Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.926
Peer-review started: June 8, 2014
First decision: June 27, 2014
Revised: July 30, 2014
Accepted: September 18, 2014
Article in press: September 19, 2014
Published online: January 21, 2015
Processing time: 227 Days and 1 Hours
AIM: To determine the prognostic significance of deficient mismatch repair (dMMR) and BRAF V600E in Thai sporadic colorectal cancer (CRC) patients.
METHODS: We studied a total of 211 out of 405 specimens obtained from newly diagnosed CRC patients between October 1, 2006 and December 31, 2007 at Siriraj Hospital, Mahidol University. Formalin-fixed paraffin-embedded blocks of CRC tissue samples were analyzed for dMMR by detection of MMR protein expression loss by immunohistochemistry or microsatellite instability using polymerase chain reaction (PCR)-DHPLC. BRAF V600E mutational analysis was performed in DNA extracted from the same archival tissues by two-round allele-specific PCR and analyzed by high sensitivity DHPLC. Associations between patient characteristics, MMR and BRAF status with disease-free survival (DFS) and overall survival (OS) were determined by Kaplan-Meier survival plots and log-rank test together with Cox’s proportional hazard regression.
RESULTS: dMMR and BRAF V600E mutations were identified in 31 of 208 (14.9%) and 23 of 211 (10.9%) tumors, respectively. dMMR was more commonly found in patients with primary colon tumors rather than rectal cancer (20.4% vs 7.6%, P =0.01), but there was no difference in MMR status between the right-sided and left-sided colon tumors (20.8% vs 34.6%, P = 0.24). dMMR was associated with early-stage rather than metastatic disease (17.3% vs 0%, P = 0.015). No clinicopathological features such primary site or tumor differentiation were associated with the BRAF mutation. Six of 31 (19.3%) samples with dMMR carried the BRAF mutation, while 17 of 177 (9.6%) with proficient MMR (pMMR) harbored the mutation (P = 0.11). Notably, patients with dMMR tumors had significantly superior DFS (HR = 0.30, 95%CI: 0.15-0.77; P = 0.01) and OS (HR = 0.29, 95%CI: 0.10-0.84; P = 0.02) compared with patients with pMMR tumors. By contrast, the BRAF V600E mutation had no prognostic impact on DFS and OS.
CONCLUSION: The prevalence of dMMR and BRAF V600E in Thai sporadic CRC patients was 15% and 11%, respectively. The dMMR phenotype was associated with a favorable outcome.
Core tip: This study is the first report of prevalence and outcome in sporadic colorectal cancer that habour deficient mismatch repair (dMMR) and BRAF gene mutation in Thai population. The prevalence of dMMR and BRAF V600E mutation was 15% and 11%, respectively. This study confirmed the favorable outcome in patients with dMMR tumors, which is consistent to the results of previous reports in Caucasian population. The method we used to detect BRAF mutation is allele specific polymerase chain reaction which has the highest sensitivity to detect this mutation when compared to previously reported methods.
- Citation: Korphaisarn K, Pongpaibul A, Limwongse C, Roothumnong E, Klaisuban W, Nimmannit A, Jinawath A, Akewanlop C. Deficient DNA mismatch repair is associated with favorable prognosis in Thai patients with sporadic colorectal cancer. World J Gastroenterol 2015; 21(3): 926-934
- URL: https://www.wjgnet.com/1007-9327/full/v21/i3/926.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.926
Colorectal tumorigenesis is a multistep process that arises from the accumulation of genetic alterations, including chromosomal abnormalities, gene mutations, and epigenetic changes[1]. With regards to genetic abnormalities, defective DNA mismatch repair (dMMR) is a type of genomic instability in tumor tissue caused by a failure to correct errors during normal DNA replication[2]. dMMR can be identified either by the presence of microsatellite instability (MSI) analyzed by polymerase chain reaction (PCR) amplification of microsatellite foci in tumor tissue, or lack of protein expression for any of the MMR genes, MLH1, MSH2, MSH6, and PMS2, detected by immunohistochemistry (IHC). Tumors with dMMR have been reported in 15%-20% of sporadic colorectal cancers (CRC), and dMMR is associated with distinct clinicopathological features such as proximal tumor site, high grade, early stage, and better prognosis[3].
The BRAF gene has 18 exons and encodes a serine/threonine protein kinase belonging to the RAS-RAF-MEK-ERK kinase pathway that is involved in CRC development[4,5]. The most common activating mutation is found in exon 15 at nucleotide position 1799, whereby a thymine (T) to adenine (A) transversion within codon 600 leads to substitution of valine by glutamate at the amino acid level. This leads to the oncogenic BRAF V600E mutation[6]. KRAS and BRAF mutations have been reported to be mutually exclusive events within tumors[6,7]. Several studies reported BRAF mutations in 5%-20% of patients with sporadic CRC, with a high frequency in dMMR tumors[4,8-10]. However, BRAF mutations are very rare in CRC patients with hereditary nonpolyposis colorectal cancer (HNPCC)[11].
Recently, the correlation between dMMR and BRAF mutation and CRC prognosis has been widely studied[8,12,13]. Patients with tumors harboring dMMR were associated with a more favorable survival than those with proficient MMR (pMMR)[3,14,15]. By contrast, patients with the BRAF mutation were associated with a worse clinical outcome, especially patients with pMMR tumors[12,13,16].
These data, however, all pertain to a Caucasian population, and there is only scarce information available for Asian populations. In this study, we systematically determined the prevalence of dMMR and BRAF mutations in Thai patients with sporadic CRC and established correlations with various clinicopathological features to determine their prognostic impact on clinical outcome.
Formalin-fixed paraffin-embedded tissue blocks from patients diagnosed with primary colon or rectal adenocarcinoma who underwent surgery between October 1, 2006 and December 31, 2007 were obtained for this study. We excluded patients with a known family history of CRC, those suspected to have hereditary or familial CRC, and those who did not receive treatment and follow-up at our institution. The study protocol was approved by the Siriraj Institutional Review Board, Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand. This study was supported by the Siriraj Research Development Fund.
Demographic information regarding age, gender, primary tumor site, date of diagnosis, date of surgery, stage at diagnosis, date of disease recurrence, date of last follow-up, and date of death were collected. Staging was classified by AJCC/UICC TMN stage (v.3 2010). Disease-free survival (DFS) is defined as the interval between the date of diagnosis and the date of disease recurrence or death, while overall survival (OS) is the interval between the date of diagnosis and the date of death from any cause. The primary objective of this study was to determine the prevalence of dMMR and BRAF V600E mutations in sporadic CRC patients; the secondary objectives were to examine correlations between MMR status and BRAF mutations, and the association of each marker with various clinicopathological characteristics and their prognostic impact on DFS and OS.
MMR status was determined by analysis of MMR protein expression by IHC or MSI testing. dMMR was defined by the presence of either high-level MSI (MSI-H) or loss of MMR protein expression. pMMR was defined by the presence of either microsatellite stable (MSS)/low-level MSI (MSI-L) or the presence of normal MMR protein expression.
IHC for four MMR proteins, MLH1, MSH2, MSH6 and PMS2, was performed on tissue microarray slides (TMAs). TMAs were assembled from paraffin-embedded tissues using a manual tissue microarrayer (UNITMA Quick-Ray 2 mm-diameter tissue cores). Hematoxylin and eosin stained slides were prepared from paraffin blocks and areas of neoplastic tissue were identified by a gastrointestinal pathologist (A.P.) who selected samples for TMA construction. Duplicated IHC for each MMR protein was performed for each patient sample. Staining for MMR proteins was performed using the following primary antibodies: mouse anti-human MLH-1 (clone G168-728; Cell Marque Corporation, Rocklin, CA), mouse anti-human MSH-2 (clone G219-1129; Cell Marque Corporation, Rocklin, CA), mouse anti-human MSH-6 (clone BC/44; Biocare, Concord, CA), and rabbit anti-human PMS2 (clone EPR3947; Cell Marque Corporation, Rocklin, CA). Loss of MMR protein was defined as the absence of nuclear staining of tumor cells in the presence of positive nuclear staining in normal epithelial cells and lymphocytes. Assessment of IHC staining was performed by the same pathologist (A.P.).
MSI was analyzed by PCR amplification of microsatellite foci from the five-marker Bethesda panel, which includes two mononucleotide (BAT-25 and BAT-26) and three dinucleotide (D5S346, D2S123, and D17S250) repeats. Samples with instability in two or more of these markers were defined as MSI-H, whereas those with one unstable marker were designated as MSI-L. Samples with no detectable alterations were defined as MSS.
Formalin-fixed paraffin-embedded tissues with tumorous regions were macroscopically dissected into 10 μm-thick sections using microtome blades, and then placed into separate tubes for DNA extraction using a standard phenol/chloroform extraction protocol. Probes were designed to target the most common BRAF mutation, a valine to glutamate transition at amino acid position 600 (V600E). BRAF V600E was detected using a previously-described two-round allele specific-polymerase chain reaction (AS-PCR)[17]. The primary AS-PCR reaction was performed using common-primer pairs (CF; 5′-TAATGCTTGCTCTGATAGGA-3′, CR; 5′ GGAAAAATAGCCTCAATTCT-3′) and a BRAF V600E-specific primer (Mt; 5′-AAATAGGTGATTTTGGTCTGGCTACGGA-3′), which is located between common-primer pairs, and CR was used as a reverse primer. The final concentration of AS-PCR reactions comprised 100 ng genomic DNA, 0.02 U/μL of Immolase DNA polymerase (Bioline, Taunton, MA), 1 × buffer, 1.5 mmol/L MgCl2, 0.2 μmol/L dNTP, 0.4 μmol/L of each common primer and 0.8 umol/L Mt primer. The reaction was amplified with a Mastercycler pro S (Eppendorf, Eppendorf AG, Hamburg, Germany) using the following protocol: activation at 95 °C for 10 min followed by 35 cycles at 95 °C for 30 s, 60 °C for 30s, 72 °C for 30 s and a final extension at 72 °C for 10 min. Secondary AS-PCR was amplified under the same conditions as the primary reaction, but genomic DNA template was replaced with 1 μL of the primary AS-PCR product. Secondary AS-PCR product was analyzed by high-sensitivity denaturing high performance liquid chromatography (HS-DHPLC) (Transgenomic Inc., Foster city, CA) equipped with WAVE Optimized HS Staining Solution I and a fluorescence detector in sizing mode. This assay has been shown to have a detection limit of at least 0.5% V600E-positive tumor cells.
Sample size determination was based on a BRAF mutation and dMMR estimated prevalence of 15% with a 95%CI of 5%: accounting for 5% of possible cases with no paraffin-embedded tumors tissue blocks, a total sample size of 210 patients was required.
Patient characteristics were described by descriptive statistics. Pearson’s χ2 test was applied to evaluate associations between BRAF/MMR status and clinicopathological variables. The association between patient characteristics and either BRAF status or MMR status with OS and DFS was explored by Kaplan-Meier estimation and log-rank test together with Cox’s proportional hazard regression. Calculations were carried out using SPSS-version 18 software. P values of less than 0.05 were considered statistically significant.
A total of 405 patients were diagnosed with colon and rectal adenocarcinoma between October 1, 2006 and December 31, 2007. We investigated 211 patients for whom tissue blocks were available. The median age was 63 years (33-95 years). The ratio of males to females was 1:1. One-hundred and fifty patients (71.1%) had stage II and III disease while 29 patients (13.7%) were diagnosed with stage IV disease. The majority of primary site tumors were rectal (91 patients, 43.1%), followed by left-sided colon tumors (73 patients, 34.6%). Patient and tumor characteristics are shown in Table 1.
| Variables | Value |
| No. of patients | 211 (100) |
| Median age (yr, range) | 63 (33-95) |
| Age (yr) | |
| ≤ 50 | 30 (14.2) |
| > 50 | 181 (85.8) |
| Sex | |
| Female | 105 (49.8) |
| Male | 106 (50.2) |
| Site | |
| Right-sided | 43 (20.8) |
| Left-sided | 73 (34.6) |
| Rectum | 91 (43.1) |
| Synchronous lesions | 4 (1.9) |
| Stage | |
| I | 32 (15.2) |
| II | 65 (30.8) |
| III | 85 (40.3) |
| IV | 29 (13.7) |
| Bowel wall invasion | |
| pT1 | 4 (1.9) |
| pT2 | 44 (20.8) |
| pT3 | 146 (69.2) |
| pT4 | 17 (8.1) |
| Lymph node metastasis | |
| pN0 | 105 (49.8) |
| pN1 | 60 (28.4) |
| pN2 | 46 (21.8) |
| Distant metastasis | |
| No | 182 (86.3) |
| Yes | 29 (13.7) |
| Invasion | |
| NO | 118 (55.9) |
| LVI | 47 (22.3) |
| PNI | 15 (7.1) |
| Both LVI/PNI | 31 (14.7) |
| Differentiation | |
| Well | 32 (15.2) |
| Moderately | 171 (81) |
| Poorly | 8 (3.8) |
Of the 211 patients, IHC for MMR proteins and MSI detection was analyzed in 164 and 47 tumors, respectively. dMMR was identified in 10 out of 164 tumors and 21 out of 44 tumors; therefore dMMR was detected in a total of 31 out of 208 tumors (14.9%). We were unable to analyze the results in three patients because of unamplified DNA by PCR. Of the 10 patients with dMMR, interpretation of the IHC staining was as follows: MLH-1 expression was absent in four tumors, while six tumors were negative for MSH-2 expression. Interpretation parameters for IHC of MMR status is shown in Table 2.
| MMR mutation | IHC staining | |||
| MLH-1 | MSH-2 | MSH-6 | PMS-2 | |
| MLH-1 | - | + | + | - |
| MSH-2 | + | - | - | + |
| MSH-6 | + | + | - | + |
| PMS-2 | + | + | + | - |
The BRAF V600E mutation was identified in 23 of 211 patients (10.9%). The prevalence of the BRAF mutation and dMMR with specific types of mutations is shown in Table 3.
| Variables | All cases |
| MMR status | n = 211 |
| IHC method | 164 (77.73) |
| pMMR | 154 |
| dMMR | 10 |
| MLH-1 | 4 |
| MSH-2 | 6 |
| MSH-6 | 0 |
| PMS-2 | 0 |
| MSI method | 44 (20.85) |
| MSI-H | 21 |
| MSI-L/MSI-S | 23 |
| Unknown | 3 (1.42) |
| BRAF status | |
| Wild type | 188 (89.1) |
| Mutation | 23 (10.9) |
The association of dMMR and BRAF mutations with clinicopathological factors was evaluated by Pearson’s χ2 test. These factors included gender, age, site of the primary tumor, UICC stage, lymphovascular or/and perineural invasion, histologic grade, pT, pN, and M stage. dMMR was more commonly found in patients with primary colon tumors rather than rectal cancer (20.4% vs 7.6%, P = 0.01) but no difference in MMR status was found between right-sided and left-sided colon tumors (P = 0.24). dMMR was associated with early stage rather than metastatic disease (17.3% vs 0%, P = 0.015). None of the abovementioned factors were found to be significantly associated with the BRAF mutation (Table 4).
| MMR status(n = 208) | BRAF status(n = 211) | |||||
| Variable | pMMR | dMMR | P value | Wild type | Mutant | P value |
| Gender | ||||||
| Female | 87 | 16 | 0.800 | 94 | 11 | 0.844 |
| Male | 90 | 15 | 94 | 12 | ||
| Age (yr) | ||||||
| ≤ 50 | 24 | 6 | 0.397 | 29 | 1 | 0.151 |
| > 50 | 153 | 25 | 159 | 22 | ||
| Site | ||||||
| Right-sided | 31 | 11 | 0.037a | 36 | 7 | 0.510 |
| Left-sided | 59 | 12 | 67 | 6 | ||
| Rectum | 84 | 7 | 81 | 10 | ||
| Synchronous lesions | 3 | 1 | 4 | 0 | ||
| Stage | ||||||
| I | 25 | 7 | 0.053 | 30 | 2 | 0.552 |
| II | 55 | 8 | 58 | 7 | ||
| III | 68 | 16 | 73 | 12 | ||
| IV | 29 | 0 | 27 | 2 | ||
| Invasion | ||||||
| No | 99 | 17 | 0.910 | 106 | 12 | 0.701 |
| LVI or PNI or both | 78 | 14 | 82 | 11 | ||
| Differentiation | ||||||
| Well-moderately | 172 | 28 | 0.067 | 180 | 23 | 0.313 |
| Poorly | 5 | 3 | 8 | 0 | ||
| Bowel wall invasion | ||||||
| pT1 | 3 | 1 | 0.511 | 4 | 0 | 0.410 |
| pT2 | 34 | 9 | 39 | 5 | ||
| pT3 | 126 | 18 | 128 | 18 | ||
| pT4 | 14 | 3 | 17 | 0 | ||
| Lymph node metastasis | ||||||
| pN- | 88 | 15 | 0.891 | 96 | 9 | 0.280 |
| pN+ | 89 | 16 | 92 | 14 | ||
| Distant metastasis | ||||||
| No | 148 | 31 | 0.015a | 161 | 21 | 0.456 |
| Yes | 29 | 0 | 21 | 2 | ||
Two-hundred and eight tumor samples with data for both MMR and the BRAF mutation were available: 25/31 cases (80.6%) with dMMR were negative for BRAF V600E, and 6/31 (19.4%) were positive for BRAF mutation; whereas 160/177 cases (90.4%) with pMMR were negative for BRAF mutation, and 17/177 (9.6%) were positive for BRAF mutation (P = 0.11) (Table 5).
| MMR status | |||
| BRAF | dMMR | pMMR | P value |
| Normal | 25 | 160 | 0.11 |
| V600E | 6 | 17 | |
The median follow-up time was 56.7 mo. At the last follow-up (December 5, 2012), there were 133 patients alive and 78 patients deceased. Estimated 5-year survival for the entire population was 64.5%.
