Published online May 21, 2015. doi: 10.3748/wjg.v21.i19.5856
Peer-review started: November 19, 2014
First decision: December 26, 2014
Revised: January 15, 2015
Accepted: February 11, 2015
Article in press: February 11, 2015
Published online: May 21, 2015
Processing time: 183 Days and 4.4 Hours
AIM: To investigate the effect of hepatocyte nuclear factor 4α (HNF4α) on the differentiation and transformation of hepatic stellate cells (HSCs).
METHODS: By constructing the recombinant adenovirus vector expressing HNF4α and HNF4α shRNA vector, and manipulating HNF4α expression in HSC-T6 cells, we explored the influence of HNF4α and its induction capacity in the differentiation of rat HSCs into hepatocytes.
RESULTS: With increased expression of HNF4α mediated by AdHNF4α, the relative expression of Nanog was downregulated in HSC-T6 cells (98.33 ± 12.33 vs 41.33 ± 5.67, P < 0.001). Consequently, the expression of G-P-6 and PEPCK was upregulated (G-P-6: 14.34 ± 3.33 vs 42.53 ± 5.87, P < 0.01; PEPCK: 10.10 ± 4.67 vs 56.56 ± 5.25, P < 0.001), the expression of AFP and ALB was positive, and the expression of Nanog, Type I collagen, α-SMA, and TIMP-1 was significantly decreased. HNF4α also downregulated vimentin expression and enhanced E-cadherin expression. The ultrastructure of HNF4α-induced cells had more mitochondria and ribosomes compared with the parental cells. After silencing HNF4α expression, EPCK, E-cadherin, AFP, and ALB were downregulated and α-SMA and vimentin were upregulated.
CONCLUSION: HNF4α can induce a tendency of differentiation of HSCs into hepatocyte-like cells. These findings may provide an effective way for the treatment of liver diseases.
Core tip: Hepatocyte nuclear factor 4α (HNF4α) is an important transcription factor in liver differentiation. When enhancing HNF4α expression in hepatic stellate cell line hepatic stellate cells (HSCs)-T6, the expression of G-P-6, PEPCK, and E-cadherin was upregulated, the expression of Type I collagen, α-SMA, TIMP-1, and vimentin was downregulated, and the induced cells were positive for AFP and ALB. When silencing HNF4α expression with shRNA vector, EPCK and E-cadherin were downregulated and α-SMA and vimentin were upregulated. The results demonstrated that HNF4α can induce a tendency of differentiation of HSCs into hepatocyte-like cells. These findings may provide an effective method for treating liver diseases.
- Citation: Liu K, Guo MG, Lou XL, Li XY, Xu Y, Ji WD, Huang XD, Yang JH, Duan JC. Hepatocyte nuclear factor 4α induces a tendency of differentiation and activation of rat hepatic stellate cells. World J Gastroenterol 2015; 21(19): 5856-5866
- URL: https://www.wjgnet.com/1007-9327/full/v21/i19/5856.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i19.5856
Hepatic stellate cells (HSCs) are stem-like cells that have recently been described as a liver-resident mesenchymal stem cell (MSC) population. This is due to their MSC-related expression profile, which expresses a variety of stem cell markers such as nestin, CD24, CD105, CD133, and c-kit, but can also serve as a progenitor cell population with hepatobiliary characteristics[1,2]. HSCs play crucial roles in liver repair and regeneration after liver injury[3-5]. Transplanted HSCs can home to the injured liver and contribute to tissue regeneration by developing into putative progenitor cells, epithelial cells, and mesenchymal tissues[2]. Under incubation of different cytokines in the culture medium, selected CD133+ cells from fresh rat HSCs have been shown to differentiate into stromal cells, endothelial cells, and hepatocyte-like cells in vitro[6,7]. In the glial fibrillary acidic protein (GFAP)-Cre/green fluorescent protein (GFP) transgenic mouse liver injury animal model, HSCs displayed the capacity to develop into albumin-expressing hepatocytes[5]. Following liver injury, activated HSCs secret cytokines, such as hepatocyte growth factors (HGF), activate hedgehog receptors to promote liver repair and regeneration[8,9]. In contrast, Foxf1+/- mice exhibited abnormal liver repair, diminished HSC activation, and aggravated liver tissue damage following CCl4 injury[10]. Therefore, differentiated HSCs can be used as seed cells in hepatocyte transplantation, and can also secrete cytokines to promote liver repair and regeneration. However, the differentiation capacity of HSCs and related molecular mechanisms remain unclear.
Genetic engineering techniques can regulate important genes in stem cell differentiation. How to directionally induce the differentiation of stem cells into hepatic cells and enhance their biological function by genetic techniques has become a central topic in the treatment of end-stage liver disease by cell transplantation[11,12]. The hepatocyte nuclear factor (HNF) family is a group of important transcription factors in the regulation of liver differentiation. Members of the HNF family include HNF1, HNF3, HNF4, HNF6, and CCAAT/enhancer-binding protein (C/EBP). Of these, HNF4 is a vital transcriptional regulator in the differentiation of liver function, and consists of three types: HNF4α, HNF4β, and HNF4γ. HNF4α regulates the differentiation of hepatocytes, preserves their biological function, and is highly expressed in mature hepatic cells, where it plays a vital role in maintaining the epithelial phenotype of hepatocytes.
The expression of HNF4α in HSCs has been reported to significantly decrease in hepatocyte injury and chronic liver disease[13]. Activated HSCs transform into myofibroblasts and secrete extracellular matrix (ECM)[14]. If the HNF4α expression in HSCs is rescued by transfection, the biological character of HSCs can be reversed, indicating that HNF4α is an important regulatory factor in maintaining the epithelial phenotype of hepatocytes. Upregulated expression of HNF4α can inhibit transformation of HSCs into stromal cells, and promote cell differentiation and regeneration into hepatocytes[15]. All of these findings indicate that HNF4α is a vital regulator that maintains the endothelial cell state of HSCs. Because of the importance of HSCs in the progression of liver fibrosis, we intend to clarify the functions of HNF4α and the mechanism by which it regulates the participation of HSCs in liver fibrosis. The results of this investigation will provide a new direction in which the pathogenesis and prevention of liver fibrosis can be studied. Therefore, in this study, we constructed a recombinant adenovirus expression vector (AdHNF4α) capable of carrying the full-length cDNA of HNF4α. By manipulating HNF4α expression using AdHNF4α, we explored the influence of HNF4α and its induction capacity in the differentiation of HSCs into hepatic cells in the rat HSC-T6 cell line.
The recombinant adenovirus vector AdHNF4α, containing the human HNF4α gene (GenBank: NM_000457.4) expression cassette, and the control adenovirus vector AdGFP, containing the green fluorescent protein (GFP) gene, were recombined as previously based on the recombinant system of adenovirus vector AdEasy and kept in the Department of Gastroenterology, Shanghai Changzheng Hospital (Shanghai, China)[16,17]. The adenovirus AdHNF4α was demonstrated to efficiently express HNF4α factor with biological functions on both human and rat cells[16]. Human embryonic kidney 293 cells (HEK293, Shanghai Institute of Cell biology, Chinese Academy of Sciences) were used as the virus carrier to amplify the recombinant adenovirus, and the adenovirus vector was purified by cesium chloride density gradient centrifugation. The virus titer was measured by the tissue culture infectious dose (TCID50) method (Q Biogene Inc.). The AdHNF4α and AdGFP titers were 1 × 1010 pfu/mL and 3 × 1010 pfu/mL, respectively.
The rat hepatic stellate cell line HSC-T6 was established by Scott L Friedman and William S Blaner’s research group (Department of Medicine, College of Physicians and Surgeons of Columbia University, NY, United States)[18]. The cell line was kindly gifted by Scott L Friedman[16,19] and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/mL of streptomycin, and 100 units/mL of penicillin. These cells were cultured at 37 °C with 5% CO2. The medium was changed once every 1-2 d and the cells were passaged every 2-3 d. For trypsinization, the cells were treated with 0.25% trypsin with 1 mmol/L ethylenediaminetetraacetic acid (EDTA) solution and incubated at 37 °C for 5 min. The reaction was stopped via the addition of Hank’s solution and the cells were collected for subsequent passage. HSC-T6 cells (1 × 105) were transferred into a well of a 6-well plate. After 24 h, the cells adhered to the well and the culture medium was replaced by a serum-free medium. The cells were incubated with AdHNF4α containing supernatant at multiplicities of infection (MOIs) of 50, 100, 200, 400, and 600 pfu/mL for 2 h. The control groups were treated with virus-free supernatant and supernatant containing AdGFP. After the medium was replaced by serum-containing medium, the cells were cultured for an additional 72 h and collected from both the test and control groups.
To calculate the efficiency of virus transfection, the GFP-positive cells in the AdGFP group were visualized by microscopy, and fluorescence antibodies were used to detect the expression of HNF4α in the AdHNF4α and virus-free groups. 4′,6′-Diamidino-2-phenylindole (DAPI) was used for nuclear staining. Goat anti-human HNF4α antibody (1:200), mouse anti-rat Nanog antibody (1:500), FITC-labeled goat anti-mouse IgG (1:500), and Cy3-labeled donkey anti-goat IgG (1:500) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA, United States).
Total RNA was isolated with TRIzol reagent. HNF4α were quantified by RT-PCR. β-actin was used as the control for equal cDNA inputs. Primer sequences for HNF4α are as follows: forward primer, 5′-AAATGTGCAGGTGTTGACCA-3′ and reverse primer, 5′-CACGCTCCTCCTGAAGAATC-3′. The expression of HNF4α at the protein level was quantified by Western blot analysis. Whole-cell extracts were isolated by incubation with 40 μL cell lysis buffer/well for 10 min. The cell lysate was collected and centrifuged, and the supernatant was transferred to an Eppendorf tube and boiled for 10 min. After measuring the protein concentration, 10 μg of the protein was separated by electrophoresis on 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a polyvinylidene fluoride (PVDF) membrane. Horseradish peroxidase (HRP)-labeled donkey anti-goat secondary antibodies (1:2000) were purchased from Rockland Immunochemicals Inc. (Gilbertsville, PA, United States).
To evaluate the effect of HNF4α on directional differentiation, immune phenotype, cell function, and epithelial-mesenchymal transition (EMT) index after transfection, RT-PCR was used to detect expression genes, such as stem cell markers, hepatocyte differentiation markers, EMT-specific markers, and ECM synthesized molecules. The primers used in this study are listed in Table 1. Products of RT-PCR were identified by electrophoresis on 1.5% gel. The gels were scanned by a UV transilluminator. The optical densities of the bands were analyzed by Multi-Analyst software. The expression of G-6-P, PEPCK, Collagen I, α-SMA, and TIMP-1 were detected by Western blotting. Primary antibodies were purchased from Santa Cruz Biotechnology Inc. The cells were fixed in 4% paraformaldehyde and 1% glutaraldehyde, and the EPON 812-embedded ultra-thin sections were prepared for observing cell ultrastructure under transmission electron microscope.
| Classification | Molecules | Sequence (Primer sequences) |
| Stem cell-related | CD133 | F: 5′-TTAATGCAGCACCAGGTACATC-3′ |
| R: 5′-TCGTTGAGCAGGTAGGGAGTAT-3′ | ||
| CD105 | F: 5′-ATCCCTCTGACCAGTGATGTCT-3′ | |
| R: 5′-CTTTTTCCGAAGTGGTGGTAAG-3′ | ||
| Nestin | F: 5′-GAGTGTCGCTTAGAGGTGCAA-3′ R: 5′-TGTCACAGGAGTCTCAAGGGTA-3′ | |
| Hepatocyte | ALB | F: 5′-TGCAGGCTTGCTGTGATAAG-3′ |
| differentiation-related | R: 5′-AGTAATCGGGGTGCCTTCTT-3′ | |
| AFP | F: 5′-TACGTCCCTCCACCATTCTC-3′ | |
| R: 5′-ATCCTGGTCTTTGCAGCACT-3′ | ||
| G-6-P | F: 5′-AAGAGGGCATAGCCCAGACT-3′ | |
| R: 5′-TTGGAAGCTTCGTTGGTCTT-3′ | ||
| PEPCK | F: 5′-CAGGTTCCCAAAGGTCTGAA-3′ | |
| R: 5′-TTCACTAGGGCCTGCTTGAT-3′ | ||
| Fibroblast cell-related | Collagen I | F: 5′-CCGTGACCTCAAGATGTGCC-3′ |
| R: 5′-GCTCATACCTTCGCTTCCAA-3′ | ||
| α-SMA | F: 5′-CCGAGATCTCACCGACTACC-3′ | |
| R: 5′-TCCAGAGCGACATAGCACAG-3′ | ||
| TIMP-1 | F: 5′-TCCCCAGAAATCATCGAGAC-3′ | |
| R: 5′-TCAGATTATGCCAGGGAACC-3′ | ||
| EMT index | Snail | F: 5′-GAGGACAGTGGCAAAAGCTC-3′ |
| R: 5′-TCGGATGTGCATCTTCAGAG-3′ | ||
| Vimentin | F: 5′-AGATCGATGTGGACGTTTCC-3′ | |
| R: 5′-CACCTGTCTCCGGTATTCGT-3′ | ||
| E-cadherin | F: 5′-GGGTTGTCTCAGCCAATGTT-3′ | |
| R: 5′-CACCAACACACCCAGCATAG-3′ | ||
| Target | HNF4α | F: 5′-AAATGTGCAGGTGTTGACCA-3′ |
| R: 5′-CACGCTCCTCCTGAAGAATC-3′ | ||
| Control | β-actin | F: 5′-ACCCACACTGTGCCCATCTATG-3′ |
| R: 5′-AGAGTACTTGCGCTCAGGAGGA-3′ |
Based on the HNF4α sequence (GenBank: NM_000457.4), a specific 19-bp shRNA (5′-CTGTAGCCACACTTTATGA-3′) was designed to bind with exon 3 of HNF4α. The shRNA was carried in the pGensil1.1-shHNF4α vector, which was transfected into HSC-T6 cells, with Western blotting then being used to measure indices that may have been altered by HNF4α interference.
Results were expressed as mean ± SD. Significance was established using analysis of variance by SPSS 11.0 software. Differences were considered significant when P value < 0.05 and very significant when P < 0.01.
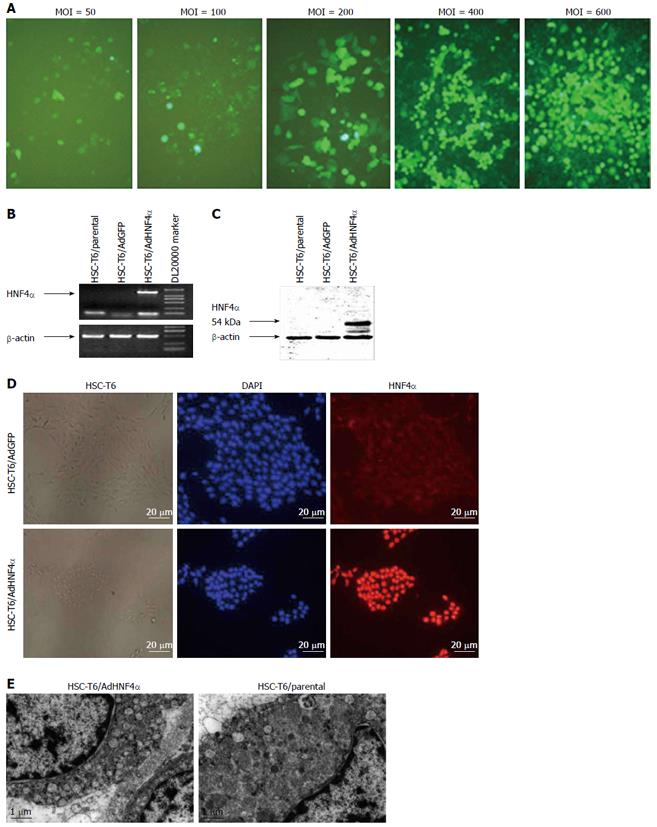
To optimize the transfection efficiency of recombinant adenovirus vector, HSC-T6 cells were transfected with the recombinant adenovirus vector AdGFP at MOIs of 50, 100, 200, 400, and 600 pfu/mL. After 72 h, the transfection efficiency was considered proportional to the ratio of GFP-positive cells to the total number of cells. The transfection efficiencies for the different MOIs used were 20%, 42%, 59%, 78%, and 90%, respectively (Figure 1A). Based on these results, 600 pfu/mL was the MOI used for further experiments.
After HSC-T6 cells were transfected with adenovirus AdHNF4α, RT-PCR and Western blot analysis were used to measure the expression of HNF4α. The results revealed that AdHNF4α can mediate highly efficient expression of HNF4α in HSC-T6 (Figures 1B and C). In order to determine the specificity of the adenovirus vector, HSC-T6 cells were transfected with two vectors: AdHNF4α and AdGFP. Immunostaining results showed that HNF4α was only expressed in the nuclei of cells in the AdHNF4α group, and not in the AdGFP group (Figure 1D). Under electron microscope, HNF4α-induced cells had more mitochondria and ribosomes when compared with the parental cells (Figure 1E).
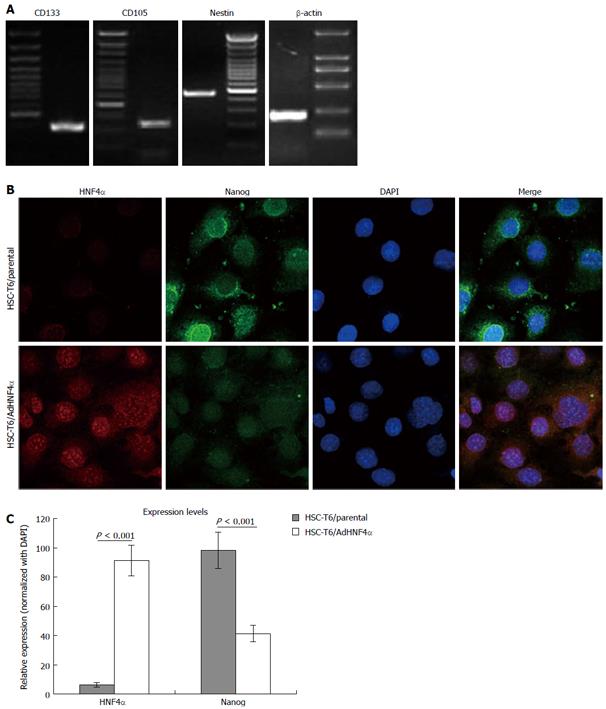
In order to measure the stemness of HSCs, the expression levels of stem cell-related genes such as CD133, CD105, and nestin were measured using RT-PCR. The results revealed that the three molecules were positively expressed (Figure 2A), indicating that the rat HSCs were progenitor cells in the liver.
After transfection of adenovirus AdHNF4α, the expression of HNF4α and traditional stem cell marker Nanog was observed by co-focal immunofluorescent staining. With the increased expression of HNF4α, the relative expression of Nanog was downregulated from 98.33 ± 12.33 to 41.33 ± 5.67 (P < 0.001; Figures 2B and C).
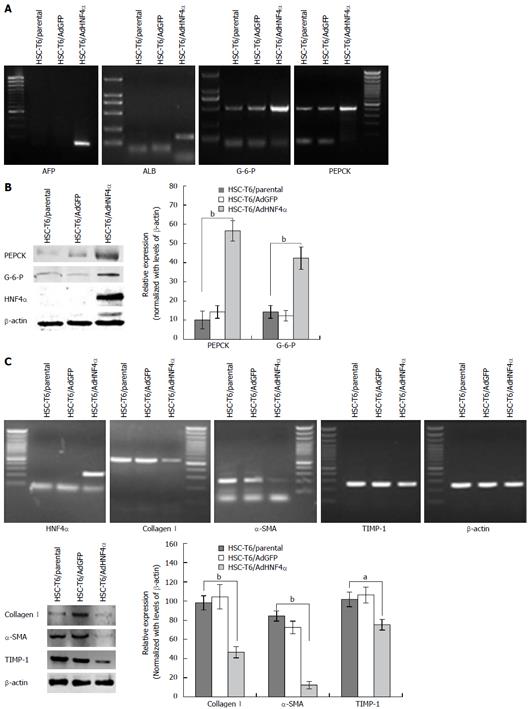
To investigate the role of HNF4α in HSC cell differentiation, we detected molecular markers related to the differentiation of HSCs into hepatocytes by RT-PCR and Western blot analysis. These molecules included the functional genes involved in the differentiation of HSCs to hepatocytes and fibroblasts. The functional genes of hepatocytes (G-P-6 and PEPCK) were expressed at much higher levels in the AdHNF4α group than in the control group. The expression of AFP and ALB was detected in the AdHNF4α group (Figures 3A and B). Furthermore, the expression levels of Collagen I, α-SMA, and TIMP-1 were significantly decreased in the AdHNF4α group compared with the control group (Figure 3C).
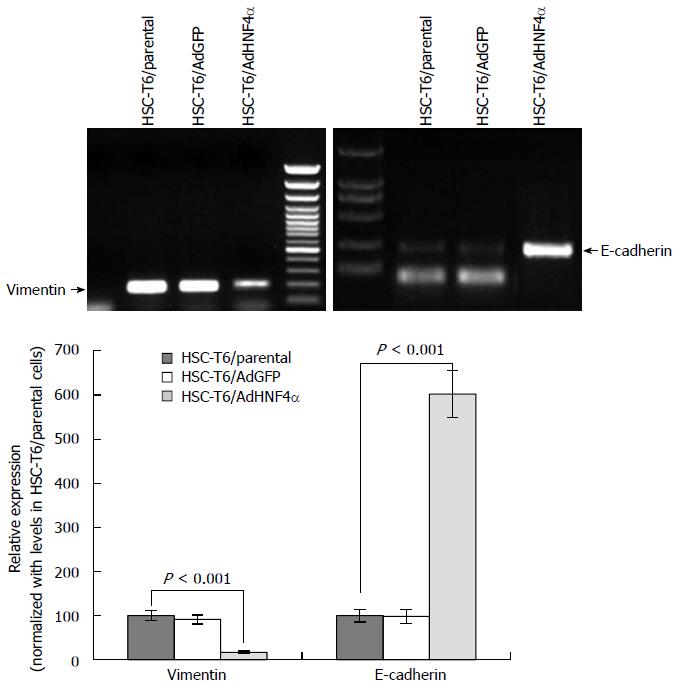
To investigate the phenotypic character of HSCs after HNF4α transfection, we tested the EMT indicators by RT-PCR. As compared with the AdGFP control group, HNF4α obviously downregulated the expression of the mesenchymal phenotypic gene vimentin and significantly enhanced the expression of the epithelial phenotypic gene E-cadherin (Figure 4).
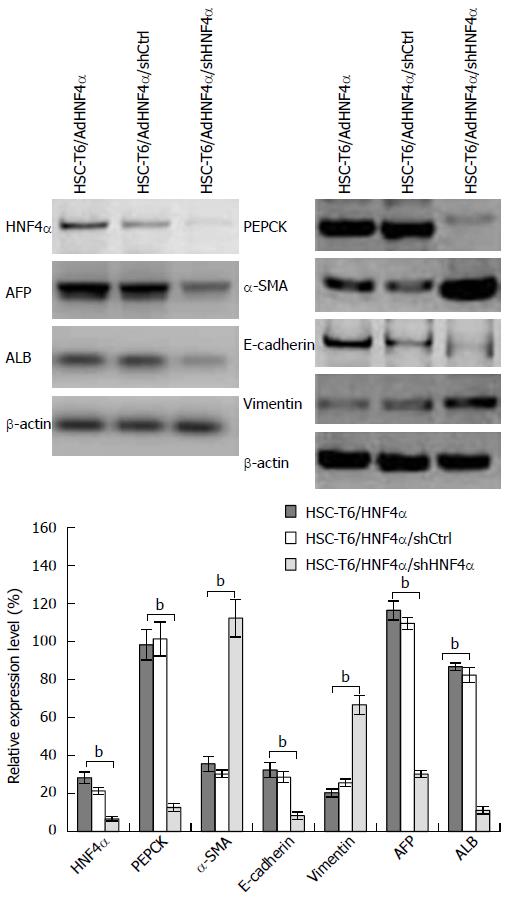
To investigate the biological characteristics of HSCs in HNF4α knockdown, the pGensil1.1-shHNF4α vector was transfected into HNF4α-positive HSC-T6 cells. Western blotting analysis revealed that silencing of HNF4α expression resulted in obvious changes to many genes. With the decrease of HNF4α expression, AFP, ALB, PEPCK, and E-cadherin were downregulated, while α-SMA and vimentin were upregulated (Figure 5).
HSCs play an important role in the regulation of liver injury repair and in the development of liver fibrosis. Despite intensive research into the biological and pathophysiological role of HSCs in fibrogenesis, the states of HSCs in the different stages of fibrogenesis are still a matter of debate. In the quiescent state, HSCs exhibit properties of stem cells in the liver. Following liver injury, HSCs become activated. Their potential to differentiate into epithelial or hepatocyte lineages demonstrates their important functions during liver regeneration. HSC activation may be stimulated by most causes of liver injury, with injured hepatocytes and activated Kupffer cells being considered as the leading cause of HSC activation. Injured hepatocytes release a wide array of soluble mediators, including lipid peroxide, hepatotoxin, and reactive oxygen species (ROS). These mediators can strongly activate HSCs and stimulate the potential of these cells in fibrogenesis[20]. Meanwhile, the homeostatic states between the activation and quiescence of HSCs can be regulated by HNF4α.
HNF4α is a nuclear transcription factor that binds to DNA as a homodimer. It can activate the expression of target genes by adjusting the structure of chromosomes and depolymerizing them. The results of chromatin immunoprecipitation (ChIP) showed that HNF4α could combine with the promoter regions of up to 12% of intracellular genes, 80% of which are combined with RNA polymerase II. Thus, we can infer that HNF4α controls a large proportion of active transcriptional genes in the liver[21,22]. The gain or loss of HNF4α function can lead to the inhibition of many genes at different stages of liver development. By comparing the different gene expression profiles between HNF4α-knocked out mice and normal mice with a gene chip array, it was found that silencing HNF4α expression results in a decrease of liver function. Possible mechanisms for this may be through causing liver developmental disorders by destroying cellular close connections, adhesion connections, and gap junctions, as well as affecting the adhesion molecules between desmosomes and the cell matrix and affecting the polarity of epithelial cells and cytoskeleton proteins[23]. Additionally, loss of HNF4α function can result in cell phenotypic abnormalities, thereby affecting liver cell phenotypes and important liver functions such as liver cell metabolism, albumin synthesis, and drug detoxification[22-24]. Inducible expression of HNF4α by oncostatin M (OSM) can promote differentiation of hepatocytes and enhance the functions of hepatocytes[25]. Moreover, upregulated HNF4α can induce hepatoma stem cells to differentiate into mature hepatocytes, inhibit the proliferation of cancer cells, and reverse the differentiation of cancer cells into a differentiated state. These results demonstrate that upregulation of HNF4α is a promising candidate for the treatment of liver cancer[26].
In the mature liver, HNF4α expression is induced when oval cells differentiate into hepatocytes, suggesting its pivotal role in the differentiation and proliferation of hepatocytes from oval cells. However, very few studies have investigated the regulation and function of HNF4α in HSCs. Previous studies have shown that the expression of HNF4α is significantly decreased in liver injury and chronic liver diseases of different causes (e.g., viral hepatitis)[27]. Decreased HNF4α expression can induce EMT in hepatocytes and HSCs[28,29]. EMT is a phenotypic change of epithelial cells induced by various cytokines, such as transforming growth factor TGF-β, following which the epithelial cells exhibit properties of mesenchymal cells. Following EMT, HSCs proliferate rapidly, transform to myofibroblast cells, generate ECM, eliminate lipid droplets, and positively induce the expression of α-SMA and Snail[18]. When HNF4α is rescued by exogenous gene transduction, EMT can be reversed to mesenchymal-epithelial transition (MET)[14,30,31]. This observation tells us that HNF4α is an important regulator for maintaining the epithelial phenotype of HSCs. HNF4α not only inhibits the mesenchymal phenotype of HSCs, but also promotes the differentiation of liver stem cells and the regeneration of hepatocytes[15]. Because of the importance of HSCs in liver fibrosis, understanding the function of HNF4α in regulating HSCs to participate in liver fibrosis will provide a new approach to studying the pathogenesis and prevention of liver fibrosis.
In this study, we sorted and cultured the HSC-T6 cell line. In a quiescent state, the HSCs showed stem cell characteristics, as evidenced by the expression of stem cell markers (CD133, Nanog, nestin, and CD105). To investigate the regulatory role of HNF4α in hepatocyte differentiation, we transfected HNF4α gene in HSCs and upregulated its expression. After transfection with AdHNF4α, the expression levels of HNF4α and E-cadherin was increased while vimentin expression levels decreased. Moreover, the HNF4α-induced HSC-T6 cells showed morphological changes that led to more mitochondria and ribosomes. These results suggested that HNF4α is an important transcriptional factor in maintaining the epithelial phenotype and facilitating the EMT of HSCs. In addition, HNF4α obviously upregulated the expression of genes related to hepatocyte function, such as ALB, AFP, G-6-P, and PEPCK, illustrating that HNF4α can induce a tendency of differentiation of HSCs to hepatocyte-like cells. Meanwhile, the transduction of HNF4α downregulated the expression of α-SMA, type I collagen, and TIMP-1, demonstrating that HNF4α inhibits the differentiation of HSCs to fibroblast cells.
In conclusion, HSCs have a high capacity of proliferation and a low level of differentiation. HNF4α can induce the expression of important epithelial cell genes in HSCs, promote HSC differentiation to hepatocyte-like cells, and inhibit HSC differentiation to the mesenchymal phenotype. All these observations suggest that HNF4α can induce a tendency of differentiation of HSCs into hepatocyte-like cells. The findings of this research may provide an effective method for treating liver diseases.
Hepatic stellate cells (HSCs) are stem-like cells that play a crucial role in liver repair and regeneration. However, the differentiation capacity of HSCs and the related molecular mechanisms remain unclear. Hepatocyte nuclear factor 4α (HNF4α) is highly expressed in mature hepatic cells, lowly expressed in HSCs, and the upregulated expression of HNF4α can maintain the endothelial cell state of HSCs, thereby demonstrating that HNF4α may play a vital role in promoting the differentiation and regeneration of HSCs into hepatocytes.
The HNF family is a group of important transcription factors in the regulation of liver differentiation, in which HNF4α can regulate the differentiation of hepatocytes and preserve their biological function. Due to the importance of HSCs in liver fibrosis, this study has clarified the functions of HNF4α and the mechanism by which it regulates the participation of HSCs in liver repair. By constructing an HNF4α-expressing adenovirus vector and manipulating HNF4α expression in rat HSC-T6 cells, the influence of HNF4α and its induction capacity in the differentiation of HSCs into hepatic cells was explored.
HNF4α is a nuclear transcription factor that controls a large proportion of active transcriptional genes in the liver. HNF4α not only inhibits the mesenchymal phenotype of HSCs, but also promotes the differentiation of liver stem cells and the regeneration of hepatocytes. To investigate the regulatory role of HNF4α in hepatocyte differentiation, the authors upregulated HNF4α expression in HSCs by transfection of HNF4α gene. The results showed that HNF4α can promote differentiation of HSCs to hepatocyte-like cells and inhibit differentiation of HSCs to the mesenchymal phenotype by regulating some target genes involved in HSC differentiation, such as Nanog, α-SMA, collagen I, TIMP-1, E-cadherin, and vimentin.
HNF4α can induce a tendency of differentiation of HSCs into mature hepatocytes, which may provide an effective method for treating liver diseases.
HSCs were positive for the stem cell-related markers (CD133, CD105, Nanog, and nestin), indicating that the HSCs were progenitor cells in the liver. The increased expression of G-P-6, PEPCK, AFP, ALB, and E-cadherin indicated that the HNF4α-induced cells had a tendency of differentiation into hepatocytes, and the decreased expression of collagen I, α-SMA, TIMP-1, and vimentin demonstrated that the HNF4α-induced cells lost the capacity to differentiate towards mesenchymal cells.
The authors presented interesting results suggesting that a mesenchymal to epithelial transition occurs in hepatic stellate cells following forced expression of HNF4α after infection with an adenovirus vector. Their conclusion was that novel donor cells should be provided for cell transplantation.
P- Reviewer: Kojima T, Kwon S, Michalopoulos GK, Mizuguchi T S- Editor: Ma YJ L- Editor: Rutherford A E- Editor: Zhang DN
| 1. | Yovchev MI, Xue Y, Shafritz DA, Locker J, Oertel M. Repopulation of the fibrotic/cirrhotic rat liver by transplanted hepatic stem/progenitor cells and mature hepatocytes. Hepatology. 2014;59:284-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | Kordes C, Sawitza I, Götze S, Herebian D, Häussinger D. Hepatic stellate cells contribute to progenitor cells and liver regeneration. J Clin Invest. 2014;124:5503-5515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 3. | Swiderska-Syn M, Syn WK, Xie G, Krüger L, Machado MV, Karaca G, Michelotti GA, Choi SS, Premont RT, Diehl AM. Myofibroblastic cells function as progenitors to regenerate murine livers after partial hepatectomy. Gut. 2014;63:1333-1344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 4. | Chan EP, Wells RG. Today’s hepatic stellate cells: not your father’s sternzellen. Hepatology. 2007;45:1326-1327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 5. | Yang L, Jung Y, Omenetti A, Witek RP, Choi S, Vandongen HM, Huang J, Alpini GD, Diehl AM. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells. 2008;26:2104-2113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 175] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 6. | Qiu Q, Hernandez JC, Dean AM, Rao PH, Darlington GJ. CD24-positive cells from normal adult mouse liver are hepatocyte progenitor cells. Stem Cells Dev. 2011;20:2177-2188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 7. | Kordes C, Sawitza I, Müller-Marbach A, Ale-Agha N, Keitel V, Klonowski-Stumpe H, Häussinger D. CD133+ hepatic stellate cells are progenitor cells. Biochem Biophys Res Commun. 2007;352:410-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 176] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 8. | Hu Z, Evarts RP, Fujio K, Marsden ER, Thorgeirsson SS. Expression of hepatocyte growth factor and c-met genes during hepatic differentiation and liver development in the rat. Am J Pathol. 1993;142:1823-1830. [PubMed] |
| 9. | Yang L, Wang Y, Mao H, Fleig S, Omenetti A, Brown KD, Sicklick JK, Li YX, Diehl AM. Sonic hedgehog is an autocrine viability factor for myofibroblastic hepatic stellate cells. J Hepatol. 2008;48:98-106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 164] [Cited by in RCA: 162] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 10. | Kalinichenko VV, Bhattacharyya D, Zhou Y, Gusarova GA, Kim W, Shin B, Costa RH. Foxf1 +/- mice exhibit defective stellate cell activation and abnormal liver regeneration following CCl4 injury. Hepatology. 2003;37:107-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 108] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 11. | Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 456] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 12. | Ishizaka S, Shiroi A, Kanda S, Yoshikawa M, Tsujinoue H, Kuriyama S, Hasuma T, Nakatani K, Takahashi K. Development of hepatocytes from ES cells after transfection with the HNF-3beta gene. FASEB J. 2002;16:1444-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 82] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 13. | Cicchini C, Filippini D, Coen S, Marchetti A, Cavallari C, Laudadio I, Spagnoli FM, Alonzi T, Tripodi M. Snail controls differentiation of hepatocytes by repressing HNF4alpha expression. J Cell Physiol. 2006;209:230-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Jiang JX, Török NJ. Liver Injury and the Activation of the Hepatic Myofibroblasts. Curr Pathobiol Rep. 2013;1:215-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Walesky C, Gunewardena S, Terwilliger EF, Edwards G, Borude P, Apte U. Hepatocyte-specific deletion of hepatocyte nuclear factor-4α in adult mice results in increased hepatocyte proliferation. Am J Physiol Gastrointest Liver Physiol. 2013;304:G26-G37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 16. | Yue HY, Yin C, Hou JL, Zeng X, Chen YX, Zhong W, Hu PF, Deng X, Tan YX, Zhang JP. Hepatocyte nuclear factor 4alpha attenuates hepatic fibrosis in rats. Gut. 2010;59:236-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 141] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 17. | Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, Deng X, Zhang JP, Han ZG. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology. 2008;48:1528-1539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 165] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 18. | Vogel S, Piantedosi R, Frank J, Lalazar A, Rockey DC, Friedman SL, Blaner WS. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882-893. [PubMed] |
| 19. | Hu PF, Zhu YW, Zhong W, Chen YX, Lin Y, Zhang X, Yin C, Yue HY, Xie WF. Inhibition of plasminogen activator inhibitor-1 expression by siRNA in rat hepatic stellate cells. J Gastroenterol Hepatol. 2008;23:1917-1925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Nieto N. Oxidative-stress and IL-6 mediate the fibrogenic effects of [corrected] Kupffer cells on stellate cells. Hepatology. 2006;44:1487-1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 189] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 21. | Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1027] [Cited by in RCA: 1035] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 22. | Parviz F, Matullo C, Garrison WD, Savatski L, Adamson JW, Ning G, Kaestner KH, Rossi JM, Zaret KS, Duncan SA. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34:292-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 479] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 23. | Hertz R, Kalderon B, Byk T, Berman I, Za’tara G, Mayer R, Bar-Tana J. Thioesterase activity and acyl-CoA/fatty acid cross-talk of hepatocyte nuclear factor-4{alpha}. J Biol Chem. 2005;280:24451-24461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Watt AJ, Garrison WD, Duncan SA. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology. 2003;37:1249-1253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 234] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 25. | Lázaro CA, Croager EJ, Mitchell C, Campbell JS, Yu C, Foraker J, Rhim JA, Yeoh GC, Fausto N. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology. 2003;38:1095-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 146] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 26. | Naiki T, Nagaki M, Shidoji Y, Kojima H, Moriwaki H. Functional activity of human hepatoma cells transfected with adenovirus-mediated hepatocyte nuclear factor (HNF)-4 gene. Cell Transplant. 2004;13:393-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Nagy P, Bisgaard HC, Thorgeirsson SS. Expression of hepatic transcription factors during liver development and oval cell differentiation. J Cell Biol. 1994;126:223-233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 180] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 28. | Friedman SL. Seminars in medicine of the Beth Israel Hospital, Boston. The cellular basis of hepatic fibrosis. Mechanisms and treatment strategies. N Engl J Med. 1993;328:1828-1835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 793] [Cited by in RCA: 886] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 29. | Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis. 2001;21:311-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 562] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 30. | Yang M, Li SN, Anjum KM, Gui LX, Zhu SS, Liu J, Chen JK, Liu QF, Ye GD, Wang WJ. A double-negative feedback loop between Wnt-β-catenin signaling and HNF4α regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J Cell Sci. 2013;126:5692-5703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 31. | Yao D, Peng S, Dai C. The role of hepatocyte nuclear factor 4alpha in metastatic tumor formation of hepatocellular carcinoma and its close relationship with the mesenchymal-epithelial transition markers. BMC Cancer. 2013;13:432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |