Published online May 28, 2015. doi: 10.3748/wjg.v21.i20.6215
Peer-review started: November 28, 2014
First decision: December 26, 2014
Revised: January 14, 2015
Accepted: February 12, 2015
Article in press: February 13, 2015
Published online: May 28, 2015
Processing time: 184 Days and 1.6 Hours
AIM: To investigate the mechanism underlying the promoting role of CD97 in gastric cancer cell proliferation and invasion.
METHODS: Two types of exosomes released by gastric cancer cells with high (SGC/wt) or low (SGC/kd) CD97 expression were isolated by ultracentrifugation and identified by electron microscopy and western blot analysis. The influences of the two exosomes on gastric cancer cell proliferation and invasion were investigated by proliferation and Matrigel invasion assays. Exosomal miRNAs were subsequently isolated from the two samples and their miRNA profiles were compared via microarray assay analysis. Reverse transcription-quantitative real-time polymerase chain reaction was used to validate the microarray assay. Target genes of the differently expressed microRNAs were predicted based on five independent algorithms and were then subjected to gene oncology enrichment and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis. After identifying the pathway that was the most likely altered, tumor cells were treated with the two exosomes at different concentrations, and the pathway activation was identified through western blot analysis.
RESULTS: Exosomes isolated from SGC/wt cells significantly promoted tumor cell proliferation in a dose-dependent manner in vitro. SGC/wt exosomes also significantly elevated the invasiveness of both SGC/wt (129.67 ± 8.327 vs 76.00 ± 5.292, P < 0.001) and SGC/kd (114.52 ± 9.814 vs 45.73 ± 4.835, P < 0.001) cells as compared to the exosomes released by SGC/kd cells. Microarray assay of the two exosomes revealed that 62 miRNAs were differently regulated with a signal intensity of > 500 and a false discovery rate < 0.05. The following KEGG analysis defined the MAPK signaling pathway as the most likely candidate pathway that regulated tumor cell proliferation and invasion. Through western blot analysis, significant up-regulations of phosphorylated MAPKs, including extracellular signal-regulated kinase, Jun NH2-terminal kinase, and p38 mitogen-activated protein kinase, were detected in a dose-dependent manner in the SGC/wt exosomes treated groups, confirming activation of the MAPK signaling pathway stimulated by SGC/wt exosomes.
CONCLUSION: CD97 promotes gastric cancer cell proliferation and invasion in vitro through exosome-mediated MAPK signaling pathway, and exosomal miRNAs are probably involved in activation of the CD97-associated pathway.
Core tip: CD97, belongs to the epidermal growth factor-seven-transmembrane subfamily, and has been found to promote proliferation and invasion of gastric cancer cells. However, the underlying mechanism is poorly understood. In this study, we found that exosomes isolated from gastric cancer cells with high CD97 expression promoted tumor cell proliferation and invasion. Furthermore, through microarray and western blot analyses, MAPK signaling pathway activation was observed when cells were treated with those exosomes. These results indicated that CD97 promotes gastric cancer cell proliferation and invasion in vitro, at least in part, through the exosome-mediated MAPK signaling pathway, and exosomal miRNAs are probably involved in activation of the CD97-associated pathway.
- Citation: Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang W, Wu YL, Chen L. CD97 promotes gastric cancer cell proliferation and invasion through exosome-mediated MAPK signaling pathway. World J Gastroenterol 2015; 21(20): 6215-6228
- URL: https://www.wjgnet.com/1007-9327/full/v21/i20/6215.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i20.6215
CD97 is a member of the epidermal growth factor-seven-transmembrane subfamily, which belongs to the class B G-protein-coupled receptors[1,2]. Originally, CD97 was found to be expressed by hematopoietic cells[3,4], then abundantly detected in numerous carcinomas including gastric, colorectal, thyroid, esophageal, pancreatic, and oral squamous cell carcinomas[5-10]. In gastric cancer, previous studies have demonstrated that enhanced CD97 expression is associated with the dedifferentiation and aggressiveness of tumor cells and directly correlates with the clinicopathological parameters such as TNM stage[11,12]. Recently, we revealed that CD97 small isoform was associated with increased invasiveness in vitro as well as elevated local growth and metastatic spread of gastric cancer in vivo[13]. However, the mechanisms underlying these promoting roles of CD97 are still poorly understood.
Exosomes are spherical and bilayer vesicles with a diameter of 30-100 nm, which are released extracellularly upon fusion of multivesicular bodies with the plasma membrane[14,15]. Many types of cells including tumor cells, lymphocytes, epithelial cells, and stem cells produce exosomes[15-18]. During the past few years, an increasing number of studies have demonstrated that tumor-derived exosomes play important roles in tumor formation and progression. In breast and gastric cancer, it was observed that tumor exosomes could enhance tumor cell proliferation[19,20]. Moreover, exosomes from breast and pancreatic cancer were found to contribute to the formation of a niche which promoted tumor metastasis[21,22]. Based on these observations, we speculate that tumor exosomes may be involved in the CD97-dependent promotion of biological behaviors of gastric cancer cells.
In addition, exosomes are enriched in proteins, lipids and nucleic acids including miRNA, mRNA and other non-coding RNAs[23,24]. When internalized by recipient cells, exosomes deliver these biological molecules, which still maintain their bioactivity, to other cells, thus mediating intercellular communication[18,25]. One topic of considerable interest is that these transferred exosomal miRNAs may participate in the regulation of biological behaviors of target cells. Yang et al[26] found that macrophages regulated the invasiveness of breast cancer cells through exosome-mediated miRNA delivery. Similarly, Rana et al[27] reported that transferred exosomal miRNAs predominantly modulated pre-metastatic organ cells, thus facilitating metastasis of pancreatic cancer. In gastric cancer, tumor cells also produce a large number of exosomes that contain abundant miRNAs[20,28]. Thus, it will be of great significance to further investigate the possible role of exosomal miRNAs in the CD97-dependent regulation of tumor cell behavior.
In the present study, by employing ultracentrifugation, two types of exosomes from gastric cancer cells with high CD97 expression and cells with stable CD97 knockdown were isolated, and their effects on cell proliferation and invasion were investigated. Moreover, miRNA profiles of the two exosomes were compared by microarray assay and significantly regulated signaling pathways related to exosomes were predicted and verified.
The stomach adenocarcinoma cell line SGC-7901 was purchased from ATCC (http://www.atcc.org). SGC-7901 cells were cultured in RPMI-1640 medium (Genom Biologic, Hangzhou, China) supplemented with 10% fetal bovine serum (FBS, Sijiqing Biologic, Hangzhou, China) and 1% penicillin/streptomycin (Genom Biologic). Cells were incubated in a standard humidified incubator in 5% CO2 at 37 °C and passaged every 3-5 d using trypsin-EDTA (Genom Biologic).
Four miRNA candidate target sequences of the human CD97 gene and the non-silencing sequence (Table 1) were designed and cloned into the pcDNA6.2-GW/EmGFP-miR vector (Invitrogen, Shanghai, China). The SGC-7901 cells were transfected and selected using 4 μg/mL of Blasticidin S HCl (Life Technologies) as described previously[13]. Down-regulation of CD97 was confirmed by reverse transcription polymerase chain reaction (RT-PCR) and western blot analysis.
| Oligo | Sequence (5’-3’) |
| 1F | TGCTGATGACATTCTGGATGGTGACCGTTTTGGCCACTGACTGACGGTCACCACAGAATGTCAT |
| 1R | CCTGATGACATTCTGTGGTGACCGTCAGTCAGTGGCCAAAACGGTCACCATCCAGAATGTCATC |
| 2F | TGCTGTATCTTCAAGGTTTGAGAGCAGTTTTGGCCACTGACTGACTGCTCTCACCTTGAAGATA |
| 2R | CCTGTATCTTCAAGGTGAGAGCAGTCAGTCAGTGGCCAAAACTGCTCTCAAACCTTGAAGATAC |
| 3F | TGCTGAAGAAAGTAGAGCTCCAGGCCGTTTTGGCCACTGACTGACGGCCTGGATCTACTTTCTT |
| 3R | CCTGAAGAAAGTAGATCCAGGCCGTCAGTCAGTGGCCAAAACGGCCTGGAGCTCTACTTTCTTC |
| 4F | TGCTGAAGATGAACAGGCCAAAGACCGTTTTGGCCACTGACTGACGGTCTTTGCTGTTCATCTT |
| 4R | CCTGAAGATGAACAGCAAAGACCGTCAGTCAGTGGCCAAAACGGTCTTTGGCCTGTTCATCTTC |
| Negative control | |
| Negative-F | TGCTGAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT |
| Negative-R | CCTGAAATGTACTGCGTGGAGACGTCAGTCAGTGGCCAAAACGTCTCCACGCGCAGTACATTTC |
Total RNAs from gastric cancer SGC-7901 wild-type cells (SGC/wt), non-silencing vector bearing cells (SGC/ns) and stable CD97 knockdown cells (SGC/kd) cells were extracted using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA synthesis and PCR assay were performed as previously described using primers suitable for amplification of all three CD97 isoforms[13]. PCR products were visualized on a 1% agarose gel with 0.05% ethidium bromide.
Total RNAs including miRNA were extracted from exosomes of SGC/wt and SGC/kd using the miRNeasy Mini kit (Qiagen) following the manufacturer’s instructions. cDNA was synthesized using the miScript II RT Kit (Qiagen). The reverse transcription reaction system included 4 μL of 5 × miScript HiSpec buffer, 2 μL of 10 × miScript nucleics mix, 2 μL of miScript reverse transcriptase mix, 5 μL of template RNA and 7 μL of RNase-free water. The mixture was incubated for 60 min at 37 °C and followed by 95 °C for 5 min to inactivate the reaction. qPCR was performed using the miScript SYBR Green PCR Kit (Qiagen) in an ABI PRISM Stepone Plus Sequence Detection System (Applied Biosystems, Foster City, CA, United States) in accordance with the manufacturer’s protocol. Briefly, 12.5 μL of 2 × QuantiTect SYBR Green PCR master mix, 2.5 μL of 10 × miScript universal primer, 2.5 μL of 10 × miScript primer assay, 2 ng of diluted template cDNA and RNase-free water added to 20 μL were mixed. The mixture were then incubated in a 96-well plate at 95 °C for 15 min, followed by 40 cycles of 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s. The differences in expression of miRNAs between SGC/wt and SGC/kd exosomes were analyzed using the ΔΔCT method and were normalized by RNU6B expression[29].
Cells were cultured until 80% confluent, the growth medium was replaced by serum-free medium, and the medium was collected after 2 d of incubation. Exosomes were then isolated by differential centrifugation as previously described[30]. Briefly, the conditioned medium was centrifuged at 300 g for 10 min at 4 °C to pellet the cells and then 16500 g for 20 min to further remove cells and cell debris. The supernatants were then filtered through 0.20 μm filters to remove particles larger than 200 nm. The exosomes were pelleted by ultracentrifugation (Beckman Coulter, Fullerton, CA, United States) at 120000 g for 70 min at 4 °C, washed in PBS and pelleted by ultracentrifugation at 120000 g for 70 min at 4 °C. The final exosome pellets were resuspended in PBS and stored at -80 °C until use. The total exosomal protein concentration was measured using the Enhanced BCA Protein Assay Kit (Beyotime, China).
Exosomes obtained via differential centrifugation were resuspended in 1% glutaraldehyde in PBS (pH 7.4). A 20 μL drop of suspension was pipetted onto an electron-microscopy grid coated with formvar carbon and allowed to stand for 1 min at room temperature. Excess fluid was removed with a piece of Whatman filter paper. The sample was then stained with 2% phosphotungstic acid for 1 min, allowed to dry under an electric incandescent lamp for 10 min and viewed using Philips Tecnai 10 transmission electron microscopy (Philips, Netherlands). Exosome size was measured by scale bar.
Tumor cell proliferation was determined by 3-(4,5-dimethyl-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays. Cells were plated in 96-well plates at 5 × 103 cells/well and cocultured with exosomes at different concentration for 24 h. At the appropriate time-point, the medium was replaced by 100 μL serum-free medium, and 20 μL MTS solution (Promega, Madison, WI, United States) was added to each well. After incubated in a humidified incubator for another 2 h, the plates were measured at 490 nm using a Microplate Reader (Bio-rad, Hercules, CA, United States). Cell proliferation was expressed using the optical densities obtained at each concentration.
Invasion assays were performed in 24-well TranswellTM chambers (Costar), which were separated by polycarbonate filters with 8 μm pore size between the upper and lower culture compartments. For tumor cell invasion, the upper chamber was coated with Matrigel matrix (0.8 mg/mL, BD Biosciences) before seeding the cells. 1.0 × 105 exosome-treated cells in RPMI-1640 medium were added to the upper chamber and the lower chamber was filled with medium containing 10% FBS. After 36 h incubation in a 5% CO2 atmosphere at 37 °C, the non-invading cells were removed using cotton swabs and the invading cells were fixed and stained with 0.2% crystal violet (Sigma). Invaded cells were counted by light microscopy (Leica) in four separate high-power fields per filter.
Proteins were harvested and resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to a PVDF membrane (Millipore, Billerica, MA, United States). After blocking with 5% non-fat milk for 2 h, the membranes were incubated overnight at 4 °C with antibodies specific against CD97 (Abnova), β-actin, CD9, hsp70, total-p38, -ERK, -JNK or phosphorylated -p38, -ERK, and -JNK (all from Santa Cruz), respectively. Horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit IgG was applied as secondary antibody for 1 h at room temperature. The immunoreactive protein bands were identified by luminescent visualization using an ECL kit (Millipore). Signal intensity was measured using a Bio-Rad XRS chemiluminescence detection system (Bio-Rad).
Total RNA from exosomes was isolated using the miRNeasy Mini Kit (Qiagen, Germany). MicroRNA microarray analysis following the miRbase v20.0 was performed by LC Sciences (Hangzhou, China; http://www.lc-bio.com/). Briefly, total RNA samples were 3’-extended with a poly(A) tail and hybridized overnight at 34 °C on a µParaflo microfluidic chip using 100 L 6 × SSPE buffer (0.90 mol/L NaCl, 60 mmol/L Na2HPO4, 6 mmol/L EDTA, pH 6.8) containing 25% formamide. After hybridization, tag-conjugating Cy3 dye was circulated through the microfluidic chip for dye staining. Fluorescence images were then collected and digitized using Array-Pro image analysis software (Media Cybernetics, Bethesda, MD, United States). Data analysis was started by first subtracting the background and then normalizing the signals using a LOWESS filter (Locally-weighted Regression). The normalized microarray data were validated by reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR).
Five computational prediction algorithms (TargetScan, miRanda, PITA, RNAhybrid and microTar) were used to predict targets of the significant changed miRNAs identified in the microarray analysis. Following a comparison of all datasets, a subset of genes that were targeted by more than four algorithms was generated.
To comprehensively describe the properties of the targets, the putative genes were subjected to gene oncology (GO) enrichment and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis based on DAVID 6.7 software (http://david.abcc.ncifcrf.gov/home.jsp). The Fisher’s exact test and χ2 test were used to select the significant GO categories and signaling pathways. The threshold of significance was defined by the P value, with P < 0.001 or P < 0.05 regarding as significance for GO and KEGG analysis, respectively.
Statistical analysis was performed with the SPSS 19.0 software. Differences were examined using Student’s t-test or one-way analysis of variance. All experiments were performed at least in triplicate and the data are presented as mean ± SD with a P value of 0.05 or less considered statistically significant.
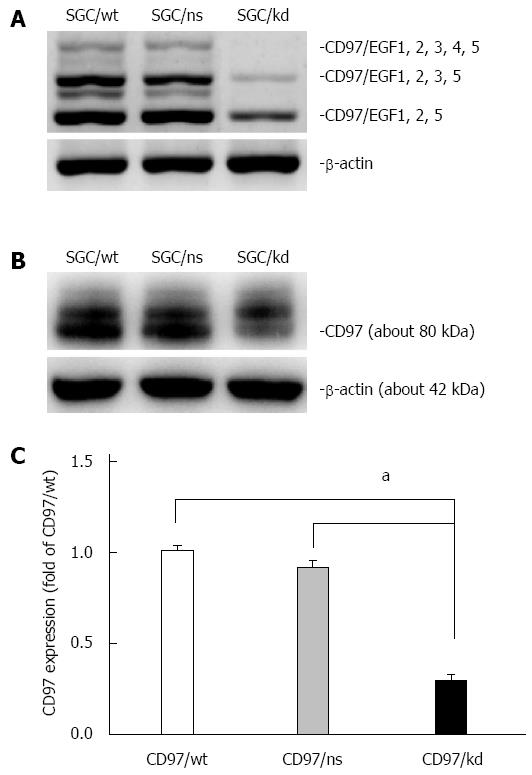
For this purpose, the human gastric carcinoma cell line SGC-7901, which shows relatively high CD97 expression[13,31], was selected for stable transfection with miR-vectors targeting the site of CD97. After transfection, stable clones were selected and CD97 levels were evaluated by RT-PCR and western blot analysis. As shown in Figure 1, the CD97 knockdown cells (SGC/kd), which were generated from candidate 4, displayed a significant loss of CD97 in both mRNA and protein level compared to the wild-type cells (SGC/wt) or the non-silencing vector bearing cells (SGC/ns). Therefore, these cells were selected for subsequent experiments.
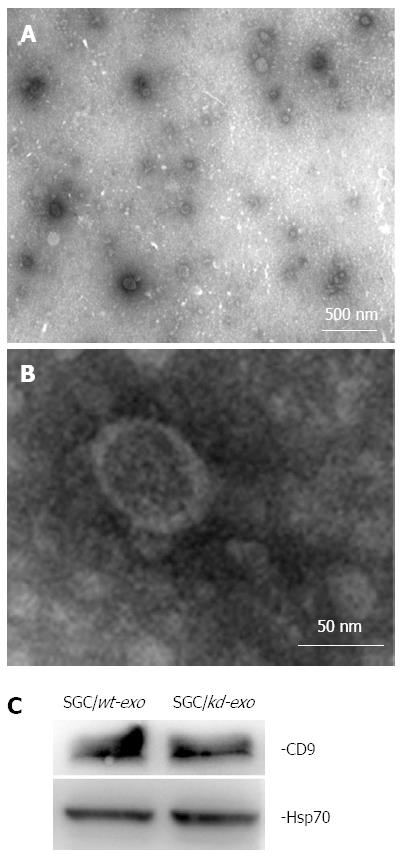
Based on their unique size and density, exosomes released by SGC/wt and SGC/kd cells were isolated by ultracentrifugation and observed by electron microscopy. The isolated exosomes were small closed vesicles limited by a lipid bilayer, and their diameters ranged from 30 to 100 nm under electron microscopy (Figure 2A and B), which was consistent with the reported size of exosomes[15]. In addition, Hsp70 and CD9, the published exosomal markers[32,33], were detected in these membrane vesicles (Figure 2C), further confirming the successful isolation of tumor exosomes.
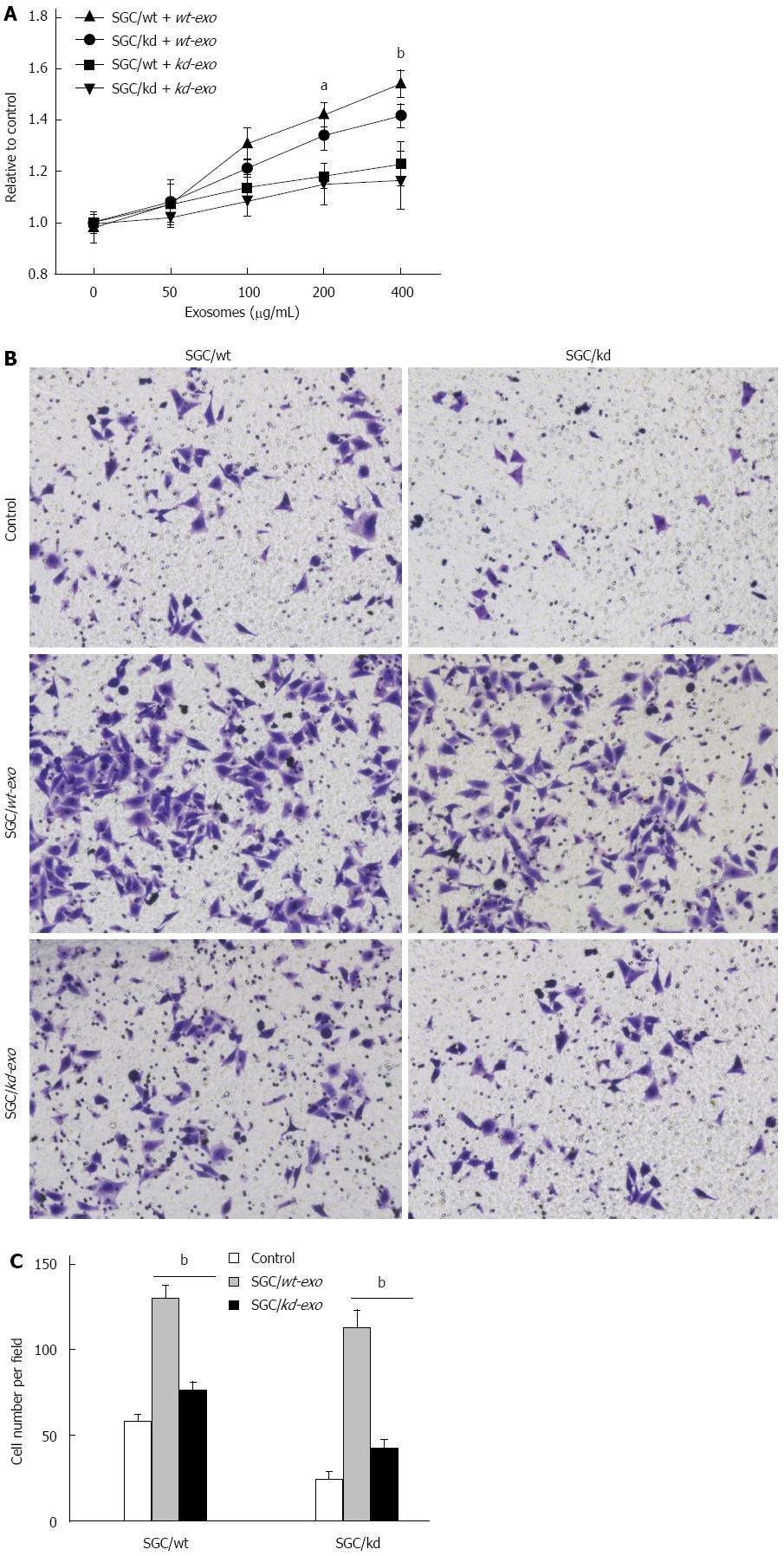
To investigate the effects of the two exosomes on proliferation of gastric cancer cells, we measured the viability of cells treated with different concentrations of exosomes (0, 50, 100, 200 and 400 μg/mL) for 24 h in vitro. The MTS assay showed that SGC/wt-derived exosomes (wt-exo) increased proliferation of both SGC/wt and SGC/kd cells in a dose-dependent manner. However, in the SGC/kd-derived exosomes (kd-exo)-treated groups, no significant difference was observed. It is noteworthy that the promoting effect of wt-exo on SGC/wt cells was not as significant as on SGC/kd clones, indicating that SGC/kd cells were more sensitive to wt-exo stimulation than SGC/wt cells (Figure 3A).
To evaluate the effects of the two types of exosomes on gastric cancer cell invasion, SGC/wt and SGC/kd cells were incubated for 4 h with serum-free medium containing 200 μg/mL of the indicated exosomes or an equal amount of bull serum albumin (BSA) as a control, then seeded on chambers coated with Matrigel matrix. After 36 h incubation in the upper chamber, the cells that had invaded through the Matrigel to the undersurface of Transwell filters were stained and counted. When compared with kd-exo or BSA control, wt-exo significantly enhanced the invasiveness of both SGC/wt and SGC/kd cells, especially SGC/kd cells where invasiveness was 2.5-fold greater than that in SGC/wt cells. However, in the kd-exo stimulated groups, cell invasiveness in SGC/wt cells and SGC/kd cells was not significantly altered (Figure 3B and C).
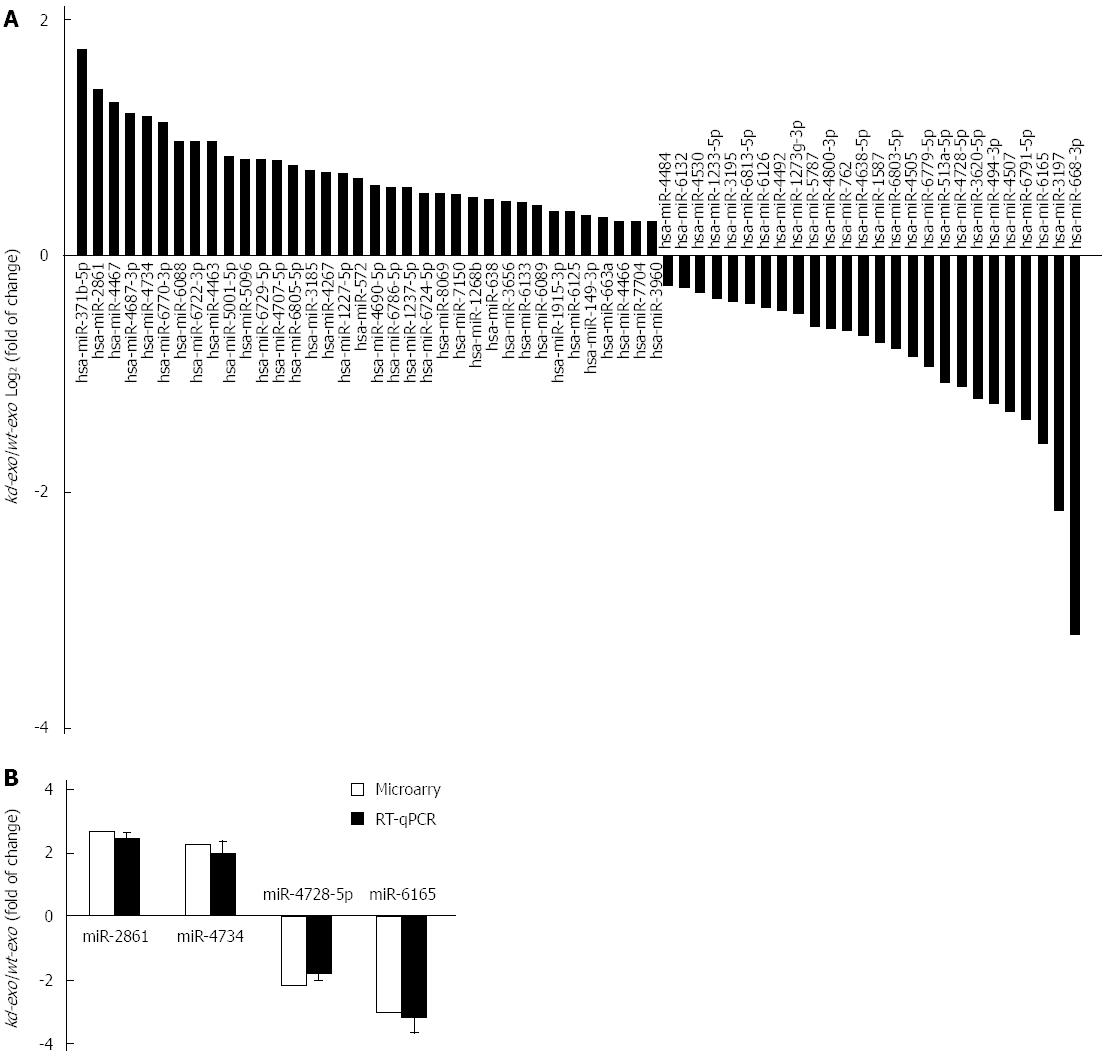
Exosomes are abundant in miRNAs and can be transferred from one cell to another via exosome secretion and internalization. These transferred miRNAs maintain their biological functions in the recipient cells. To determine whether exosomal miRNAs were relevant in cell biological behavior, we compared their miRNA profiles across SGC/wt and SGC/kd exosomes via microarray assay. Analysis of microarray data indicated that a total of 265 detectable miRNA transcripts were present in SGC/wt and SGC/kd exosomes, of which 83 miRNAs had a signal intensity of > 500. In order to reduce the false negative rate, we considered differently regulated miRNAs as those with a fold change of more than 1.5 and a false discovery rate (FDR) of less than 0.05 according to the microarray data. As shown in Figure 4A, 62 miRNAs were differently regulated, of which 36 were up-regulated and 26 were down-regulated in SGC/wt exosomes compared to SGC/kd exosomes. To verify the microarray assay data, four miRNAs displaying either increased (miR-2861, miR-4734) or decreased (miR-4728-5p, miR-6165) expression were selected to perform RT-qPCR analysis. The outcomes confirmed the microarray data and indicated a positive correlation between the quantity of transcripts measured by microarray and the RT-qPCR assay (Figure 4B).
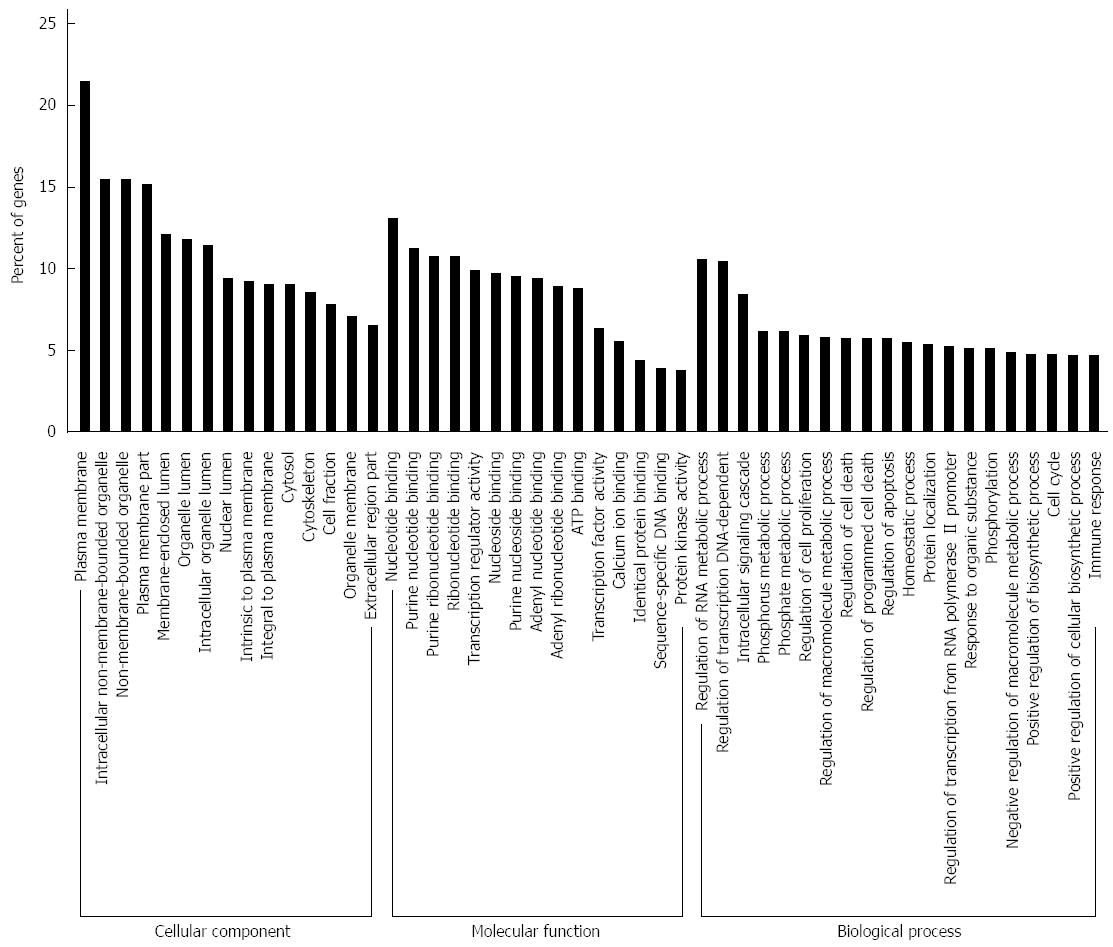
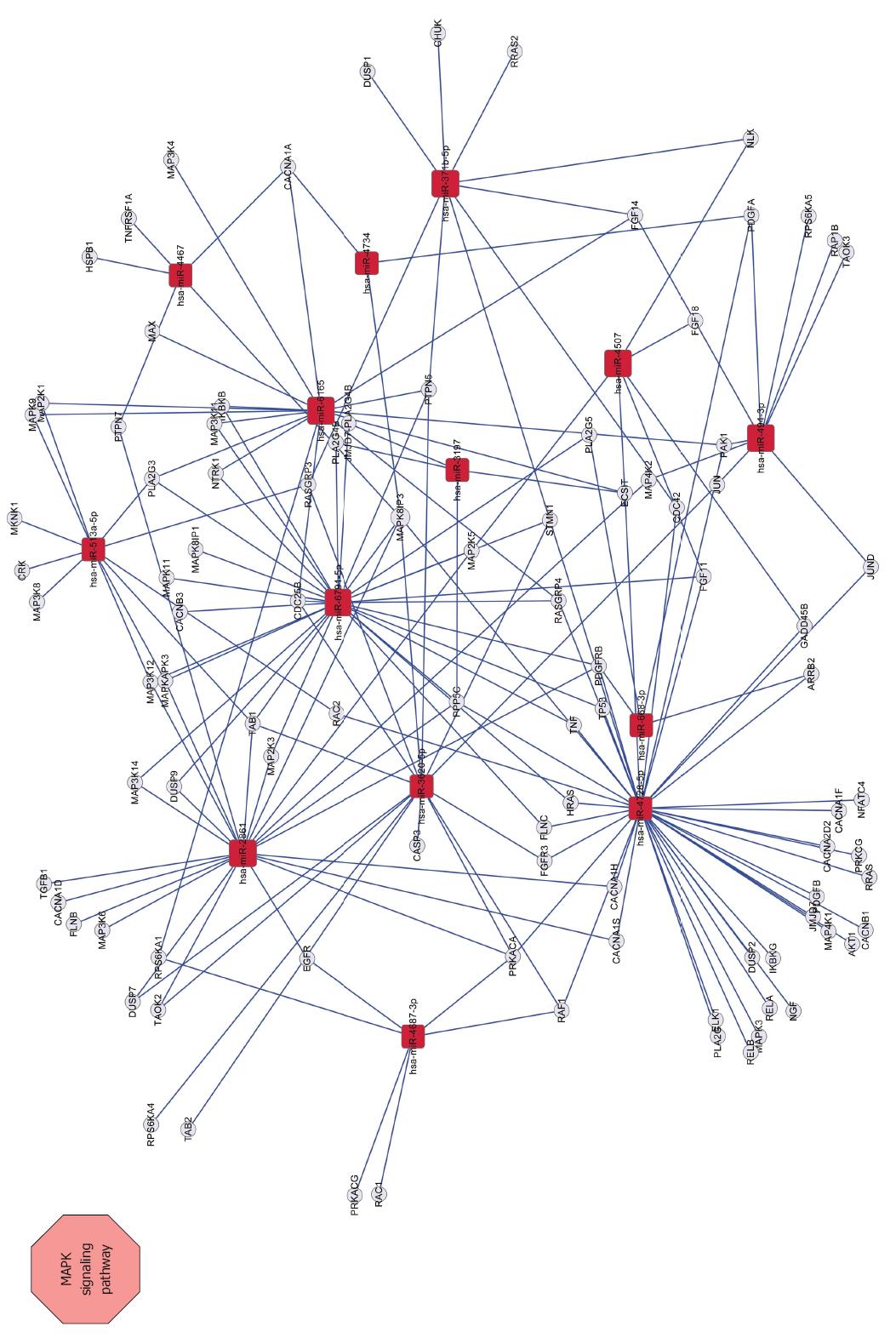
To identify potential target genes of the 62 miRNAs differently expressed between SGC/wt and SGC/kd exosomes, we performed target gene prediction using five established algorithms. A total of 5746 putative targets were identified (data not shown). To better understand the functions of the predicted genes, those targets were subjected to GO functions from three ontologies: cellular component, molecular function and biological process. As shown in Figure 5, the high-enriched GO categories were nucleotide binding, plasma membrane, regulation of RNA metabolic process, regulation of transcription, and intracellular signaling cascade. These categories were mainly involved in cell metabolism, proliferation, signal transduction, apoptosis, and homeostatic processes. Furthermore, these targets were classified according to KEGG functional annotations to identify pathways that might be actively regulated. The results suggested 38 relevant KEGG pathways, and the top three pathways enriched were pathways in cancer, cytokine-cytokine receptor interaction and the MAPK signaling pathway (Table 2). It is worth noting that, among the list of highly-enriched pathways, pathways in cancer were prominent, especially the MAPK signaling pathway. Thus, it is necessary to further investigate whether CD97 promotes tumor cell proliferation and invasion through an exosome-mediated MAPK signaling pathway.
| No. | Pathway ID | Pathway description | Target genes with pathway annotation, n (%) | P value |
| 1 | hsa05200 | Pathways in cancer | 179 (2.37) | 5.50 ×10-3 |
| 2 | hsa04060 | Cytokine-cytokine receptor interaction | 152 (2.01) | 4.00 × 10-4 |
| 3 | hsa04010: | MAPK signaling pathway | 146 (1.93) | 1.13 × 10-2 |
| 4 | hsa04080 | Neuroactive ligand-receptor interaction | 139 (1.84) | 1.81 × 10-2 |
| 5 | hsa04510 | Focal adhesion | 121 (1.60) | 2.00 × 10-4 |
| 6 | hsa04810 | Regulation of actin cytoskeleton | 116 (1.54) | 3.79 × 10-2 |
| 7 | hsa04062 | Chemokine signaling pathway | 107 (1.42) | 5.60 × 10-3 |
| 8 | hsa04020 | Calcium signaling pathway | 95 (1.26) | 5.88 × 10-2 |
| 9 | hsa04142 | Lysosome | 72 (0.95) | 2.3 × 10-3 |
| 10 | hsa04670 | Leukocyte transendothelial migration | 68 (0.90) | 2.36 × 10-3 |
To verify MAPK signaling pathway activation, the expression of three best-characterized MAPKs, including extracellular signal-regulated kinase ERK), Jun NH2-terminal kinase (JNK), and p38 mitogen-activated protein kinase (p38), were determined by western blot analysis. The results revealed that SGC/wt exosomes caused significant up-regulation of phosphorylated ERK, JNK and p38 expression in a dose-dependent manner. On the other hand, although SGC/kd exosomes enhanced the expression of these three phosphorylated proteins, their expression was much weaker compared to the SGC/wt exosomes stimulated groups (Figure 6). These observations demonstrated that exosomes derived from SGC/wt cells, which show high CD97 expression, could activate the MAPK signaling pathway, and activation of this pathway may lead to elevated proliferation and invasion of tumor cells.
Although it is well studied that overexpressed CD97 plays important roles in gastric cancer formation and progression, the mechanisms underlying these functions are poorly investigated. In order to identify the possible mechanism involved, we isolated exosomes from wild-type gastric cancer SGC-7901 cells with high CD97 expression and from SGC-7901 cells with stable CD97 knockdown. We demonstrated that exosomes from wild-type cells, but not from CD97 knockdown cells, significantly promoted tumor cell proliferation and invasion. Furthermore, through microarray and western blot analyses, we found that enhanced proliferation and invasion ability may be induced by exosome-mediated activation of the MAPK signaling pathway.
During the past few years, many studies have demonstrated that tumor-derived exosomes are involved in a wide range of tumor pathophysiological processes in numerous ways, such as promoting tumor growth and metastasis, activating certain signaling pathways, and exerting detrimental effects on the anti-tumor immune system[19-21,34,35]. The present study further suggested that the effects of exosomes on tumor biological behavior depend on their parent cells. It was shown that exosomes from SGC/wt cells with high CD97 expression, significantly increased gastric cancer cell proliferation and invasion; whereas, exosomes from SGC/kd cells with low CD97 expression did not show similar effects. From these observations, it is arguable that CD97 increases the proliferation and invasion of tumor cells, at least in part, by releasing CD97-related exosomes. We speculate that those two exosomes contain different biological molecules, thus biomaterials in SGC/wt exosomes have higher malignant-promoting capacities as compared to that of SGC/kd exosomes; and upon internalization, these biomaterials are transferred to the recipient cells, resulting in elevated malignant behavior.
miRNAs are small, non-coding RNA molecules that regulate the activity of complementary mRNAs and play important roles in a wide range of physiologic and pathologic processes[36]. Microarray analysis of the two exosomes showed significant differences in their exosomal miRNA profiles. The GO enrichments showed that the predicted targets were mainly located in the cell membrane and organelles and significantly involved in cell metabolism, proliferation and signal transduction. The results of KEGG analysis suggested that the MAPK signaling pathway was prominent. Subsequent western blotting verified that MAPK signaling pathway activation was induced by SGC/wt exosomes. Thus, it is reasonable that the promoting role of CD97-related exosomes depends on activation of the MAPK signaling pathway and this activation is probably indirectly induced by the exosomal miRNAs.
In addition to nucleic acids, exosomes contain abundant proteins. In a study conducted by Liang et al[37], thousands of proteins were identified in exosomes released by two ovarian cancer cell lines, OVCAR-3 and IGROV1, and some proteins were tissue specific or associated with tumorigenesis or metastasis. Furthermore, exosomes from human breast and colorectal cancer cells were reported to contain full-length, signaling-competent epidermal growth factor receptor (EGFR) ligands[38]. In that study, the researchers found that exosomes expressing individual EGFR ligands displayed different bioactivities. Specifically, exosomes with higher amphiregulin levels exhibited greater invasive potential than other EGFR ligand-containing exosomes, indicating that exosomes participate in diverse tumor biological activities via certain exosomal proteins. In the present study, although microarray analysis showed that the exosomal miRNAs were involved in MAPK signaling pathway activation, the possibility that exosomal proteins directly regulated the signaling pathway and/or tumor biological behavior could not be excluded. Thus, further studies focused on the relationships between exosomal proteins and the diverse biological behaviors of gastric cancer related to CD97 are required.
Apart from the effects exerted on proliferation and invasion, tumor-derived exosomes were also proved to contribute to the establishment of a pre-metastatic niche, which is a suitable microenvironment in distant metastatic organs generated by primary tumor prior to arrival of metastatic cells[22,39]. Jung et al[22] reported that depending on CD44v6, tumor exosomes prepared the pre-metastatic niche in distant organs, which allowed embedding and growth of highly metastatic pancreatic ASML cells. Recently, Rana et al[27] suggested that exosomal miRNAs were recovered in pre-metastatic organs, where they significantly influenced mRNA translation and predominantly modulated pre-metastatic organs to prepare stromal cells for tumor cell hosting. Thus, tumor exosomes play important roles in the cross-talk between primary tumor and metastatic organs. Our previous study demonstrated that CD97 small isoform facilitated gastric cancer metastasis in an orthotopically implanted mouse model[13]. Therefore, the contribution of CD97 to metastasizing tumor cell settlement is probably facilitated by the formation of the pre-metastatic niche, which may depend on CD97-related exosomes or exosomal miRNAs. However, to the best of our knowledge, there are few studies describing the relationship between CD97-related exosomes and tumor metastasis, which also merits investigation in future in vivo studies.
In conclusion, using exosomes isolated from cells with high and low CD97 expression, we demonstrate that CD97 promotes gastric cancer cell proliferation and invasion in vitro, at least in part, through exosome-mediated MAPK signaling pathway, and that exosomal miRNAs are probably involved in activation of the CD97-associated pathway. Although shown in vitro, these findings should be confirmed in other gastric cancer cell types as well as in animal models to have translational relevance.
Gastric cancer is one of the most common and lethal malignancies worldwide. Understanding gastric cancer formation and progression is of significant importance in the treatment of this carcinoma. As a member of the epidermal growth factor-seven-transmembrane subfamily, CD97 has been found to be overexpressed in the majority of gastric carcinoma tissues and has been demonstrated to promote gastric cancer cell proliferation and invasion. However, the underlying mechanism remains unclear.
During the past few years, an increasing number of studies have demonstrated that tumor-derived exosomes play important roles in tumorigenesis and tumor progression. In gastric cancer, it was observed that tumor exosomes could enhance tumor cell proliferation.
Previous studies have demonstrated that CD97 is involved in gastric cancer progression, but the underlying mechanism is unknown. In the present study, the authors combined CD97 and exosomes, and found that CD97 promotes gastric cancer cell proliferation and invasion in vitro through exosome-mediated MAPK signaling pathway. In addition, they suggested that exosomal miRNAs are involved in activation of the CD97-associated pathway. These findings will be helpful in understanding the role of CD97 in gastric cancer formation and progression.
In understanding the mechanism of CD97-dependent promotion of proliferation and invasion in gastric cancer, the results of this study may provide a future strategy for CD97 as a diagnostic biomarker and/or a way to improve clinical treatment of gastric cancer.
Exosomes are spherical and bilayer vesicles with a diameter of 30-100 nm, which are released extracellularly upon fusion of multivesicular bodies with the plasma membrane. Many types of cells including tumor cells, lymphocytes, epithelial cells, and stem cells can produce exosomes.
It is a well-written and a well-designed study, which showed, in an elegant manner, the potential role of CD97 and MAPK signaling in gastric cancer cell proliferation and invasion. This is thought to be interesting and excellent novel study. These findings must be demonstrated in vivo to have translational relevance.
Biostatistics statement: The statistical methods used in this study were reviewed by Guogang Li from Second Affiliated Hospital, College of Medicine, Zhejiang University.
P- Reviewer: Aoyagi K, Jacome AAA S- Editor: Qi Y L- Editor: Webster JR E- Editor: Ma S
| 1. | Galle J, Sittig D, Hanisch I, Wobus M, Wandel E, Loeffler M, Aust G. Individual cell-based models of tumor-environment interactions: Multiple effects of CD97 on tumor invasion. Am J Pathol. 2006;169:1802-1811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 2. | Aust G, Steinert M, Schütz A, Boltze C, Wahlbuhl M, Hamann J, Wobus M. CD97, but not its closely related EGF-TM7 family member EMR2, is expressed on gastric, pancreatic, and esophageal carcinomas. Am J Clin Pathol. 2002;118:699-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 3. | Martens GJ. Molecular biology of G-protein-coupled receptors. Prog Brain Res. 1992;92:201-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Eichler W, Aust G, Hamann D. Characterization of an early activation-dependent antigen on lymphocytes defined by the monoclonal antibody BL-Ac(F2). Scand J Immunol. 1994;39:111-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Veninga H, Becker S, Hoek RM, Wobus M, Wandel E, van der Kaa J, van der Valk M, de Vos AF, Haase H, Owens B. Analysis of CD97 expression and manipulation: antibody treatment but not gene targeting curtails granulocyte migration. J Immunol. 2008;181:6574-6583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 59] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 6. | Han SL, Xu C, Wu XL, Li JL, Liu Z, Zeng QQ. The impact of expressions of CD97 and its ligand CD55 at the invasion front on prognosis of rectal adenocarcinoma. Int J Colorectal Dis. 2010;25:695-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Steinert M, Wobus M, Boltze C, Schütz A, Wahlbuhl M, Hamann J, Aust G. Expression and regulation of CD97 in colorectal carcinoma cell lines and tumor tissues. Am J Pathol. 2002;161:1657-1667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 8. | Aust G, Eichler W, Laue S, Lehmann I, Heldin NE, Lotz O, Scherbaum WA, Dralle H, Hoang-Vu C. CD97: a dedifferentiation marker in human thyroid carcinomas. Cancer Res. 1997;57:1798-1806. [PubMed] |
| 9. | Hoang-Vu C, Bull K, Schwarz I, Krause G, Schmutzler C, Aust G, Köhrle J, Dralle H. Regulation of CD97 protein in thyroid carcinoma. J Clin Endocrinol Metab. 1999;84:1104-1109. [PubMed] |
| 10. | Jaspars LH, Vos W, Aust G, Van Lier RA, Hamann J. Tissue distribution of the human CD97 EGF-TM7 receptor. Tissue Antigens. 2001;57:325-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Liu Y, Chen L, Peng SY, Chen ZX, Hoang-Vu C. Role of CD97(stalk) and CD55 as molecular markers for prognosis and therapy of gastric carcinoma patients. J Zhejiang Univ Sci B. 2005;6:913-918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Liu Y, Chen L, Peng S, Chen Z, Gimm O, Finke R, Hoang-Vu C. The expression of CD97EGF and its ligand CD55 on marginal epithelium is related to higher stage and depth of tumor invasion of gastric carcinomas. Oncol Rep. 2005;14:1413-1420. [PubMed] |
| 13. | Liu D, Trojanowicz B, Ye L, Li C, Zhang L, Li X, Li G, Zheng Y, Chen L. The invasion and metastasis promotion role of CD97 small isoform in gastric carcinoma. PLoS One. 2012;7:e39989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871-881. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 621] [Cited by in RCA: 616] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 15. | Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1477] [Article Influence: 113.6] [Reference Citation Analysis (0)] |
| 16. | Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 697] [Cited by in RCA: 754] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 17. | Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 408] [Cited by in RCA: 513] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 18. | Zhu W, Huang L, Li Y, Zhang X, Gu J, Yan Y, Xu X, Wang M, Qian H, Xu W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012;315:28-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 314] [Cited by in RCA: 380] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 19. | Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kuroki S, Tanaka M. Purification, characterization and biological significance of tumor-derived exosomes. Anticancer Res. 2005;25:3703-3707. [PubMed] |
| 20. | Qu JL, Qu XJ, Zhao MF, Teng YE, Zhang Y, Hou KZ, Jiang YH, Yang XH, Liu YP. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41:875-880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 251] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 21. | Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 242] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 22. | Jung T, Castellana D, Klingbeil P, Cuesta Hernández I, Vitacolonna M, Orlicky DJ, Roffler SR, Brodt P, Zöller M. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia. 2009;11:1093-1105. [PubMed] |
| 23. | Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1601] [Cited by in RCA: 1792] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 24. | Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248-7259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 750] [Cited by in RCA: 813] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 25. | Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581-593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2703] [Cited by in RCA: 3109] [Article Influence: 194.3] [Reference Citation Analysis (0)] |
| 26. | Yang M, Chen J, Su F, Yu B, Su F, Lin L, Liu Y, Huang JD, Song E. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 491] [Cited by in RCA: 557] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 27. | Rana S, Malinowska K, Zöller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15:281-295. [PubMed] |
| 28. | Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 424] [Cited by in RCA: 481] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 29. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133975] [Article Influence: 5582.3] [Reference Citation Analysis (1)] |
| 30. | Lässer C, Eldh M, Lötvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;e3037. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 293] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 31. | Liu D, Trojanowicz B, Radestock Y, Fu T, Hammje K, Chen L, Hoang-Vu C. Role of CD97 isoforms in gastric carcinoma. Int J Oncol. 2010;36:1401-1408. [PubMed] |
| 32. | Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente-Poirot S. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res. 2010;51:2105-2120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 506] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 33. | Cho JA, Lee YS, Kim SH, Ko JK, Kim CW. MHC independent anti-tumor immune responses induced by Hsp70-enriched exosomes generate tumor regression in murine models. Cancer Lett. 2009;275:256-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 111] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176:1375-1385. [PubMed] |
| 35. | Taylor DD, Gerçel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer. 2005;92:305-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 271] [Cited by in RCA: 305] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 36. | Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1065] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 37. | Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, Yang J, Li H, Gui T, Li X. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80:171-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 252] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 38. | Higginbotham JN, Demory Beckler M, Gephart JD, Franklin JL, Bogatcheva G, Kremers GJ, Piston DW, Ayers GD, McConnell RE, Tyska MJ. Amphiregulin exosomes increase cancer cell invasion. Curr Biol. 2011;21:779-786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 278] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 39. | Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 956] [Article Influence: 59.8] [Reference Citation Analysis (0)] |