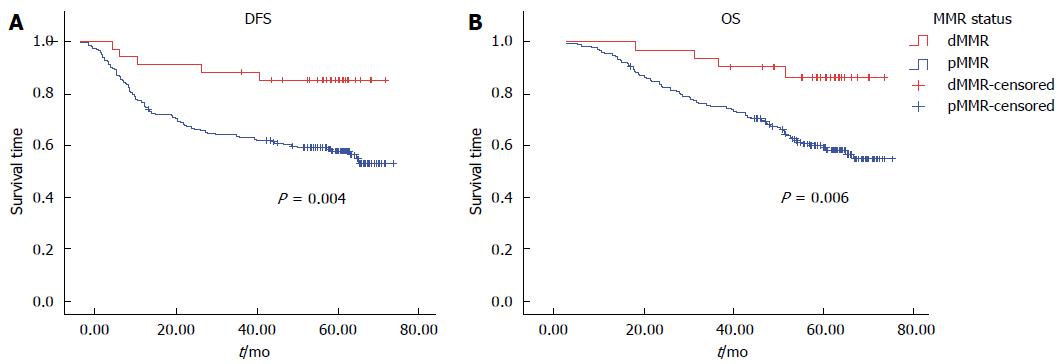
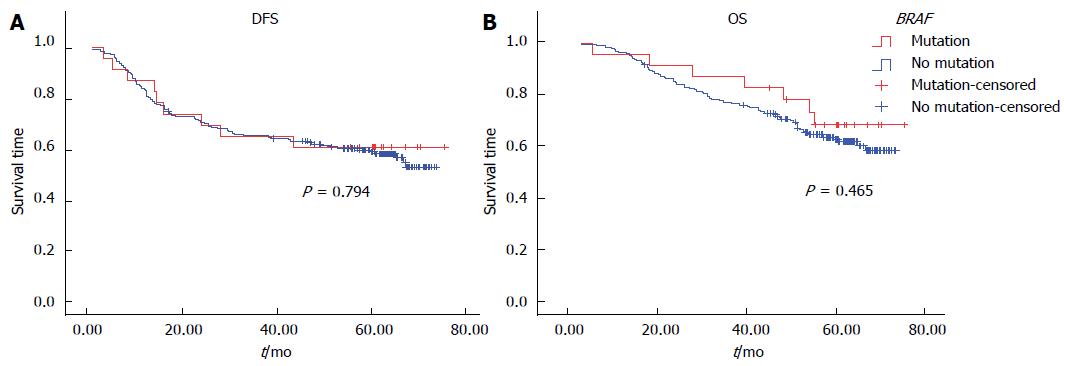
Univariate analysis by Kaplan-Meier survival analysis and log rank test was performed using clinical parameters and known prognostic factors to evaluate their significance with DFS and OS. These factors included age, gender, primary tumor site, UICC stage, T stage, regional lymph node involvement, distant metastasis, histological grade, angiolymphatic (LVI) and/or perineural invasion (PNI), MMR status, and the BRAF V600E mutation. Factors with statistical significance for DFS were UICC stage (P < 0.01), pT3 (P = 0.01), regional lymph node involvement (P < 0.01), and LVI and/or PNI (P < 0.01). Factors that were statistically significant for OS were UICC stage, pT3, pT4, regional lymph node involvement, and LVI and/or PNI (P < 0.01 for all) (Table 6). Patients with dMMR tumors were found to have significantly better DFS (P < 0.01) and OS (P < 0.01) (Figure 1). There was no significant difference in either DFS or OS with respect to adjuvant chemotherapy in dMMR patients (P = 0.35, 0.21, respectively). However, there was no significant difference in either DFS or OS with respect to BRAF mutation (Figure 2).
| Variable | n | DFS | OS | ||
| Median survival (mo) | P value | Median survival (mo) | P value | ||
| Gender | |||||
| Female | 105 | NR | NR | ||
| Male | 106 | NR | 0.528 | NR | 0.640 |
| Age (yr) | |||||
| ≤ 50 | 30 | NR | NR | ||
| > 50 | 181 | NR | 0.695 | NR | 0.424 |
| Site | |||||
| Right-sided | 43 | NR | NR | ||
| Left-sided | 73 | NR | NR | ||
| Rectum | 91 | NR | NR | ||
| Synchronous lesions | 4 | NR | 0.682 | NR | 0.788 |
| Stage | |||||
| I | 32 | NR | NR | ||
| II | 65 | NR | < 0.0001a | NR | < 0.0001a |
| III | 85 | 66.7 | < 0.0001a | NR | < 0.0001a |
| IV | 29 | 10.4 | < 0.0001a | 21.6 | < 0.0001a |
| Invasion | |||||
| No invasion | 118 | NR | NR | ||
| LVI or PNI or both | 93 | 52.8 | 0.002a | 57.13 | < 0.0001a |
| Differentiation | |||||
| Well-moderately | 203 | NR | NR | ||
| Poorly | 8 | 29.83 | 0.575 | 40.67 | 0.306 |
| Bowel wall invasion | |||||
| pT1-T2 | 48 | NR | NR | ||
| pT3 | 146 | 62.23 | 0.011a | NR | < 0.0001a |
| pT4 | 17 | 66.73 | 0.122 | 66.733 | < 0.0001a |
| Lymph node metastasis | |||||
| pN- | 105 | NR | NR | ||
| pN+ | 106 | 27.57 | < 0.0001a | 54 | < 0.0001a |
| Distant metastasis | |||||
| No | 182 | - | - | NR | < 0.0001a |
| Yes | 29 | - | - | 21.6 | |
| BRAF status | |||||
| Wild type | 188 | NR | 0.794 | NR | 0.465 |
| Mutation | 23 | NR | NR | ||
| MMR status | |||||
| pMMR | 177 | NR | 0.004a | NR | 0.006a |
| dMMR | 31 | NR | NR | ||
Multivariate Cox proportional hazard regression analysis of factors influencing DFS and OS was performed using the factors mentioned previously. The independent risk factor for worse DFS was stage III (HR = 4.03, 95%CI: 1.57-10.32, P < 0.01) and risk factors for worse OS were stage III (HR = 4.94, 95%CI: 1.50-16.27, P < 0.01), stage IV (HR = 32.64, 95%CI: 9.57-112.27, P < 0.01) and poor differentiation (HR = 3.78, 95%CI: 1.27-11.21, P = 0.02). dMMR remained a significant prognostic factor for longer DFS (HR = 0.30, 95%CI: 0.15-0.77, P = 0.01) and OS (HR = 0.29, 95%CI: 0.10-0.84, P = 0.02) (Table 7).
| Adjusted analysis for DFS | Adjusted analysis for OS | |||||
| Variable | HR | 95%CI | P value | HR | 95%CI | P value |
| Stage | ||||||
| Stage I1 | ||||||
| Stage II | 1.23 | 0.43-3.62 | 0.702 | 1.47 | 0.40-5.40 | 0.559 |
| Stage III | 4.03a | 1.57-10.32 | 0.004a | 4.94a | 1.50-16.27 | 0.009a |
| Stage IV | - | 32.64a | 9.57-111.27 | < 0.001a | ||
| Invasion | ||||||
| No invasion1 | ||||||
| LVI/PNI/both | 1.17 | 0.75-1.81 | 0.486 | 1.36 | 0.85-2.19 | 0.204 |
| Differentiation | ||||||
| Well-moderate1 | ||||||
| Poorly | 2.57 | 0.87-7.43 | 0.083 | 3.78 | 1.27-11.21 | 0.017a |
| MMR status | ||||||
| pMMR1 | ||||||
| dMMR | 0.30 | 0.15-0.77 | 0.013a | 0.29a | 0.10-0.84 | 0.023a |
In this study, we performed an analysis of MMR status and the BRAF V600E mutation in sporadic CRC using archival paraffin-embedded tissue blocks from patients diagnosed between 2006 and 2007. All patients included in the study underwent resection of their primary tumor to ensure sufficient tumor tissues would be available for the analysis. Therefore, the incidence of stage IV disease in this study (14%) was lower than previously reported from our institution in the same period (32%)[18]. Because we only selected patients who had undergone primary tumor resection, selection bias might have been introduced by not using all of the patients. The primary objective was to determine the prevalence of each biomarker. Sample size determination was based on BRAF mutation and dMMR estimated incidences of 15% as reported in previous studies[3,4,8-10].
IHC is an excellent method to determine MMR status. This method is inexpensive, technically simple with demonstrably good correlation with MSI-H[9,14]. Review of the literature suggested that IHC has an overall sensitivity of approximately 90% and specificity of greater than 99% in detecting MSI in sporadic CRC[19]. In our study, we used antibodies against all four MMR proteins to increase detection sensitivity for specific loss of protein expression other than MLH-1 and MSH-2, the two most common dMMR genes. TMA is an approach that allows high-throughput IHC to be performed on large numbers of small punched-out tissue cores from different tumors[20], thus providing time- and cost-saving benefits. TMA-IHC for MMR protein expression was validated in tumors from HNPCC patients and found to have concordant results when compared to MSI analysis and conventional IHC on whole slides[21].
Approximately 15%-20% of sporadic CRC carried dMMR. The proportion of samples with dMMR was higher in tumors located in or above the splenic flexure, stage II rather than stage III disease, poorly differentiated tumors, and those of mucinous subtype[9]. Several studies, including a meta-analysis, found that patients with dMMR tumors had significantly better survival compared with that of pMMR patients[9,14,22].
In this study, we performed IHC staining of 211 cases. MSI analysis was completed in 47 cases with uninformative IHC staining results. The final result indicated 14.9% (31/208) of sporadic CRC patients harbored dMMR tumors detected by both TMA-IHC and MSI analysis (10/164 and 21/47 cases, respectively), which was comparable with 11.3% (36 of 318) found in sporadic CRC patients in South Korea[23] and in that reported in the literature[3]. dMMR was more commonly found in patients with primary colon tumors rather than rectal cancer, but there was no difference in the prevalence of dMMR in right-sided and left-sided colon tumors. This might be related to the small number of patients included in the study and the fact that the majority (78%) of patients had left-sided tumors. We did not find any association between dMMR and high grade tumors (P = 0.067). This result should be interpreted carefully because the majority were moderately differentiated tumors, accounting for 79.1% of cases, while only 15.1 and 3.8% were well and poorly differentiated tumors, respectively. However, this study confirmed that dMMR was independently associated with favorable outcome in both DFS and OS.
Recently BRAF mutations have been widely studied with respect to their prognostic and predictive value in CRC. Studies have shown that the BRAF mutation was associated with a poor outcome in CRC patients with pMMR but not with dMMR[9,10]. The prevalence of the BRAF mutation in this study was 10.9%, which was comparable with Caucasian patients, but the result was higher than that reported by studies in Taiwan (1.7%), South Korea (4.5%) and Japan (6.5%)[24-26]. The main difference between our study and others is that our data were analyzed using allele-specific PCR to detect BRAF V600E, which is more highly sensitive in detecting this mutation than other methods. Several other approaches are suitable for evaluating this BRAF mutation, including direct sequencing, real-time PCR with melt curve analysis, and AS-PCR[27]. The BRAF mutation was strongly associated with dMMR, right-sided and poorly differentiated tumors[9]. A recent study showed that the BRAF V600E mutation was independently associated with worse DFS in patients with stage III colon cancer receiving adjuvant chemotherapy[15]. In this study, there was no association between the BRAF mutation and any pathological features, no significant difference in the prevalence of the BRAF mutation in dMMR and pMMR tumors, and also no prognostic impact for either DFS or OS. This is likely a result of too few patients analyzed to determine the prognostic impact.
In conclusion, we found that the prevalence of dMMR and the BRAF V600E mutation in Thai sporadic CRC patients was 15 and 11%, respectively. However, only the dMMR phenotype was associated with favorable DFS and OS in our study cohort.
The authors are grateful to Professor Peter Hokland for his kind help in critically reading our manuscript.
Colorectal tumorigenesis is a multistep process, which can arise from the accumulation of genetic alterations. DNA mismatch repair (dMMR) and BRAF gene have been widely studied in this disease. The correlation between dMMR and BRAF mutation and colorectal cancer (CRC) prognosis has been reported. Patients with tumors harboring dMMR were found to be associated with a more favorable survival than those having proficient MMR (pMMR). In contrast, BRAF mutation was found to be associated with worse clinical outcome especially in patients with pMMR tumors. Unfortunately, most of these data have been studied in western country, and there is only scare information in the Asian population.
In this study, the authors have systematically determined the prevalence of dMMR and BRAF mutation in sporadic CRC patients and established the correlations between MMR status and BRAF mutation, association with various clinicopathological features, and prognostic impact on the outcome of Thai patients with sporadic CRC.
The method the authors used to detect BRAF mutation is allele specific polymerase chain reaction which has the highest sensitivity to detect this mutation when compared to previously reported methods. This study is the first reported data in Thai population.
This study confirmed the favorable outcome in patients with dMMR tumors, which is consistent to the results of previous reports in Caucasian population.
This is a good retrospective study in which this is the first reported data in Thai population. The method that used to detected BRAF mutation has the highest sensitivity to detect this mutation when compare to previously reported studies and the result of the study can imply that patients with dMMR tumors had a significantly better survival in both Asian and Caucasian population.
P- Reviewer: Wang YD S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | Worthley DL, Whitehall VL, Spring KJ, Leggett BA. Colorectal carcinogenesis: road maps to cancer. World J Gastroenterol. 2007;13:3784-3791. [PubMed] |
| 2. | Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447-2454. [PubMed] |
| 3. | Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1346] [Article Influence: 67.3] [Reference Citation Analysis (1)] |
| 4. | Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 938] [Cited by in RCA: 951] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 5. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7459] [Cited by in RCA: 7633] [Article Influence: 331.9] [Reference Citation Analysis (0)] |
| 6. | Ikenoue T, Hikiba Y, Kanai F, Tanaka Y, Imamura J, Imamura T, Ohta M, Ijichi H, Tateishi K, Kawakami T. Functional analysis of mutations within the kinase activation segment of B-Raf in human colorectal tumors. Cancer Res. 2003;63:8132-8137. [PubMed] |
| 7. | Fransén K, Klintenäs M, Osterström A, Dimberg J, Monstein HJ, Söderkvist P. Mutation analysis of the BRAF, ARAF and RAF-1 genes in human colorectal adenocarcinomas. Carcinogenesis. 2004;25:527-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 211] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 8. | Maestro ML, Vidaurreta M, Sanz-Casla MT, Rafael S, Veganzones S, Martínez A, Aguilera C, Herranz MD, Cerdán J, Arroyo M. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol. 2007;14:1229-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261-1270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 528] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 10. | MacDonald CM, Boursier L, D’Cruz DP, Dunn-Walters DK, Spencer J. Mathematical analysis of antigen selection in somatically mutated immunoglobulin genes associated with autoimmunity. Lupus. 2010;19:1161-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 11. | Deng G, Bell I, Crawley S, Gum J, Terdiman JP, Allen BA, Truta B, Sleisenger MH, Kim YS. BRAF mutation is frequently present in sporadic colorectal cancer with methylated hMLH1, but not in hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:191-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 301] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 12. | Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90-96. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 633] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 13. | Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063-6069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 598] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 14. | Lanza G, Gafà R, Santini A, Maestri I, Guerzoni L, Cavazzini L. Immunohistochemical test for MLH1 and MSH2 expression predicts clinical outcome in stage II and III colorectal cancer patients. J Clin Oncol. 2006;24:2359-2367. [PubMed] |
| 15. | Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466-474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 801] [Cited by in RCA: 917] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 16. | Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856-862. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 319] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 17. | Kannim S, Thongnoppakhun W, Auewarakul CU. Two-round allele specific-polymerase chain reaction: a simple and highly sensitive method for JAK2V617F mutation detection. Clin Chim Acta. 2009;401:148-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 18. | Techawathanawanna S, Nimmannit A, Akewanlop C. Clinical characteristics and disease outcome of UICC stages I-III colorectal cancer patients at Siriraj Hospital. J Med Assoc Thai. 2012;95 Suppl 2:S189-S198. [PubMed] |
| 19. | Shia J, Ellis NA, Klimstra DS. The utility of immunohistochemical detection of DNA mismatch repair gene proteins. Virchows Arch. 2004;445:431-441. [PubMed] |
| 20. | Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195:72-79. [PubMed] |
| 21. | Hendriks Y, Franken P, Dierssen JW, De Leeuw W, Wijnen J, Dreef E, Tops C, Breuning M, Bröcker-Vriends A, Vasen H. Conventional and tissue microarray immunohistochemical expression analysis of mismatch repair in hereditary colorectal tumors. Am J Pathol. 2003;162:469-477. [PubMed] |
| 22. | Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45:1890-1896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 216] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 23. | Park JW, Chang HJ, Park S, Kim BC, Kim DY, Baek JY, Kim SY, Oh JH, Choi HS, Park SC. Absence of hMLH1 or hMSH2 expression as a stage-dependent prognostic factor in sporadic colorectal cancers. Ann Surg Oncol. 2010;17:2839-2846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 24. | Shen H, Yuan Y, Hu HG, Zhong X, Ye XX, Li MD, Fang WJ, Zheng S. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol. 2011;17:809-816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 25. | Lee S, Cho NY, Choi M, Yoo EJ, Kim JH, Kang GH. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int. 2008;58:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 115] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 26. | Yokota T, Shibata N, Ura T, Takahari D, Shitara K, Muro K, Yatabe Y. Cycleave polymerase chain reaction method is practically applicable for V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS)/V-raf murine sarcoma viral oncogene homolog B1 (BRAF) genotyping in colorectal cancer. Transl Res. 2010;156:98-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Sharma SG, Gulley ML. BRAF mutation testing in colorectal cancer. Arch Pathol Lab Med. 2010;134:1225-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |