stroma[52]. Amato et al[53] used next-generation sequencing to assess mutations in 48 surgically resected IPMNs and identified GNAS (79%) and KRAS (50%) to be most commonly affected genes. These studies using surgical specimens are susceptible to selection bias and the detection of these genetic mutations is not ideally suited to be applied to the diagnostic algorithm of cysts that do not meet currently established criteria for surgical resection.
Our understanding of the progression model of pancreatic cancer has improved significantly over the past decade. The term pancreatic intraepithelial neoplasia (PanIN) was introduced in 1999 to describe ductal lesions that are precursors of invasive cancer[54]. Although high-grade ductal precursor lesions (PanIN-3) have a greater malignant potential, low-grade PanINs (PanIN-1 and PanIN-2) are more common; especially in older adults and do not always progress to cancer. Thus, biomarkers targeting PanIN-3 would be of greatest clinical utility for early detection. The overexpression of HER-2/neu and point mutations in the K-ras gene appear to be early events that discriminate low-grade ductal premalignant lesions (PanIN-1) from normal ductal epithelial cells[54]. Inactivation of the p16 gene appears to be more common in higher-grade PanINs compared to low-grade PanINs and probably occurs at an intermediate stage of tumorigenesis[54]. Inactivation of p53, DPC4, and BRCA2 are relatively terminal events in this neoplastic progression[54]. Gene expression profiling analyses of cells from normal pancreatic ducts, PanINs and PDAC have revealed more than 1,000 molecules including S100P, MMP7, MUC4, FSCN1, and MUC5AC that are preferentially expressed both in PDAC and precursor lesions[55]. More work is needed both at the discovery level and with subsequent validations to make meaningful progress in identification of useful markers for PanIN-3.
Recent studies have focused on analysis of pancreatic juice sampled endoscopically from the duodenum following intravenous administration of secretin to stimulate pancreatic excretion and facilitate juice collection. In one study KRAS mutations were detected in pancreatic juice of 73% of patients with PDAC[56]. However, KRAS mutations were detected in pancreatic juice from 19% of controls and attributed to the presence of small pancreatic intraepithelial neoplasia (PanIN) lesions[56]. Detection of mutant TP53 has been studied in pancreatic juice from patients with both cancer and precancerous lesions in an enriched patient population. Although the sensitivity of this test was only 67.1% in patients with invasive PDAC, TP53 mutations were not identified in any of the 58 controls or in patients with PanIN-1 lesions[57]. In our recent early study using non-optimized techniques on archival pancreatic juice samples, we identified a panel of novel methylated DNA markers and mutant KRAS in patients with PDAC. At specificity cutoffs of 90% and 95%: this combined marker panel achieved sensitivities of 88% and 77% for diagnosis of PDAC; ADCY1, was the most sensitive single methylation marker. Overall discrimination between PDAC and controls by area under ROC curve was 0.91 for the panel which was significantly higher than by any single marker (P < 0.05), except ADCY1. Positivity rates were substantially lower in patients with chronic pancreatitis compared to those with PDAC for all markers (P < 0.0001)[58]. With optimization of marker selection, sample processing, and assay techniques, analysis of pancreatic juice has potential to characterize indeterminate pancreatic lesions with high accuracy.
Although considerable attention has been devoted to discovery of biomarkers in pancreatic juice and cyst fluid, the acquisition of these specimens are dependent on invasive procedures and would not be suitable for general screening. In a recent study we explored the possibility of detecting DNA markers in stool as an approach to the early detection of PDAC[59]. Nine target genes were assayed comparing stool samples from patients with PDAC compared to controls with normal colons. BMP3 was the most discriminant methylation marker in stool. At 90% specificity, the combination of methylated BMP3 and mutant KRAS in stool detected 67% of PDACs. AUC for the combination in stool was 0.85, which was better than the AUC for either test in isolation. Further studies are necessary to improve the discriminatory accuracy of stool methylation markers in patients with early PDAC.
Several additional approaches have recently surfaced which offer creative strategies to early detection of this lethal cancer.
Exosomes are extracellular vesicles containing proteins and nucleic acids surrounded by a lipid-bilayer wall. Isolation of specific cancer-cell derived exosomes has been studied as a tool for early diagnosis. In a recently published study, circulating cancer-cell derived exosomes enriched in glypican-1 were identified as a marker for diagnosis of PDAC with reported 100% specificity and 100% sensitivity (AUC of 1.0) for detecting PDAC[60].
The pancreas has a major role in regulation of metabolism in healthy individuals. It follows intuitively that metabolomic profiling of individuals affected by PDAC could reveal diagnostic clues. A study using serum assay in a p48-Cre/LSL-KrasG12D mouse model identified a distinct metabolic pattern that distinguished animals with early stage PDAC from wild-type controls with an accuracy of 82%[61]. Metabolomic markers will require further research to determine their potential value in the early detection of PDAC.
Core fucosylation (CF) is a form of glycosylation mediating post-translational modification of cellular protein. CF-glycosylation of an isoform of α-fetoprotein (AFP-L3) is a sensitive biomarker of hepatocellular carcinoma (HCC). Tan et al[62] recently published an elegant study describing differentially expressed CF peptides distinguishing between serum of healthy control and patients with pancreatic cancer. Research in this field is at a nascent stage and carries great future promise. Another approach that has recently emerged as an exciting new avenue for biomarker discovery in PDAC is the detection of discriminatory cytokine biomarker panels[63].
These and other novel molecular approaches on the discovery horizon warrant further study in the search for better methods of to improve early detection of PDAC.
The number of promising biomarkers for early diagnosis of pancreatic cancer has risen dramatically in recent years. However despite the large number of studies and new discoveries, no molecular approach has been rigorously validated or ascended to application for routine clinical use to date. Mutational analysis points to the heterogeneity in acquired genetic changes in PDAC and underscores the complexity and obstacles inherent to the use of mutations as candidate markers for detection. Other classes of markers, such as aberrantly methylated DNA or miRNA, may be more informative and practical at this time. Carefully designed clinical validation studies are now needed to sort out which molecular markers measured, which assay platforms, and in which biological media will prove to be of greatest value in screening and surveillance applications.
P- Reviewer: Jorg K, Zoller M S- Editor: Yu J L- Editor: A E- Editor: Zhang DN
| 1. | Available from: http://seer.cancer.gov/statfacts/html/pancreas.html. |
| 2. | Yeo TP. Demographics, epidemiology, and inheritance of pancreatic ductal adenocarcinoma. Semin Oncol. 2015;42:8-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 3. | Smith BD, Smith GL, Hurria A, Hortobagyi GN, Buchholz TA. Future of cancer incidence in the United States: burdens upon an aging, changing nation. J Clin Oncol. 2009;27:2758-2765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1195] [Cited by in RCA: 1314] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 4. | Eheman C, Henley SJ, Ballard-Barbash R, Jacobs EJ, Schymura MJ, Noone AM, Pan L, Anderson RN, Fulton JE, Kohler BA. Annual Report to the Nation on the status of cancer, 1975-2008, featuring cancers associated with excess weight and lack of sufficient physical activity. Cancer. 2012;118:2338-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 345] [Cited by in RCA: 333] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 5. | Sohn TA, Yeo CJ, Cameron JL, Koniaris L, Kaushal S, Abrams RA, Sauter PK, Coleman J, Hruban RH, Lillemoe KD. Resected adenocarcinoma of the pancreas-616 patients: results, outcomes, and prognostic indicators. J Gastrointest Surg. 2000;4:567-579. [PubMed] |
| 6. | Okano K, Suzuki Y. Strategies for early detection of resectable pancreatic cancer. World J Gastroenterol. 2014;20:11230-11240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 32] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 7. | Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114-1117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2041] [Cited by in RCA: 1936] [Article Influence: 129.1] [Reference Citation Analysis (0)] |
| 8. | Chari ST, Kelly K, Hollingsworth MA, Thayer SP, Ahlquist DA, Andersen DK, Batra SK, Brentnall TA, Canto M, Cleeter DF. Early detection of sporadic pancreatic cancer: summative review. Pancreas. 2015;44:693-712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 221] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 9. | Correa-Gallego C, Ferrone CR, Thayer SP, Wargo JA, Warshaw AL, Fernández-Del Castillo C. Incidental pancreatic cysts: do we really know what we are watching? Pancreatology. 2010;10:144-150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 169] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 10. | Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 351] [Reference Citation Analysis (3)] |
| 11. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1110] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 12. | Berger AC, Garcia M, Hoffman JP, Regine WF, Abrams RA, Safran H, Konski A, Benson AB, MacDonald J, Willett CG. Postresection CA 19-9 predicts overall survival in patients with pancreatic cancer treated with adjuvant chemoradiation: a prospective validation by RTOG 9704. J Clin Oncol. 2008;26:5918-5922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 273] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 13. | Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol. 2007;33:266-270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 539] [Cited by in RCA: 609] [Article Influence: 32.1] [Reference Citation Analysis (1)] |
| 14. | Duraker N, Hot S, Polat Y, Höbek A, Gençler N, Urhan N. CEA, CA 19-9, and CA 125 in the differential diagnosis of benign and malignant pancreatic diseases with or without jaundice. J Surg Oncol. 2007;95:142-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 15. | Pan S, Brentnall TA, Kelly K, Chen R. Tissue proteomics in pancreatic cancer study: discovery, emerging technologies, and challenges. Proteomics. 2013;13:710-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Grønborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 152] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Brand RE, Nolen BM, Zeh HJ, Allen PJ, Eloubeidi MA, Goldberg M, Elton E, Arnoletti JP, Christein JD, Vickers SM. Serum biomarker panels for the detection of pancreatic cancer. Clin Cancer Res. 2011;17:805-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 18. | Jenkinson C, Elliott V, Menon U, Apostolidou S, Fourkala OE, Gentry-Maharaj A, Pereira SP, Jacobs I, Cox TF, Greenhalf W. Evaluation in pre-diagnosis samples discounts ICAM-1 and TIMP-1 as biomarkers for earlier diagnosis of pancreatic cancer. J Proteomics. 2015;113:400-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Wang X, Li Y, Tian H, Qi J, Li M, Fu C, Wu F, Wang Y, Cheng D, Zhao W. Macrophage inhibitory cytokine 1 (MIC-1/GDF15) as a novel diagnostic serum biomarker in pancreatic ductal adenocarcinoma. BMC Cancer. 2014;14:578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 20. | Hu H, Zhang Q, Huang C, Shen Y, Chen X, Shi X, Tang W. Diagnostic value of S100P for pancreatic cancer: a meta-analysis. Tumour Biol. 2014;35:9479-9485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 21. | Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801-1806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3216] [Cited by in RCA: 3023] [Article Influence: 177.8] [Reference Citation Analysis (0)] |
| 22. | Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2130] [Cited by in RCA: 1982] [Article Influence: 198.2] [Reference Citation Analysis (1)] |
| 23. | Koorstra JB, Hustinx SR, Offerhaus GJ, Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8:110-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 24. | Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549-554. [PubMed] |
| 25. | Däbritz J, Preston R, Hänfler J, Oettle H. Follow-up study of K-ras mutations in the plasma of patients with pancreatic cancer: correlation with clinical features and carbohydrate antigen 19-9. Pancreas. 2009;38:534-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Fuccio L, Hassan C, Laterza L, Correale L, Pagano N, Bocus P, Fabbri C, Maimone A, Cennamo V, Repici A. The role of K-ras gene mutation analysis in EUS-guided FNA cytology specimens for the differential diagnosis of pancreatic solid masses: a meta-analysis of prospective studies. Gastrointest Endosc. 2013;78:596-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, Parker AR, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. LigAmp for sensitive detection of single-nucleotide differences. Nat Methods. 2004;1:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Kinugasa H, Nouso K, Miyahara K, Morimoto Y, Dohi C, Tsutsumi K, Kato H, Matsubara T, Okada H1, Yamamoto K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 181] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 29. | Rhim AD, Thege FI, Santana SM, Lannin TB, Saha TN, Tsai S, Maggs LR, Kochman ML, Ginsberg GG, Lieb JG. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology. 2014;146:647-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 182] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 30. | Fukushima N, Walter KM, Uek T, Sato N, Matsubayashi H, Cameron JL, Hruban RH, Canto M, Yeo CJ, Goggins M. Diagnosing pancreatic cancer using methylation specific PCR analysis of pancreatic juice. Cancer Biol Ther. 2003;2:78-83. [PubMed] |
| 31. | Ueki T, Walter KM, Skinner H, Jaffee E, Hruban RH, Goggins M. Aberrant CpG island methylation in cancer cell lines arises in the primary cancers from which they were derived. Oncogene. 2002;21:2114-2117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Matsubayashi H, Sato N, Fukushima N, Yeo CJ, Walter KM, Brune K, Sahin F, Hruban RH, Goggins M. Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin Cancer Res. 2003;9:1446-1452. [PubMed] |
| 33. | Fukushima N, Sato N, Sahin F, Su GH, Hruban RH, Goggins M. Aberrant methylation of suppressor of cytokine signalling-1 (SOCS-1) gene in pancreatic ductal neoplasms. Br J Cancer. 2003;89:338-343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 100] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 34. | Sato N, Fukushima N, Maehara N, Matsubayashi H, Koopmann J, Su GH, Hruban RH, Goggins M. SPARC/osteonectin is a frequent target for aberrant methylation in pancreatic adenocarcinoma and a mediator of tumor-stromal interactions. Oncogene. 2003;22:5021-5030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 221] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 35. | Jansen M, Fukushima N, Rosty C, Walter K, Altink R, Heek TV, Hruban R, Offerhaus JG, Goggins M. Aberrant methylation of the 5’ CpG island of TSLC1 is common in pancreatic ductal adenocarcinoma and is first manifest in high-grade PanlNs. Cancer Biol Ther. 2002;1:293-296. [PubMed] |
| 36. | Nones K, Waddell N, Song S, Patch AM, Miller D, Johns A, Wu J, Kassahn KS, Wood D, Bailey P. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer. 2014;135:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 177] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 37. | Kisiel JB, Yab TC, Ghoz HM, Foote PH, Devens ME, Mahoney DW, Smyrk TC, Boardman LA, Petersen GM, Buttar NS, Roberts LR, Lidgard GP, Ahlquist DA. Accurate site prediction of gastrointestinal cancer by novel methylated DNA markers: Discovery & Validation. Proceedings of the American Association for Cancer Research; 2015 April 18-22; Philadelphia, PA. AACR; Cancer Res 2015; 75 (15 Suppl): Abstract nr 4252. . |
| 38. | Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848-5856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 787] [Article Influence: 49.2] [Reference Citation Analysis (0)] |
| 39. | Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 723] [Cited by in RCA: 768] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 40. | Li A, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 41. | Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Holländer NH. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 343] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 42. | Wang J, Paris PL, Chen J, Ngo V, Yao H, Frazier ML, Killary AM, Liu CG, Liang H, Mathy C. Next generation sequencing of pancreatic cyst fluid microRNAs from low grade-benign and high grade-invasive lesions. Cancer Lett. 2015;356:404-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 43. | Matthaei H, Wylie D, Lloyd MB, Dal Molin M, Kemppainen J, Mayo SC, Wolfgang CL, Schulick RD, Langfield L, Andruss BF. miRNA biomarkers in cyst fluid augment the diagnosis and management of pancreatic cysts. Clin Cancer Res. 2012;18:4713-4724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 44. | Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol. 2013;11:219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 218] [Article Influence: 18.2] [Reference Citation Analysis (1)] |
| 45. | Zöller M. Pancreatic cancer diagnosis by free and exosomal miRNA. World J Gastrointest Pathophysiol. 2013;4:74-90. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 59] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (2)] |
| 46. | Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1400] [Cited by in RCA: 1663] [Article Influence: 127.9] [Reference Citation Analysis (0)] |
| 47. | Sato N, Ueki T, Fukushima N, Iacobuzio-Donahue CA, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Aberrant methylation of CpG islands in intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology. 2002;123:365-372. [PubMed] |
| 48. | Khalid A, Zahid M, Finkelstein SD, LeBlanc JK, Kaushik N, Ahmad N, Brugge WR, Edmundowicz SA, Hawes RH, McGrath KM. Pancreatic cyst fluid DNA analysis in evaluating pancreatic cysts: a report of the PANDA study. Gastrointest Endosc. 2009;69:1095-1102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 314] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 49. | Wu J, Matthaei H, Maitra A, Dal Molin M, Wood LD, Eshleman JR, Goggins M, Canto MI, Schulick RD, Edil BH. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 594] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 50. | Hong SM, Omura N, Vincent A, Li A, Knight S, Yu J, Hruban RH, Goggins M. Genome-wide CpG island profiling of intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res. 2012;18:700-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 51. | Pan S, Chen R, Brand RE, Hawley S, Tamura Y, Gafken PR, Milless BP, Goodlett DR, Rush J, Brentnall TA. Multiplex targeted proteomic assay for biomarker detection in plasma: a pancreatic cancer biomarker case study. J Proteome Res. 2012;11:1937-1948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 52. | Pan S, Chen R, Reimel BA, Crispin DA, Mirzaei H, Cooke K, Coleman JF, Lane Z, Bronner MP, Goodlett DR. Quantitative proteomics investigation of pancreatic intraepithelial neoplasia. Electrophoresis. 2009;30:1132-1144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 53. | Amato E, Molin MD, Mafficini A, Yu J, Malleo G, Rusev B, Fassan M, Antonello D, Sadakari Y, Castelli P. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol. 2014;233:217-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 54. | Hruban RH, Goggins M, Parsons J, Kern SE. Progression model for pancreatic cancer. Clin Cancer Res. 2000;6:2969-2972. [PubMed] |
| 55. | Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LD. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 239] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 56. | Eshleman JR, Norris AL, Sadakari Y, Debeljak M, Borges M, Harrington C, Lin E, Brant A, Barkley T, Almario JA. KRAS and guanine nucleotide-binding protein mutations in pancreatic juice collected from the duodenum of patients at high risk for neoplasia undergoing endoscopic ultrasound. Clin Gastroenterol Hepatol. 2015;13:963-9.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 57. | Kanda M, Sadakari Y, Borges M, Topazian M, Farrell J, Syngal S, Lee J, Kamel I, Lennon AM, Knight S. Mutant TP53 in duodenal samples of pancreatic juice from patients with pancreatic cancer or high-grade dysplasia. Clin Gastroenterol Hepatol. 2013;11:719-30.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 133] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 58. | Kisiel JB, Raimondo M, Taylor WR, Yab TC, Mahoney DW, Sun Z, Middha S, Baheti S, Zou H, Smyrk TC. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clin Cancer Res. 2015;21:4473-4481. [PubMed] |
| 59. | Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, Ahlquist DA. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer. 2012;118:2623-2631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 60. | Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2108] [Cited by in RCA: 2186] [Article Influence: 218.6] [Reference Citation Analysis (0)] |
| 61. | LaConti JJ, Laiakis EC, Mays AD, Peran I, Kim SE, Shay JW, Riegel AT, Fornace AJ, Wellstein A. Distinct serum metabolomics profiles associated with malignant progression in the KrasG12D mouse model of pancreatic ductal adenocarcinoma. BMC Genomics. 2015;16 Suppl 1:S1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 62. | Tan Z, Yin H, Nie S, Lin Z, Zhu J, Ruffin MT, Anderson MA, Simeone DM, Lubman DM. Large-scale identification of core-fucosylated glycopeptide sites in pancreatic cancer serum using mass spectrometry. J Proteome Res. 2015;14:1968-1978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 63. | Shaw VE, Lane B, Jenkinson C, Cox T, Greenhalf W, Halloran CM, Tang J, Sutton R, Neoptolemos JP, Costello E. Serum cytokine biomarker panels for discriminating pancreatic cancer from benign pancreatic disease. Mol Cancer. 2014;13:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |