Published online Jun 7, 2015. doi: 10.3748/wjg.v21.i21.6639
Peer-review started: November 3, 2014
First decision: December 26, 2014
Revised: February 3, 2015
Accepted: February 11, 2015
Article in press: February 11, 2015
Published online: June 7, 2015
Processing time: 220 Days and 12.9 Hours
AIM: To investigate precore/basal core promoter (PC/BCP) mutants throughout hepatitis B virus (HBV) infection and to determine their relationship to hepatitis B early antigen (HBeAg) titers.
METHODS: We enrolled 191 patients in various stages of HBV infection at the Huashan Hospital and the Taizhou Municipal Hospital from 2010 to 2012. None of the patients received antiviral therapy. HBV DNA from serum, was quantified by real-time PCR. The HBV genotype was determined by direct sequencing of the S gene. We used the Simpleprobe ultrasensitive quantitative method to detect PC/BCP mutants in each patient. We compared the strain number, percentage, and the changes in PC/BCP mutants in different phases, and analyzed the relationship between PC/BCP mutants and HBeAg by multiple linear regression and logistic regression.
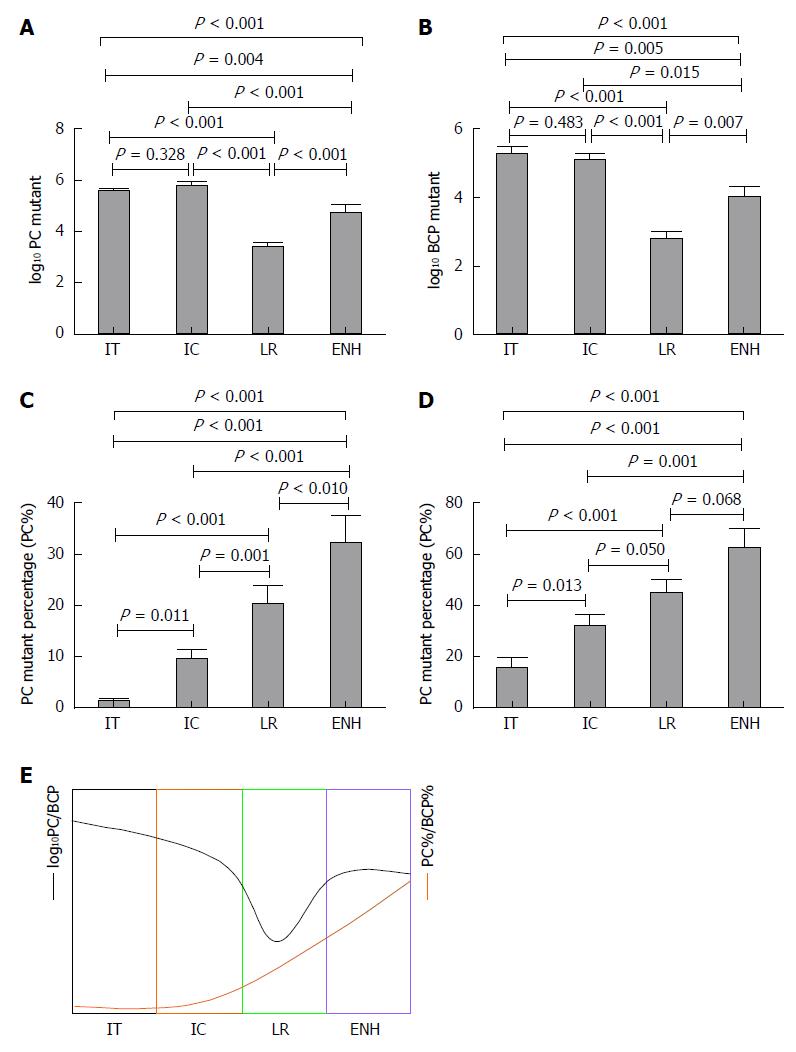
RESULTS: Patients with HBV infection (n = 191) were assigned to groups by phase: Immune tolerance (IT) = 55, Immune clearance (IC) = 67, Low-replicative (LR) = 49, and HBeAg-negative hepatitis (ENH) = 20. Of the patients (male, 112; female, 79) enrolled, 122 were HBeAg-positive and 69 were HBeAg-negative. The median age was 33 years (range: 18-78 years). PC and BCP mutation detection rates were 84.82% (162/191) and 96.86% (185/191), respectively. In five HBeAg-negative cases, we detected double mutation G1896A/G1899A. The logarithm value of PC mutant quantities (log10 PC) significantly differed in IT, IC, and LR phases, as well as in the ENH phase (F = 49.350, P < 0.001). The logarithm value of BCP mutant quantities (log10 BCP) also differed during the four phases (F = 25.530, P < 0.001). Log10 PC and log10 BCP values were high in the IT and IC phases, decreased in the LR phase, and increased in the ENH phase, although the absolute value at this point remained lower than that in the IT and IC phases. PC mutant quantity per total viral load (PC%) and BCP mutant quantity per total viral load (BCP%) differed between phases (F = 20.040, P < 0.001; F = 10.830, P < 0.001), with PC% and BCP% gradually increasing in successive phases. HBeAg titers negatively correlated with PC% (Spearman’s rho = -0.354, P < 0.001) and BCP% (Spearman’s rho = -0.395, P < 0.001). The negative correlation between PC% and HBeAg status was significant (B = -5.281, P = 0.001), but there was no such correlation between BCP% and HBeAg status (B = -0.523, P = 0.552).
CONCLUSION: PC/BCP mutants become predominant in a dynamic and continuous process. Log10 PC, log10 BCP, PC% and BCP% might be combined to evaluate disease progression. PC% determines HBeAg status.
Core tip: During the natural history of hepatitis B virus infection, no evidence for the correlation between the dynamic alteration of precore/basal core promoter (PC/BCP) mutated strains and hepatitis B early antigen titers has been obtained by qualitative analysis. Using Simpleprobe ultrasensitive quantification of the wild-type and mutated hepatitis B virus (HBV) strains, we provided new insights into the process by which PC/BCP-mutated strains become dominant during the natural course of infection. Thus, we provide important clues for the evaluation of HBV infection status and the corresponding host immune responses.
- Citation: Tu WH, Lv Y, Zhang YM, Hou W, Wang JY, Zhang YJ, Liu HY, Zhu HX, Qin YL, Mao RC, Zhang JM. Precore/basal core promoter mutants quantification throughout phases of hepatitis B virus infection by Simpleprobe. World J Gastroenterol 2015; 21(21): 6639-6648
- URL: https://www.wjgnet.com/1007-9327/full/v21/i21/6639.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i21.6639
Although the hepatitis B vaccine is in widespread use, hepatitis B virus (HBV) infection remains a serious public health problem[1]. In the general Chinese population, 7.18% of individuals are hepatitis B surface antigen (HBsAg) carriers, including 93 million individuals with chronic HBV infection and 20 million patients with chronic hepatitis B (CHB)[2]. HBV infection is the most important factor in acute and chronic hepatitis, and is associated with the occurrence of hepatocellular carcinoma (HCC)[3].
Hepatitis B early antigen (HBeAg) clearance (with or without anti-HBe) is the endpoint of treatment for HBeAg-positive CHB[4,5]; however, HBeAg clearance is insufficient to alleviate the disease process. Many patients treated with nucleotide analogs (NAs) achieve HBeAg clearance and experience recurrent HBeAg; alternatively, HBV DNA is detected after treatment is stopped[6]. Interferon therapy produces a more sustained effect of HBeAg clearance, although long-term follow-up has revealed the presence of HBV DNA in some patients[7,8]. Thus, HBeAg clearance with interferon therapy alone may produce a false impression of therapeutic efficacy. Precore/basal core promoter (PC/BCP) mutants inhibit HBeAg synthesis and may account for the apparent clearance of HBeAg. The HBV Precore G1896A mutation generates a termination codon, eliminating HBeAg synthesis[9]. The BCP A1762T/G1764A double mutation inhibits preC mRNA synthesis, and thus inhibits HBeAg production[10,11]. Precore G1896A or G1899A mutations have been associated with increased risk of HCC[12,13], more serious hepatitis disease, and acute or chronic liver failure[14,15]. The A1762T/G1764A double mutation is associated with increased virulence and disease progression to liver cirrhosis and HCC[16-18].
PC/BCP mutant detection is currently achieved by direct DNA sequencing, restriction fragment length polymorphism (RFLP) analysis, hybridization probes, and qualitative detection methods, such as the line probe assay (INNO-LiPA)[19-23]. In our previous study of 207 HBeAg-positive patients[24], RFLP analysis indicated that PC/BCP mutants also exist in HBeAg-positive patients and influence the expression of HBeAg. However, these methods do not provide precise quantification of mutated HBV isolates, thus limiting longitudinal observation of PC/BCP mutation during the natural course of hepatitis B virus infection and the dynamic evaluation of PC/BCP mutation. This has also limited our understanding of the correlation between PC/BCP mutation and HBeAg titers.
In this study, we used the Simpleprobe method for ultrasensitive quantitation of PC/BCP mutants, which allowed us to detect mutations present at a level as low as 0.001% in a wild-type background[25]. Quantitation of PC/BCP mutants throughout the natural history of HBV infection will clarify the relationship between virus mutations and the patient’s immune status.
We enrolled 191 patients with chronic HBV infection in the Huashan Hospital and the Taizhou Municipal Hospital from 2010 to 2012. Enrolled participants met the following criteria: (1) age 18-78 years; (2) HBsAg-positive for at least six months at the time of enrollment; (3) no autoimmune liver disease or concurrent hepatitis C virus (HCV), hepatitis D virus (HDV), or human immunodeficiency virus (HIV) infection; (4) no history of antiviral drug treatment; (5) no end-stage liver disease or HCC; and (6) no history of immunosuppressive therapy. Patients were assigned to one of the following groups based on their HBV infection state: immune tolerance (IT = 55 cases), immune clearance (IC = 67 cases), low replication (LR = 49 cases), and HBeAg-negative hepatitis (ENH = 20 cases). The IT phase is characterized by high HBeAg titers, high HBV DNA levels, but normal ALT, and normal liver histology. The IC phase is characterized by presence of HBeAg, high HBV DNA levels (over 20000 IU/mL), elevated ALT and necroinflammation of the liver. The LR phase is characterized by HBeAg negativity and anti-HBe positivity, low or undetectable HBV DNA (below 2000 IU/mL), persistently normal ALT, and no histologically active inflammation, with mild fibrosis. The ENH phase is characterized by negative HBeAg, positive anti-HBe, detectable HBV DNA levels (2000-20 million IU/mL), elevated ALT, and moderate to severe necroinflammation, with variable amounts of fibrosis. Blood samples were stored at -20 °C until use. Study protocols conformed to the Declaration of Helsinki principles of ethics, and all participants were enrolled with written informed consent.
HBsAg, HBeAg, anti-HBe, anti-HCV, and anti-HDV were assessed using commercial AxSYM MEI kits (Abbott Laboratories, North Chicago, IL, United States). Anti-HIV was assayed by using the Diagnostic Kit for Antibody to Human Immunodeficiency Virus Type 1 and/or 2 and HIV-1 Antigen (bioMérieux, France). HBeAg values were determined using a microparticle enzyme immunoassay and the results are expressed as signal/cutoff (S/CO), with HBeAg values > 1S/CO considered positive (Abbott Architect i2000SR). HBV DNA was quantified using quantitative real-time PCR with commercial kits (Shenzhen PG Biotech, Shenzhen, China). The assay detection limit was 100 IU/mL. HBV DNA standards were obtained from the National Institute for the Control of Pharmaceutical and Biological Products, China.
HBV DNA was isolated from 200 μL serum and eluted in 50 μL buffer EB from the QIAamp DNA Blood Mini kit (Qiagen, Shanghai, China), according to the manufacturer’s protocols. Genotyping was performed by sequencing the 1.3-kb HBsAg complete gene fragment after PCR amplification (nt 2816-886). Phylogenetic analysis was performed using Vector NTI 11.0 TreeView software. A neighbor-joining tree was constructed using Jukes-Cantor corrected distances in the MEGA2 package with 1000 bootstrap replicates. All procedures were performed as previously described[24].
PC/BCP quantitation was performed by two-step real-time PCR ,as described by Nie et al[25]. The first step included a wild-type blocker probe (WT-blocker probe) for selective inhibition PCR (siPCR), which blocked further amplification of wild-type DNA; subsequent cycles amplified mutant viral DNA by about 10000 times, making it easy to detect in the subsequent reaction. The second step involved amplification with a Simpleprobe, followed by melting curve analysis. The first step of PC region amplification was performed in a 15-μL reaction contained 0.5 μmol/L each F1 (5'-CCAAATTCTTTATACGGGTCAATGTCCATG-3', nt 1929 to 1900) and R1 (5'-CCTCCAAGCTGTGCC-3', nt 1869 to 1883) primer, 5 μL of the purified patient sample DNA as template, and 2 μmol/L WT-blocker (5’-gtccatgCcCCAAagcc-PH, nt 1906 to 1890). Amplification was carried out at 95 °C for 10 s, 59 °C for 10 s and 72 °C for 5 s, for a total of 20 cycles. The first-step PCR products were diluted to 1:32 and used as templates in the second step real-time PCR, which was composed of LightCycler480 Genotyping Master Mix, 0.1 μmol/L primer F1, 0.5 μmol/L primer R1, 3 mmol/L MgCl2, and 0.1 μmol/L Simpleprobe (5’-gtcaatgtccatgTcCTAaagcc-3’, nt 1912 to 1890). The real time PCR was performed at 95 °C for 10 min; followed by 40 cycles of 95 °C for 10 s, 66 °C for 10 s, and 72 °C for 5 s; and then followed by a melting curve analysis from 30 °C to 80 °C.
The first step PCR amplification of BCP region was performed in a 15-μL reaction mixture containing 0.5 μmol/L each F1(5'-AGGAGTTGGGGGAGGAGATTAGGTTAA-3',and R1 (5'-CTTGGAGGCTTGAACAGTAGGAC-3', nt 1881 to 1854) primers, 5 μL of the purified patient sample DNA as the template, and 2 μmol/L WT-blocker (5’-aggagattaGgttAaAGGtctttGt-PH, nt 1747 to 1771). PCR was carried out at 95 °C for 10 s, 57 °C for 10 s, and 65 °C for 5 s, for a total of 20 cycles. The second step PCR was composed of LightCycler480 genotyping master mix, 0.1 μmol/L primer F2 (5’-GATAAGTTGAGGAGTTGGGGG-3’, nt 1726 to 1746), 0.5 μmol/L primer R1, 0.1 μmol/L Simpleprobe (5’-ggagattaGgttAaTGAtct-3’, nt 1748 to 1767). The real-time PCR was performed at 95 °C for 10 min to activate the polymerase; followed by 40 cycles of 95 °C for 10 s, 55 °C for 10 s and 72 °C for 5 s. Capital letters in the WT-blocker and Simpleprobe sequences indicate Locked Nucleic Acids (LNAs), and the 3’-end “-PH” stands for phosphorylation. The fluorescent label is indicated by bold letter. The method also employed a primer-blocker-probe partial overlap design to reduce the influence of polymorphisms and improve detection sensitivity to 0.001%. The assay detection limit is 60 IU/mL.
G1896A and G1896A/G1899A variants are defined as PC mutations; A1762T/G1764A variants are defined as BCP mutations. We used the log10 PC and log10 BCP values to represent PC and BCP mutant quantities. PC% and BCP% represented PC and BCP mutant quantities per total viral load.
Data are expressed as the median. All data analyses were performed in SPSS17.0 software. The Mann-Whitney rank sum test, one-way ANOVA, Independent-Sample T test, Spearman correlation, multiple linear regression, and logistic regression analysis were used. Statistical significance was defined as P < 0.05.
Patients with HBV infection (n = 191) were assigned to groups by phase: IT = 55, IC = 67, LR = 49, and ENH = 20. Patients with HBeAg positive 122 and HBeAg negative 69. The population included 112 male and 79 female subjects, with a median age of 33 years (range: 18-78 years). Younger patient age was associated with higher total log10 HBV DNA in the IT and IC groups versus the LR and ENH groups (6.99 ± 0.97 vs 4.07 ± 1.15, P < 0.001). HBeAg titers differed significantly between the IT and IC phases (1261.27 ± 465.56 vs 825.30 ± 580.62, P < 0.001). PC% and BCP% were significantly higher in the LR and ENH phases (U = 1422.0, P < 0.001; U = 2357.0, P < 0.001; Table 1).
| IT (n = 55) | IC (n = 67) | LR (n = 49) | ENH (n = 20) | ANOVA P value | |
| Age (yr), median (range) | 27 (18-55) | 32 (18-78) | 40 (21-65) | 41 (25-65) | < 0.001 |
| Gender, n, M/F | 28/27 | 40/27 | 28/21 | 16/4 | 0.160 |
| Log10 HBV DNA, | |||||
| Log10 (IU/mL), median | 7.55 | 6.98 | 3.7 | 4.69 | < 0.001 |
| HBV genotype, n | |||||
| B | 34 | 39 | 29 | 11 | |
| C | 21 | 28 | 20 | 9 | 0.960 |
| ALT (U/mL) median | 16 | 98 | 20 | 93 | < 0.001 |
| AST (U/mL) median | 13 | 67 | 18 | 62 | < 0.001 |
| HBeAg (S/CO) median | 1376 | 899 | NA | NA | < 0.001 |
| PC%, median | 0.31 | 1.73 | 9.20 | 33.69 | < 0.001 |
| BCP%, median | 1.42 | 15.25 | 43.58 | 67.44 | < 0.001 |
We identified 157 cases with G1896A mutations, 29 cases with G1899A mutations, and five cases with the G1896A/G1899A double mutation. Twenty-seven of the cases carrying the G1899A mutation were also HBeAg-positive, while all five cases of G1896A/G1899A double mutation were HBeAg-negative. The PC mutation detection rate was 84.82% (162/191, PC mutant quantification range: 60-60 million IU/mL). We identified 96.86% (185/191, BCP mutant quantification range: 60-60 million IU/mL) cases as carriers of the BCP (A1762T/G1764A) mutation. We identified the BCP mutation alone in 26 cases, and the PC mutation alone in five cases. The PC/BCP joint mutation was identified in 159 cases, whereas the wild-type PC/BCP was found in only one case.
Log10 PC and log10 BCP distributions differed significantly between the four phases (F = 49.350, P < 0.001; F = 23.530, P < 0.001). The log10 PC and log10 BCP values were higher in IT and IC, decreased in LR, and were higher in the ENH phase, although the absolute value remained lower than in the IT and IC phases (Figure 1A and B). PC% and BCP% distributions gradually increased through each phase (F = 20.040, P < 0.001; F = 10.830, P < 0.001; Figure 1C and D). We compared PC%, BCP%, log10 PC and log10 BCP values between each group and observed significant differences (Figure 1A-D). The dynamic variation of PC%, BCP%, log10 PC and log10 BCP are shown in Figure 1E.
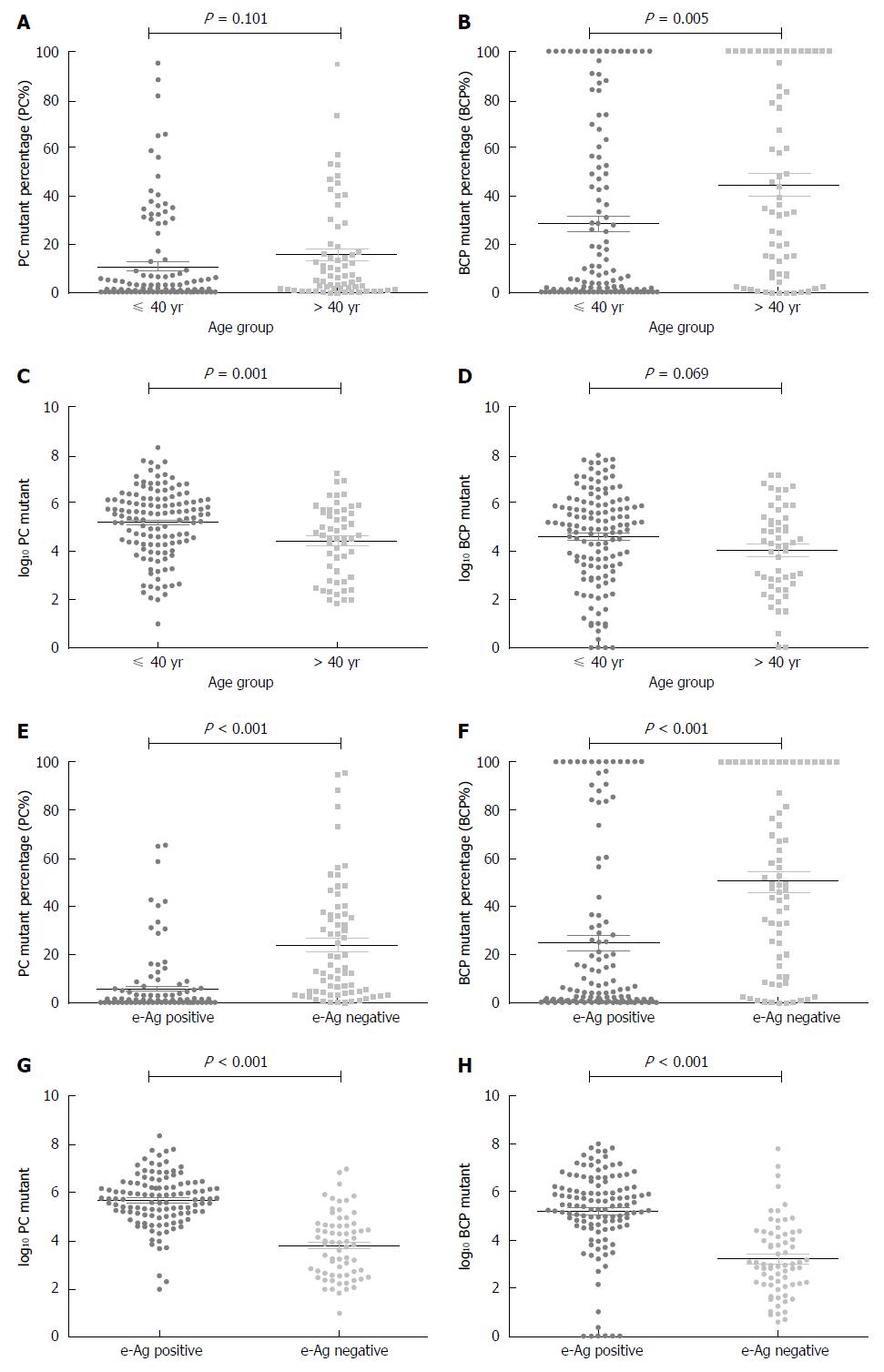
PC% increased with age (Spearman’s rho = 0.281, P < 0.001); however, the PC% distribution did not significantly differ between the ≤ 40 and > 40 age groups (t = 1.650, P = 0.101; Figure 2A); the log10 PC distribution did differ between age groups (t = 3.340, P = 0.001; Figure 2C). Similarly, BCP% also increased with age (Spearman’s rho = 0.31, P < 0.001) and its distribution significantly differed (≤ 40 vs > 40; t = 2.859, P = 0.005; Figure 2B). The log10 BCP distribution did not differ between age groups (t = 1.830, P = 0.069; Figure 2D).
When divided by HBeAg status, PC% distribution significantly differed between groups. PC% was higher in HBeAg-negative patients than in HBeAg-positive patients (23.95% vs 5.75%, P < 0.001; Figure 2E). The log10 PC value also differed between groups; values were higher in HBeAg-positive patients (5.68 ± 1.05 vs 3.81 ± 1.32, P < 0.001; Figure 2G). BCP% was higher in HBeAg-negative patients than in HBeAg-positive patients (50.33% vs 24.80%; P < 0.001; Figure 2F). The log10 BCP values were higher in the HBeAg-positive group (5.17 ± 1.77 vs 3.13 ± 1.66, P < 0.001; Figure 2H).
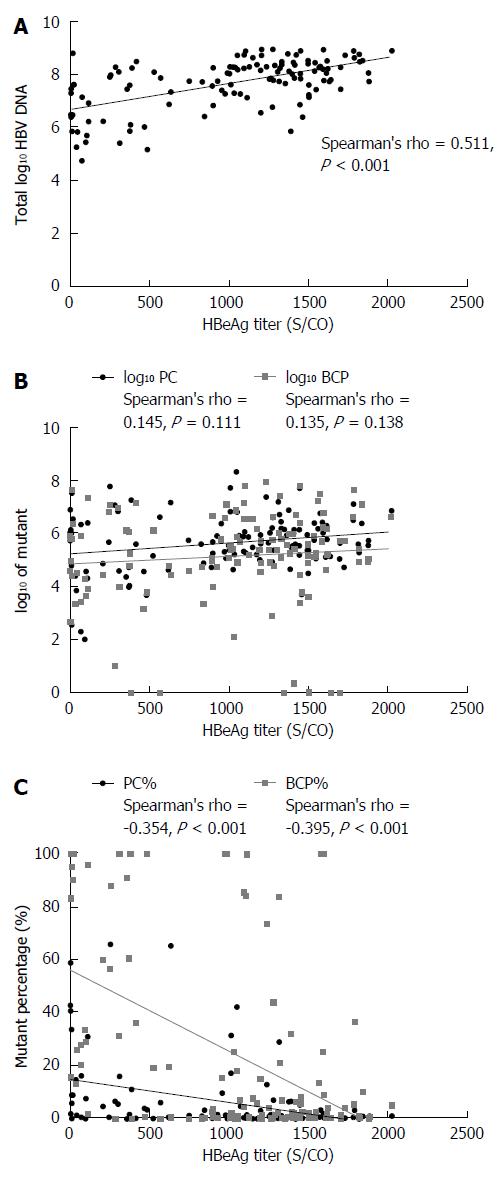
HBeAg titers and total log10 HBV DNA were positively correlated (Spearman’s rho = 0.511, P < 0.001; Figure 3A). The total log10 HBV DNA value was higher in HBeAg-positive than in HBeAg-negative patients (7.684 vs 4.769, P < 0.001). HBeAg titers and log10 PC values showed no correlation (Spearman’s rho = 0.145, P = 0.111), neither did HBeAg titers and log10 BCP value (Spearman’s rho = 0.135, P = 0.138) (Figure 3B). The log10 PC and log10 BCP value differed significantly between the HBeAg-positive and HBeAg-negative groups (5.682 vs 3.810, P < 0.001; 5.167 vs 3.129, P < 0.001).
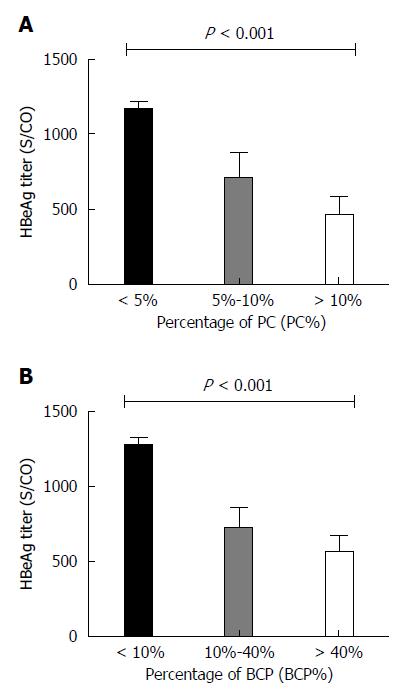
HBeAg titers negatively correlated with PC% (Spearman’s rho = -0.354, P < 0.001) and BCP% (Spearman’s rho = -0.395, P < 0.001) (Figure 3C). The PC% and BCP% significantly differed between HBeAg < 1000 S/CO and ≥ 1000 S/CO groups (t = 3.520, P < 0.001; t = 5.289, P < 0.001): PC% and BCP% were higher in the HBeAg < 1000 S/CO group. HBeAg titers significantly differed between PC% groups of < 5%, 5% to 10%, and >10% (F = 15.230, P < 0.001), as shown in Figure 4A. HBeAg titers also differed between BCP% groups of <10%, 10%-40% and > 40% (F = 26.310, P < 0.001), as shown in Figure 4B.
Logistic regression analysis revealed no correlation between HBeAg status and age (B = -0.050, P = 0.075). Total log10 HBV DNA positively correlated with HBeAg status (B = 1.837, P < 0.001). PC% and HBeAg status showed a significant negative correlation (B = -5.281, P = 0.001), while ALT and BCP% did not correlate with HBeAg status. Multiple linear regression analyses were conducted between HBeAg titer with age, log10 HBV DNA, ALT, PC% and BCP%: age and HBeAg titer showed no significant correlation (B = -3.511, P = 0.357). Total log10 HBV DNA positively correlated with HBeAg titer (B = 259.233, P < 0.001). ALT did not correlate with HBeAg titer (B = 0.048, P = 0.739). PC% negatively correlated with HBeAg titer (B = -1209.594, P < 0.001). BCP% also negatively correlated with HBeAg titer (B = -446.964, P < 0.001) (Table 2).
Multiple linear regression analyses were conducted between PC% or BCP% with age, log10 HBV DNA, ALT, HBeAg titer. The results revealed that PC% negatively correlated with HBeAg titer (B = -0.000, P < 0.001). PC% did not correlate with age, log10 HBV DNA or ALT. BCP% showed a significant negative correlation with HBeAg titer (B = -0.000, P < 0.001). BCP% positively correlated with ALT (B = 0.000, P = 0.007), while age and log10 HBV DNA did not correlate with BCP% (Table 3).
| PC%P value | BCP%P value | |
| Age (yr) | 0.280 | 0.746 |
| Log10 HBV DNA | 0.440 | 0.723 |
| (log10 IU/mL) | ||
| ALT (U/L) | 0.655 | 0.007 |
| HBeAg titer (S/CO) | < 0.001 | < 0.001 |
The high replication rate of HBV leads to production of 1012 virions per day. The lack of a proofreading function in the reverse transcriptase, combined with the high replication rate, yields a high mutation rate. Each replication cycle produces about 10-5 mismatches/base in actively replicating virus strains, yielding 1010 to 1011 point mutations per day[26]. Therefore, nucleotide polymorphisms and acquired mutations are common in the HBV virus pool, and are difficult to differentiate and detect quantitatively. Thus, it is important to find ways to overcome the influence of nucleotide polymorphisms on the detection of acquired mutations.
PC/BCP mutants are common in HBeAg-negative patients[27,28], and are observed in HBeAg-positive patients[24]. Yan et al[28] reported PC or BCP mutation in 65.1% of HBeAg positive patients (125/192), implying that PC/BCP mutations exists to varying degrees in different stages of HBV infection. In this article, we provided the first overview of the quantification and distribution of PC/BCP mutated isolates. BCP mutations were detected in 96.86% and PC mutations in 84.82% of a population of 191 patients; wild-type PC/BCP was found in only one patient. The mutation rate was significantly higher than in previous reports, suggesting that the prevalence of PC/BCP mutations has been severely underestimated. PC% and BCP% increase with age during the natural history of HBV infection. There was a significant difference in BCP% distribution between age groups (≤ 40 vs > 40), indicating a significant correlation between BCP% and age. PC% did not differ between groups, perhaps because of the different baseline natural histories of the enrolled patients. Grouping by HBeAg status showed higher PC% and BCP% in HBeAg-negative patients[24,25,28].
During the natural history of HBV infection, there is a dynamic alteration in the log10 PC and log10 BCP values. Log10 PC and log10 BCP values are high in IT and IC, decrease in the LR phase, and rise again in the ENH phase. The PC% and BCP% were much higher than in the wild-type isolates after the selection of immune responses in the LR phase, demonstrating that PC/BCP mutants may evade the host immune system, consistent with the report of Nie et al[25]. Interferon treatment in wild-type strains is an independent predictor of HBsAg disappearance[29]. Previous studies showed that wild-type strains could be eliminated by a drug-enhanced immune reaction; however, the presence and accumulation of PC/BCP mutants may reduce the efficacy of IFN antiviral therapy.
PC% and BCP% increased during the natural course of HBV infection, which provided a clue to a dynamic variation whereby the PC and BCP mutants gradually become the dominant strains. Thus, disease progression can be estimated by the combination of log10 PC, log10 BCP, PC% and BCP% (Figure 1E). High log10 PC and log10 BCP, and low PC% and BCP% indicate that the virus infection is in the early stages (IT, IC, and HBeAg-positive). If the log10 PC and log10 BCP value is low, and the PC% and BCP% are high, the infection is in the LR phase. While a high log10 PC, log10 BCP, PC% and BCP% could be considered to be a marker of ENH phase, HBeAg-negative chronic hepatitis flare may occur afterwards. Our quantitative analysis of PC/BCP mutants clarified the relationship between the PC/BCP mutant distribution and the host immune state.
The HBeAg titer correlated with PC% and BCP%. Subgroup analysis of HBeAg titers and PC%, BCP% showed a significant difference between groups, suggesting that HBeAg titers are closely correlated with PC% and BCP%, which is consistent with previous reports[24,25]. Logistic regression analysis showed that PC% and HBeAg status have a significant negative correlation, which is also consistent with previous findings regarding the molecular mechanism of HBeAg formation[9,20,23,24]. While age, serum HBV DNA, ALT and BCP% are not relevant factors in determining HBeAg status. Interestingly, our results showed that elevated ALT significantly correlated with BCP%, indicating that BCP% reflects hepatic inflammatory degradation. Nie et al[25] reported that PC% (not BCP%) positively correlated with ALT elevations among HBeAg(-) patients. Several studies have shown that PC/BCP mutants cause hepatic inflammation, acute or chronic liver failure, and liver cirrhosis. Our results suggest ALT elevation might also be a result of varied host immune responses. Previous reports suggested that the PC mutation is more commonly seen in genotype B patients, while the BCP mutation predominates in genotype C patients[30]. Our results showed no significant difference in the prevalence of PC/BCP mutation between the two genotypes.
Our findings showed that PC/BCP mutants become predominant through the natural course of HBV infection, in a dynamic and continuous manner. PC/BCP mutants have a survival advantage under host immune selection pressure and accumulate in the HBV population through the course of infection. PC/BCP mutations reduce or eliminate HBeAg production, allowing the virus to avoid cellular and humoral immune attack. Only PC% determined HBeAg status. Log10 PC, log10 BCP, PC% and BCP% might be combined to evaluate disease progression and to guide treatment of chronic hepatitis B.
Hepatitis B early antigen (HBeAg) clearance is considered the endpoint of antiviral treatment for HBeAg-positive chronic hepatitis B. However, follow-up reveals the presence of hepatitis B virus (HBV) DNA in some patients after the loss of HBeAg, since precore and basal core promoter (PC/BCP) mutations may produce a false impression of therapeutic efficacy. So far, PC/BCP mutation detection has been achieved by indirect qualitative detection methods without precise quantification. This study provided an accurate quantitation of PC/BCP mutants throughout the natural course of HBV infection using the Simpleprobe method, which clarified the relationship between PC/BCP mutations and HBeAg status.
The occurrence of PC/BCP mutations has been underestimated by qualitative analysis, which was affected by the presence of dominant wild isolates and nucleotide polymorphisms. To understand the practical significance of HBeAg seroconversion, precise quantification methods can yield profiles of the dynamic process by which PC/BCP-mutated strains become dominant during the natural course of infection.
The authors applied the Simpleprobe method for ultrasensitive quantitation of PC/BCP mutants, which overcame the impact of the dominant wild-type isolates and nucleotide polymorphisms with a two-step PCR method. The detection sensitivity of our method was 0.001%. which, to the best of our knowledge, is the most precise quantification reported to date. Quantitation of PC/BCP mutants throughout the natural history of HBV infection. We also produced a model that combines log10 PC, log10 BCP, PC% and BCP% to evaluate disease progression.
The study revealed the dynamic process by which PC/BCP-mutated strains become dominant during the natural course of infection, and established a practical model that combines log10 PC, log10 BCP, PC% and BCP% to evaluate disease progression in routine clinical practice. The method will provide a more comprehensive and accurate evaluation of disease progression to clinicians.
HBeAg is a soluble antigen secreted from HBV-infected cells, and is a sign of HBV viral replication. HBeAg seroconversion is an indicator of treatment withdrawal. PC/BCP mutants inhibit HBeAg synthesis and may account for the apparent clearance of HBeAg. The HBV Precore G1896A mutation generates a termination codon, eliminating HBeAg synthesis. The BCP A1762T/G1764A double mutation inhibits preC mRNA synthesis, and thus inhibits HBeAg production.
The authors have presented an interesting retrospective study in which they aimed to show that PC/BCP mutant levels may correlate with the natural history of chronic hepatitis B. To show this, the authors enrolled 191 patients and used a Simpleprobe quantitative method to identify PC/BCP mutations in HBV isolates from these patients. The authors concluded that PC/BCP mutations became predominant in a dynamic process through the different phases of HBV infection. They conclude that the status of log10 PC, log10 BCP, PC% and BCP% are useful to evaluate disease progression, while only PC% determines HBeAg status. Overall, the manuscript is well written and is mainly concise in its content. The design of the retrospective study is well performed.
P- Reviewer: Bock CT, Liu ZW, Panduro A, Rodriguez-Frias F, Tamori A S- Editor: Ma YJ L- Editor: Stewart G E- Editor: Wang CH
| 1. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1700] [Cited by in RCA: 1712] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 2. | Lu FM, Zhuang H. Management of hepatitis B in China. Chin Med J (Engl). 2009;122:3-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 3. | Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, Shih WL, Kao JH, Chen DS, Chen CJ. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97:265-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 421] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 4. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of chronic hepatitis B. J Hepatol. 2009;50:227-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1152] [Cited by in RCA: 1155] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 5. | Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;50:661-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2125] [Cited by in RCA: 2170] [Article Influence: 135.6] [Reference Citation Analysis (0)] |
| 6. | Reijnders JG, Perquin MJ, Zhang N, Hansen BE, Janssen HL. Nucleos(t)ide analogues only induce temporary hepatitis B e antigen seroconversion in most patients with chronic hepatitis B. Gastroenterology. 2010;139:491-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 7. | Wong VW, Wong GL, Yan KK, Chim AM, Chan HY, Tse CH, Choi PC, Chan AW, Sung JJ, Chan HL. Durability of peginterferon alfa-2b treatment at 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:1945-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 8. | Senturk H, Baysal B, Tahan V, Zerdali H, Ozaras R, Tabak F, Mert A, Canbakan B, Tabak O, Ozbay G. Long-term effect of interferon therapy in patients with HBeAg positive chronic hepatitis B infection. Dig Dis Sci. 2011;56:208-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 9. | Milich D, Liang TJ. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology. 2003;38:1075-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 297] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 10. | Scaglioni PP, Melegari M, Wands JR. Biologic properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology. 1997;233:374-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 132] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 11. | Laras A, Koskinas J, Hadziyannis SJ. In vivo suppression of precore mRNA synthesis is associated with mutations in the hepatitis B virus core promoter. Virology. 2002;295:86-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Basal core promoter T1762/A1764 and precore A1896 gene mutations in hepatitis B surface antigen-positive hepatocellular carcinoma: a comparison with chronic carriers. Liver Int. 2007;27:1356-1363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 13. | Chen CH, Changchien CS, Lee CM, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN. Combined mutations in pre-s/surface and core promoter/precore regions of hepatitis B virus increase the risk of hepatocellular carcinoma: a case-control study. J Infect Dis. 2008;198:1634-1642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 93] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 14. | Zhang D, Ma S, Zhang X, Zhao H, Ding H, Zeng C. Prevalent HBV point mutations and mutation combinations at BCP/preC region and their association with liver disease progression. BMC Infect Dis. 2010;10:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Xu Z, Ren X, Liu Y, Li X, Bai S, Zhong Y, Wang L, Mao P, Wang H, Xin S. Association of hepatitis B virus mutations in basal core promoter and precore regions with severity of liver disease: an investigation of 793 Chinese patients with mild and severe chronic hepatitis B and acute-on-chronic liver failure. J Gastroenterol. 2011;46:391-400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Fang ZL, Sabin CA, Dong BQ, Ge LY, Wei SC, Chen QY, Fang KX, Yang JY, Wang XY, Harrison TJ. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103:2254-2262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 17. | Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, Chen DS. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193:1258-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 140] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 18. | Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 328] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 19. | Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS, Luketic VA, Terrault N. Prevalence of HBV precore/core promoter variants in the United States. Hepatology. 2003;38:619-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 156] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 20. | Pang A, Yuen MF, Yuan HJ, Lai CL, Kwong YL. Real-time quantification of hepatitis B virus core-promoter and pre-core mutants during hepatitis E antigen seroconversion. J Hepatol. 2004;40:1008-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Waltz TL, Marras S, Rochford G, Nolan J, Lee E, Melegari M, Pollack H. Development of a molecular-beacon assay to detect the G1896A precore mutation in hepatitis B virus-infected individuals. J Clin Microbiol. 2005;43:254-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Ren XD, Lin SY, Wang X, Zhou T, Block TM, Su YH. Rapid and sensitive detection of hepatitis B virus 1762T/1764A double mutation from hepatocellular carcinomas using LNA-mediated PCR clamping and hybridization probes. J Virol Methods. 2009;158:24-29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Yuen MF, Sablon E, Yuan HJ, Hui CK, Wong DK, Doutreloigne J, Wong BC, Chan AO, Lai CL. Relationship between the development of precore and core promoter mutations and hepatitis B e antigen seroconversion in patients with chronic hepatitis B virus. J Infect Dis. 2002;186:1335-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 24. | Qin Y, Zhang J, Mao R, Guo H, Yin Y, Wu X, Weng X, Wands J, Tong S. Prevalence of basal core promoter and precore mutations in Chinese chronic hepatitis B patients and correlation with serum HBeAG titers. J Med Virol. 2009;81:807-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Nie H, Evans AA, London WT, Block TM, Ren XD. Quantitative dynamics of hepatitis B basal core promoter and precore mutants before and after HBeAg seroconversion. J Hepatol. 2012;56:795-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 26. | Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology. 2007;132:1574-1585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 27. | Kitab B, Essaid El Feydi A, Afifi R, Trepo C, Benazzouz M, Essamri W, Zoulim F, Chemin I, Alj HS, Ezzikouri S. Variability in the precore and core promoter regions of HBV strains in Morocco: characterization and impact on liver disease progression. PLoS One. 2012;7:e42891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Yan CH, Zhao CY, Ding H, Peng YQ, Jin PY, Yan L, Zhuang H, Li T. Hepatitis B virus basal core promoter mutations A1762T/G1764A are associated with genotype C and a low serum HBsAg level in chronically-infected HBeAg-positive Chinese patients. Antiviral Res. 2012;96:108-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 29. | Sonneveld MJ, Rijckborst V, Zeuzem S, Heathcote EJ, Simon K, Senturk H, Pas SD, Hansen BE, Janssen HL. Presence of precore and core promoter mutants limits the probability of response to peginterferon in hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2012;56:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 66] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 30. | Chen CH, Lee CM, Hung CH, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. Clinical significance and evolution of core promoter and precore mutations in HBeAg-positive patients with HBV genotype B and C: a longitudinal study. Liver Int. 2007;27:806-815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |