Published online Apr 28, 2015. doi: 10.3748/wjg.v21.i16.5039
Peer-review started: September 25, 2014
First decision: October 14, 2014
Revised: October 30, 2014
Accepted: January 16, 2015
Article in press: January 16, 2015
Published online: April 28, 2015
Processing time: 216 Days and 7 Hours
AIM: To investigate the clinical implications of hepatitis B virus (HBV) preS1 deletion.
METHODS: We developed a fluorescence resonance energy transfer-based real-time polymerase chain reaction (RT-PCR) that can detect four genotypes (wild type, 15-bp, 18-bp and 21-bp deletion). The PCR method was used in two cohorts of Korean chronic HBV subjects with genotype C infections. Cohort I included 292 chronic HBV subjects randomly selected from Cheju National University Hospital (Jeju, South Korea) or Seoul National University Hospital (Seoul, South Korea), and cohort II included 90 consecutive chronic HBV carriers recruited from Konkuk University Hospital (Seoul, South Korea); the cohort II patients did not have hepatocellular carcinoma or liver cirrhosis.
RESULTS: The method proposed in this study identified 341 of 382 samples (89.3%). Deletion variants were identified in 100 (29.3%) of the 341 detected samples. In both cohorts, the subjects with deletions had a significantly higher Hepatitis B virus e antigen (HBeAg)-positive seroprevalence [cohort I, wild (51.0%) vs deletion (75.0%), P < 0.001; cohort II, wild (69.2%) vs deletion (92.9%), P = 0.002] and higher HBV DNA levels [cohort I, wild (797.7 pg/mL) vs deletion (1678.9 pg/mL), P = 0.013; cohort II, wild (8.3 × 108 copies/mL) vs deletion (2.2 × 109 copies/mL), P = 0.049], compared to subjects with wild type HBV.
CONCLUSION: HBV genotype C preS1 deletion may affect disease progression in chronic HBV subjects through an extended duration of HBeAg seropositive status and increased HBV replications.
Core tip: Our data indicate that the hepatitis B virus (HBV) genotype C preS1 deletion might significantly contribute to disease progression in chronic HBV subjects through an extended duration of Hepatitis B virus e antigen seropositive status and increased HBV replication. This study provides novel insight into the greater infectivity and virulence of genotype C compared with other genotypes. In addition, the fluorescence resonance energy transfer-based real-time polymerase chain reaction used to detect the preS1 deletion shows promise for the earlier prediction of the risk of liver disease progression in chronic HBV subjects.
- Citation: Lee SA, Kim KJ, Kim H, Choi WH, Won YS, Kim BJ. Hepatitis B virus preS1 deletion is related to viral replication increase and disease progression. World J Gastroenterol 2015; 21(16): 5039-5048
- URL: https://www.wjgnet.com/1007-9327/full/v21/i16/5039.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i16.5039
Hepatitis B virus (HBV) infection is a global health problem, and more than 350 million people are chronic carriers of the virus[1]. The infection is associated with a wide spectrum of clinical manifestations ranging from acute or fulminant hepatitis to various forms of chronic infection, including asymptomatic carrier, chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC)[2]. Despite the recent significant decline in HBV chronic subjects thanks to a successful HBsAg vaccination program, South Korea is still recognized as an endemic area of HBV infection[3]. It has also been reported that genotype C2 is exclusively prevalent in South Korea[4], which is known to be more prone to mutations and is related to more severe liver diseases, as well as having a lower antiviral response compared with genotype B[5]. Furthermore, the high prevalence of basal core promoter (BCP) double mutations and the presence of distinct immune responses against HBV proteins in the Korean population could lead to the generation of distinct HBV variants that are not (or rarely) encountered in other areas[6-15] and that result in distinct clinical manifestations in Korean chronic HBV subjects[16].
The preS proteins might have a pivotal function in virus assembly and attachment to hepatocytes[17,18]. Infections with preS/S HBV variants, particularly deletion variants, correlated with the most progressive forms of liver disease and HCC. Both preS1 and preS2 mutations reportedly contribute to hepatocarcinogenesis by inducing an endoplasmic reticulum (ER) stress pathway that is caused by the accumulation of large hepatitis B surface (LHBs) proteins in the ER or by altering the transactivating capacity[19-21].
Recently, we introduced two types of preS1 mutations that are related to the progression of liver diseases: W4P/R[11] and preS1 start codon deletion[14]. The former proved to be exclusively detected in males and also proved to be related to liver disease progression in chronic HBV subjects with genotype C infections, according to our fluorescence resonance energy transfer (FRET)-based real-time polymerase chain reaction (RT-PCR) method[11]. The latter mutation proved to be prevalent in chronic HBV subjects with genotype C, according to our previous molecular epidemiologic study based on a direct sequencing protocol[14]. However, the clinical implications of HBV preS1 mutations have rarely been investigated. Furthermore, the direct sequencing protocol has a limitation that could underestimate the presence of quasispecies variants in a subject, which could interfere with the genuine interpretation of specific mutations from clinical and virological perspectives. To overcome this drawback, FRET-based RT-PCR could be used for molecular epidemiologic purposes. The FRET-based RT-PCR allows not only the simultaneous identification of coexisting quasispecies but also the direct identification of target mutations from primary specimens, such as serum through melting curve analyses of the amplification product[22,23].
Therefore, the present study aims to achieve the following goals: (1) to develop a FRET-based RT-PCR method to detect the HBV preS1 start codon deletion; and (2) to determine the clinical implications of this mutation using the developed methods on DNA from the sera of genotype C-infected chronic HBV subjects.
In an effort to further support the clinical implications of preS1 deletion, our developed RT-PCR method was applied to two patient cohorts. Cohort I included 292 subjects randomly selected from the chronic HBV subjects who visited Cheju National University Hospital (Jeju, Korea) in 2003 or Seoul National University Hospital (Seoul, Korea) in 2005. The cohort I study protocol was approved by the Institutional Review Board of Seoul National University Hospital (C-1007-021-322). In cohort II, 90 consecutive chronic HBsAg carriers without liver cirrhosis or HCC were recruited from January 2012 to March 2012 at Konkuk University Hospital (Seoul, Korea). The cohort II study protocol was approved by the Institutional Review Board of Konkuk University Hospital (KUH-1010544). The clinical details of cohorts I and II are presented in Table 1.
| Clinical factor | Cohort I | Cohort II |
| Patient No. | 292 | 90 |
| Age in years, mean ± SD | 46.8 ± 16.0 | 37.8 ± 9.6 |
| Male | 225 (77.1) | 46 (51.1) |
| HBeAg-positive | 162 (55.5) | 58 (64.4) |
| Liver disease No. | C : CH : LC : HCC | C : CH |
| 66 : 33 : 67 : 126 | 36 : 54 | |
| ALT [IU/liter (mean ± SD)] | 77.7 ± 153.4 | 117.4 ± 226.5 |
| Median of HBV-DNA (range) | 979.4 (0-6000)1 | 1.3 × 109 (100-15.1 × 109)2 |
The HBV DNA sera levels for cohort I and cohort II were determined using different methods. In cohort I, the serum HBV DNA levels were determined using the Digene Hybrid Capture II assay (Digene Diagnostic Inc., Gaithersburg, MD, United States), which has a lower limit of detection of 0.5 pg/mL. In cohort II, the serum HBV DNA levels were assessed using the COBAS Amplicor PCR assay, which has a lower limit of detection of 300 copies/mL (Roche Molecular Systems, Branchburg, NJ, United States).
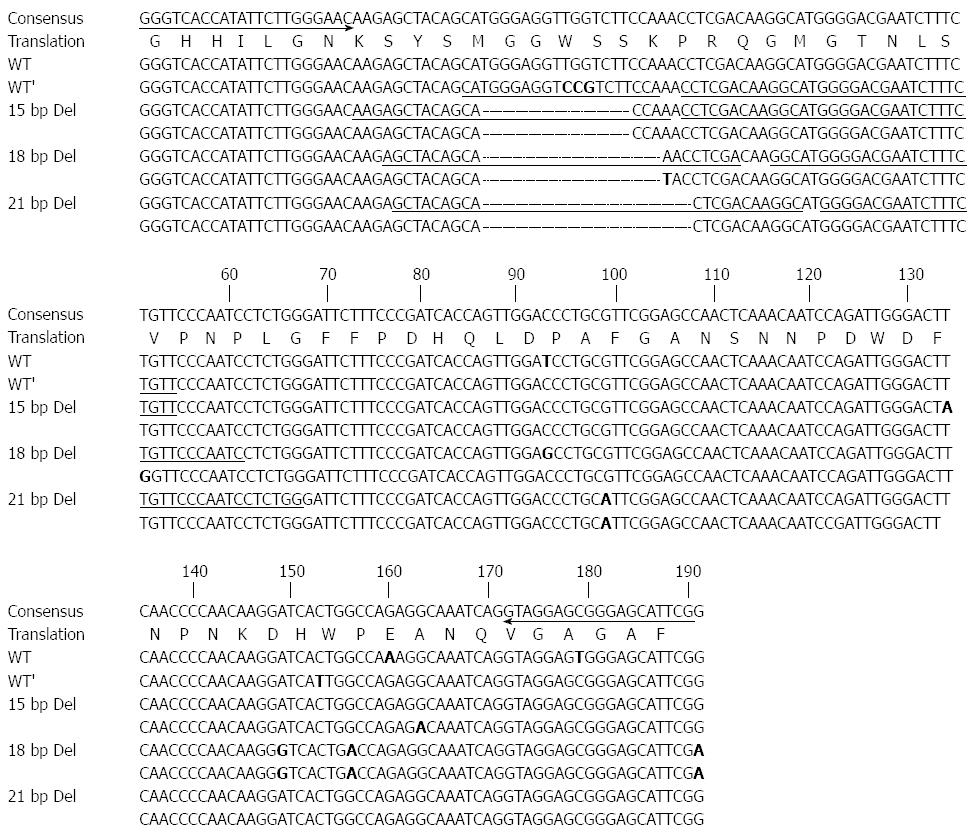
Figure 1 and Table 2 present the primers designed to amplify the HBV preS1 gene fragment and the probes designed to identify the deletion sequence variants at the start codon, respectively. A total of 99 cloned HBV genes were aligned using SeqManTM II software (DNASTAR). A primer pair for amplification of the HBV preS1 gene fragment was designed using Oligo V 6.5 (Molecular Biology Insights), which produced a 203-224 bp amplicon. We used four pairs of hybridization probes (an anchor probe and a sensor probe; HybProbe) to detect wild type at channel 640 and three types of deletion polymorphisms (15-bp deletion, 18-bp deletion, and 21-bp deletion) at three separate channels (670, 610 and 640, respectively) using the LightCycler 2.0 RT-PCR system. The same preS1 primer set and wild type probe previously developed for the W4P/R mutation detection[11] were used. The remaining probes, which were used to detect three types of deletion variants, were designed in this study.
| Name | Sequence (5'→3') | Tm (°C)1 | Target sequence | Channel | |
| Forward primer | HBV_sF | GGGTCACCATATTCTTGGGAAC | 62.5 | 203 - 224 bp of S gene | |
| Reverse primer | HBV_sR | CGAATGCTCCCRCTCCTAC | 60.5 or 63.3 | 203 - 224 bp of S gene | |
| Anchor probe | WT_A | ACAGAAAGATTCGTCCCCATGCCTTGTCGAGG -FL2 | 72.1 | ||
| Sensor probe | WT_S | LC Red6403-TGGAAGACGGACCTCCCATG-PH | 63.4 | Wild type S gene | 640 |
| Anchor probe | 18D_A | GATTGGGAACAGAAAGATTCGTCCCCATGCC-FL | 70.3 | ||
| Sensor probe | 18D_S | LC Red6104-TCGAGGTTTGCTGTAGCT-PH5 | 59.9 | 18 bp deletion | 610 |
| Anchor probe | 21D_A | CCAGAGGATTGGGAACAGAAAGATTCGTCCCC-FL | 70.5 | ||
| Sensor probe | 21D_S | LC Red640-GCCTTGTCGAGTGCTGTAGC -PH | 64.7 | 21 bp deletion | 640 |
| Anchor probe | 15D_A | ACAGAAAGATTCGTCCCCATGCCTTGTCGAGG -FL | 73.2 | ||
| Sensor probe | 15D_S | LC Red6706-TTGGTGCTGTAGCTCTT-PH | 57 | 15 bp deletion | 670 |
| Anchor probe | preS1_A | GATCCTTGTTGGGGTTGAAGTCCC-FL | 65.8 | ||
| Sensor probe | preS1_S | LC Red6104-ATCTGGATTGTTTGAGTTGGCT-PH | 62.4 | PreS1 | 610 |
A LightCycler 2.0 system was used, and its four detection channels were calibrated for color compensation and activated for the experiment. The LC Faststart DNA Master HP kit (Roche Diagnostic, Basel, Switzerland) was used to prepare the master mix, according to the kit protocol. A 10 μL reaction mixture was prepared for each sample and each target variant, as follows: 1 μL Taq buffer (containing dNTP mix and 10 mmol/L MgCl2), an additional 2 mmol/L MgCl2, 0.75 μmol/L forward primer (HBV_sF), 0.35 μmol/L reverse primer (HBV_sR), and a pair of 0.18 μmol/L HybProbes for the target variant (Table 2). Two separate reactions were performed for each sample: one to detect the wild type virus at channel 640, and the other to detect the deletion variants at channels 610, 640, and 670. For the relative quantification of the deletion variants vs the wild type in a subject, the melting temperature height values of the deletion variants vs wild type of 72 subjects with deletions in cohort I were calculated and compared with subjects with severe types of disease (HCC and liver cirrhosis) and mild types of disease (carriers and chronic hepatitis). The values of each deletion probe were normalized to those of the wild type probe.
To verify the fidelity of the developed FRET based RT-PCR, 23 samples from cohort II, 10 samples identified as mixed infections of both wild type and deletion variants and 13 samples identified as wild type-only infections, according to the RT-PCR assay, were subject to direct sequencing analyses. PCR amplification was performed using the same primer pair used for RT-PCR. Direct sequencing was used with the forward primer.
All detection items in this study were repeated at least three times, and the results were expressed as percentages, the mean ± SD, or medians (range). Differences between categorical variables were analyzed using Fisher’s exact test or the χ2 test. For continuous variables, Student’s t-test was used when the data showed a normal distribution, while the Mann-Whitney U test was used when the data were not normally distributed. SPSS version 21.0 software (Professional Statistic, Chicago, IL, United States) was used for all statistical analyses and a P value of < 0.05 (two-tailed) was considered statistically significant.
This retrospective study was reviewed and approved by the Institutional Review Boards (IRBs) at Seoul National University Hospital (IRB Grant No. C-1007-021-322) for cohort I and at Konkuk University Hospital (IRB Grant No. KUH-1010544) for cohort II. The patients’ medical records were anonymized and de-identified prior to the analyses. Experiments were mainly based on viral DNA extracted from isolates; therefore, the research study was conducted without informed consent. Informed consent waivers were granted by the IRBs.
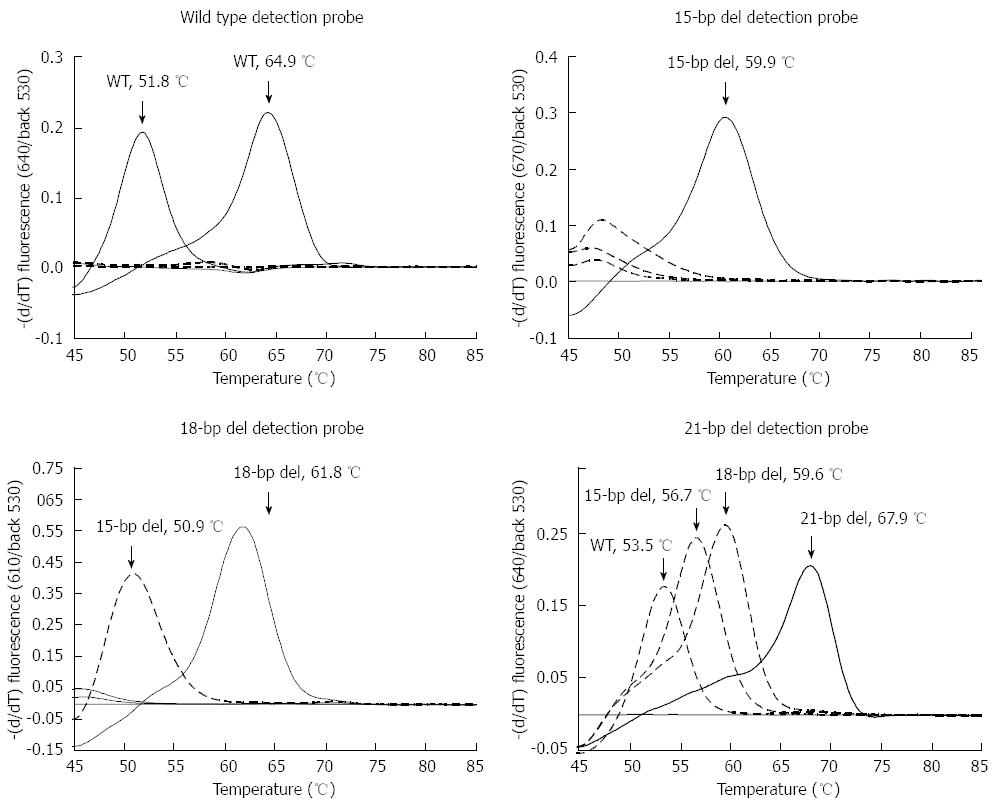
Application of the developed FRET-based RT-PCR to the wild type and three deletion types of the control plasmid DNAs cloned from the subjects demonstrated that separation between the wild type and variants was possible, in addition to separation between the three different types of variants (15-bp deletion, 18-bp deletion, and 21-bp deletion). The deletion variants exhibited distinct melting temperatures according to the respective probe. As demonstrated in the previous report[11], the wild type formed two distinct melting peaks at Ch. 640, one at 52.0 ± 0.2 °C [no mutation in codon 4 of preS1 (TGG)] and another at 65.0 ± 0.1 °C [CCG in codon 4 of preS1], which did not exhibit distinguishable melting peaks from the other variants. The 15-bp deletion variant clone formed a single distinct melting peak at 59.7 ± 0.1 °C at Ch. 670, which distinguished it from other cross-reacting melting peaks of the other variants. The 18-bp deletion variant clone formed a distinct melting peak at 67.8 ± 0.1 °C at Ch. 610, which exhibited a cross, as well as a clearly distinguishable 15-bp deletion peak and no detectable peaks of the other variants. The 21-bp deletion variant clone formed a distinct melting peak at the highest temperature (67.8 ± 0.2 °C at Ch. 640), with a cross and clearly distinguishable melting peaks from the other variants (Figure 2).
In this study, to correct the bias that results from the time and location of sample collection, we applied the developed FRET-based RT-PCR to samples from two different cohorts: cohort I (292 samples) and cohort II (90 samples). Applying this PCR method to 382 DNA samples taken from Korean chronic HBV subjects from the two cohorts resulted in the successful differentiation of 341 samples (89.3%), including 264 (90.4%) of the 292 samples in cohort I and 77 (85.6%) of the 90 samples in cohort II (Table 3). Ten types of polymorphisms were found in the 341 subjects detected from cohorts I and II. Of the 341 subjects detected, a total of 100 subjects (29.3%) [72 (27.3%) of the 264 detected samples from cohort I and 28 (36.4%) of the 77 samples from cohort II] proved to have more than one of the three deletions. Of the 100 subjects with deletions, 90 (90%) simultaneously exhibited positive signals with wild type probes, which indicated the coexistence of both wild type and deletion variants in most subjects with deletions (Table 4).
| Diseases | Number of tests positive/total | |
| Cohort I | Cohort II | |
| HCC | 109/125 (87.2) | |
| LC | 59/69 (85.5) | |
| CH | 33/33 (100) | 50/54 (92.6) |
| C | 63/66 (95.5) | 27/36 (75) |
| Total | 264/292 (90.4) | 77/90 (85.6) |
| Types | n (%) |
| WT only | 241 (70.7) |
| 15 del only | 3 (0.9) |
| 18 del only | 3 (0.9) |
| 21 del only | 4 (1.2) |
| WT + 15 del | 16 (4.7) |
| WT + 18 del | 47 (13.8) |
| WT + 21 del | 6 (1.8) |
| WT + 15 del + 18 del | 18 (5.3) |
| WT + 18 del + 21 del | 2 (0.6) |
| WT + 15 del + 18 del + 21 del | 1 (0.3) |
| Total | 341 |
In the 264 subjects detected from cohort I, no significant differences were found in age, gender, and alanine transaminase (ALT) level between the subjects with (192 subjects) and without (72 subjects) deletions. However, significant differences in Hepatitis B virus e antigen (HBeAg) serostatus and HBV DNA levels were found. The HBeAg-positive ratio was significantly higher in the subjects with wild type HBV than in those with deletions (51.0%, 98/192 subjects vs 75.0%, 54/72 subjects, P < 0.001). The median HBV DNA level was also significantly higher in the subjects with wild type HBV than in those with deletions (797.7 pg/mL vs 1678.9 pg/mL, P = 0.013) (Table 5).
| Clinical factor | Cohort I | Cohort II | ||||
| Wild type (n = 192) | Deletion (n = 72) | P value | Wild type (n = 49) | Deletion (n = 28) | P value | |
| Age in years, mean ± SD | 46.3 ± 15.7 | 44.7 ± 17.8 | 0.471 | 35.8 ± 8.9 | 39.1 ± 11.1 | 0.186 |
| Male | 148 (77.1) | 52 (72.2) | 0.423 | 20 (40.8) | 16 (57.1) | 0.235 |
| HBeAg-positive | 98 (51.0) | 54 (75.0) | < 0.001 | 29 (69.2) | 26 (92.9) | 0.002 |
| Liver disease (C : CH : LC : HCC) | 43 : 27 : 47 : 75 | 20 : 6 : 12 : 34 | 18 : 31 | 9 : 19 | ||
| ALT [IU/liter (mean ± SD)] | 87.7 ± 182.4 | 54.4 ± 33.5 | 0.295 | 125.5 ± 271.2 | 120.9 ± 160.3 | 0.935 |
| Median of HBV-DNA (range) | 797.7 (0-6000)1 | 1678.9 (0-6000)1 | 0.013 | 8.3 × 108 (100-10.1 × 109)2 | 2.2 × 109 (2.3 × 103-15.1 × 109)2 | 0.049 |
Similar to cohort I, a significant difference was also found among the 77 cohort II subjects, in terms of two clinical indicators of HBeAg serostatus (69.2%, 29/49 subjects vs 92.9%, 26/28 subjects, P = 0.002) and HBV DNA levels (8.3 × 108 copies/mL vs 2.2 × 109 copies/mL, P = 0.049) between the subjects with (49 subjects) and without (28 subjects) deletions (Table 5). This trend was also found in the combined analyses of the clinical data of the subjects (n = 341) detected in cohorts I and II. A significant difference in the HBeAg serostatus (52.7%, 127/241 subjects vs 80.0%, 80/100 subjects, P < 0.001) and HBV DNA levels (4.6 × 108 copies/mL vs 1.4 × 109 copies/mL, P = 0.014) was observed between the subjects with (241 subjects) and without (100 subjects) deletions (Table 6).
| Wild type (n = 241) | Deletion (n = 100) | 21 del (n = 13) | 18 del (n = 70) | 15 del (n = 38) | P value | |
| Age in years, mean ± SD | 44.1 ± 15.2 | 43.1 ± 16.3 | 51.6 ± 15.7 | 42.0 ± 15.9 | 40.3 ± 14.8 | 0.564 |
| Male | 168 (69.7) | 68 (68.0) | 6 (69.2) | 46 (65.7) | 24 (63.1) | 0.797 |
| HBeAg-positive | 127 (52.7) | 80 (80.0) | 8 (61.5) | 58 (82.9) | 33 (86.8) | < 0.001 |
| Liver disease (C : CH : LC : HCC) | 61 : 58 : 47 : 75 | 29 : 25 : 12 : 34 | 4 : 3 : 0 : 8 | 21 : 17 : 9 : 23 | 12 : 13 : 5 : 8 | |
| ALT [IU/liter (mean ± SD)] | 96.8 ± 207.1 | 81.1 ± 111.2 | 63.2 ± 43.3 | 85.5 ± 128.3 | 89.0 ± 122.4 | 0.476 |
Of the 100 subjects with deletions in cohorts I and II, the 18-bp deletion variants were the most prevalent (70 subjects, 70%), followed by the 15-bp deletion (38 subjects) and 21-bp deletion (13 subjects). Twenty-one subjects (21%) proved to have mixed deletion variants, with more than two types (Table 6).
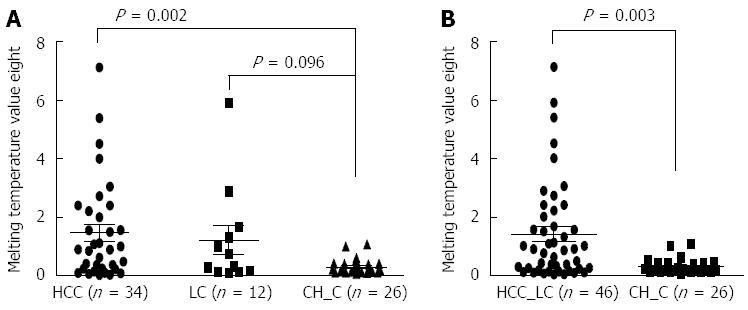
Significant differences in the prevalence of preS1-del were not found between the subjects with different clinical stages in cohort I [CH (27.1%, 26/96 subjects), liver cirrhosis (20.3%, 12/59 subjects) and HCC (31.2%, 34/109 subjects)]. It is possible to estimate the relative amounts of deletion variants vs wild type in a subject by calculating the melting temperature height values[24]. We compared the mean melting temperature height values of the variants vs the wild type of the 72 subjects with deletions in cohort I, according to the different clinical stages. Among the subjects with the three types of deletion variants, those with severe types of diseases (i.e., liver cirrhosis and HCC) exhibited significantly higher levels of melting temperature height values in deletion variants vs wild type compared with the subjects with mild types of chronic hepatitis. The latter finding suggests that increases in deletion variants in a subject may contribute to liver disease progression (Figure 3).
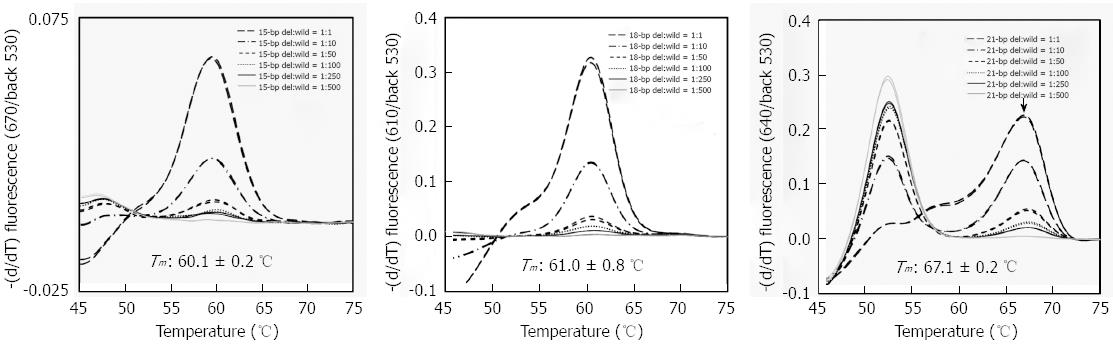
To validate the effectiveness of FRET-based RT-PCR in detecting deletion variants, 23 samples from cohort II (i.e., 10 samples identified as mixed infection of deletion and wild type and 13 samples of wild type-only infection identified by FRET-based RT-PCR) were subjected to direct sequencing analyses. In direct sequencing data, samples that exhibited the presence of mixed peaks in the electropherogram were regarded as mixed infections with wild type and deletion variants. No difference was found between the two protocols in the detection of wild type-only infections. The 13 samples identified as wild type-only infection using RT-PCR were also identified as wild type using the direct sequencing protocol. However, a discrepancy between the two protocols was found in detecting the deletion variants. While only 7 (70%) of 10 samples were identified as mixed infection using RT-PCR, mixed infections with both wild type and deletion variants were proven using direct sequencing protocols, and mixed peaks were not found in the other three samples (Table 7). This discrepancy might have resulted from the sensitivity difference between the two protocols used to detect the deletion variants in the mixed samples. To address this issue, we investigated the limits of our RT- PCR method (15-bp-, 18-bp and 21-bp deletion) for detection of deletion variants mixed with various concentrations of wild type DNA. Our analysis showed that the RT-PCR could detect deletion variants even in cases of a 100-fold higher level of wild type DNA compared with deletion DNA, irrespective of the deletion types (Figure 4).
There are clinical and virological differences between the HBV genotypes. Genotype C represents most HBV infections in Asian areas, including China, South Korea, and Japan, where HBV infections are the most prevalent worldwide. Recent studies have demonstrated that patients with genotype C are more likely to develop HCC than patients with other HBV genotypes[25-27]. Furthermore, higher levels of sustained HBeAg seropositivity and serum HBV-DNA, which are known to be HCC risk factors, were found in the genotype C subjects compared with the genotype B subjects[25,28]. In a previous study, we found that the preS1 start codon deletion is related to HCC in Korean HBV subjects[14]. Interestingly, comparisons of clinical information between subjects with the preS1 deletion and preS2 deletion indicated that the preS2 deletion (as with most HBV mutations) but not the preS1 deletion is positively related to HBeAg seronegative status[14]. This finding indicates that the preS1 deletion might be generated in the HBeAg-positive phase rather than in the HBeAg-negative phase. It also indicates that the preS1 deletion might be produced by factors other than the host immune response.
Despite the presence of overt, distinct traits in genotype C infection, the underlying mechanisms capable of explaining these distinct traits remain unknown to date. Therefore, in the present study, we used the FRET-based RT-PCR method to investigate the clinical implications of preS1 deletion in Korean chronic HBV subjects with genotype C infections. Thus, we demonstrated the significant roles of preS1 start codon deletions in liver disease progression among Korean chronic HBV subjects infected with genotype C via enhanced duration of HBeAg-positivity and HBV DNA replication. Our data might provide, at least in part, an explanation for the distinct traits of genotype C infections (higher levels of sustained HBeAg-positivity and HBV DNA replication) compared with other genotypes, particularly genotype B infections. Most HBV mutations found to date, including BCP double mutations and preC mutations, have been positively related to HBV persistent infections and HBeAg seronegative status[29,30]. However, this relationship is not true for the preS1 start codon deletions. To the best of our knowledge, the preS1 deletion is the first HBV mutation related to enhanced HBV replication and HBeAg-positive serostatus.
We applied the proposed FRET-based RT-PCR method to two different cohorts. The results from both cohorts were similar, which further strengthens our data. Our FRET-based RT-PCR method demonstrated that there were no significant differences in the prevalence of the preS1 deletion between subjects with different clinical stages (Tables 5 and 7), which does not align with the previous result indicating that the preS1 deletion is significantly more prevalent in HCC subjects than in subjects with mild types of liver disease, CH, and carriers[14]. The difference between the two studies could be related to differences in the applied protocols: FRET-based RT-PCR and nested-direct sequencing. Unlike the latter protocol, the former can detect deletion variants even if they exist in a small percentage compared to the wild type. Because three types of probes (for detecting the variants) existed in their respective independent channels, even the majority type cannot interfere with the detection of other probes. Using control plasmid DNAs, we discovered that our FRET-based RT- PCR could accurately detect the respective deletion type even in cases of mixed DNA showing a 100-fold higher level of wild type DNA than deletion type DNA (Figure 4). These differences can also explain the disparity in sensitivity found between our FRET-based RT-PCR and the direct sequencing protocol for the detection of deletion variants, demonstrating the superiority of the former protocol over the latter as a method for early prediction of liver disease progression. Although there were no differences in the prevalence of the preS1 deletion between the different clinical stages, a comparison of the melting temperature heights across the different clinical stages demonstrated increases in the deletion variants vs the wild type in subjects with severe liver disease (Figure 3), which indicates the positive effect of the variants on disease progression.
Interestingly, the most deletion variants (90%, 90/100 variants) coexisted with the wild type in the subjects with chronic hepatitis (Table 5). This finding indicates that the preS1 deletion event might occur sequentially after a wild type infection in HBeAg-positive stages instead of a de novo horizontal infection of deletion variants. Our phylogenetic study based on the quasispecies sequences demonstrated the same coherent clustering of wild type and deletion variants in a subject (data not shown), thus supporting this notion.
In particular, irrespective of the deletion types, all preS1 start codon deletions led to 11-N terminal loss from the preS1 of the otherwise 119 amino acids, as shown in genotype D, with 108 amino acids of preS1. Although the function of N-terminal region remains unknown, its absence in the genotype D strain indicates that it is dispensable in the HBV life cycle. However, together with a previous report demonstrating that deletions leading to 11 amino acids in the preS1 5’ region were observed more frequently in HCC subjects[14], our data strongly support that the N terminal loss of 11 amino acids plays a primary role in the progression of liver disease through enhanced HBeAg production and DNA replication. Alternatively, the N terminal deletion region could lead to simultaneous deletion of the overlapped polymerase region. Therefore, the molecular mechanism for enhanced HBeAg production and DNA replication induced by deletion variants should be elucidated in a future study that focuses on the modified function of the deleted LHBs or deleted polymerase.
In conclusion, the HBV genotype C preS1 deletion has the potential to lead to disease progression in chronic HBV subjects through the extended duration of HBeAg seropositive status and increases in HBV replications. This study provides novel insight into the higher infectivity and virulence of genotype C compared with other genotypes. In addition, the FRET-based RT-PCR for detection of the preS1 deletion shows promise for the earlier prediction of the risk of liver disease progression in chronic HBV subjects.
The preS1 start codon deletion proved to be prevalent in chronic hepatitis B virus (HBV) subjects with genotype C using our previous molecular epidemiologic study based on a direct sequencing protocol. However, its clinical implications have rarely been investigated. In this study, the authors developed a molecular diagnostic method to detect this mutation and gained insight into its clinical implications.
This study developed a novel fluorescence resonance energy transfer (FRET)-based real-time polymerase chain reaction (RT-PCR) method to detect the preS1 start codon deletion and applied it to 2 Korean chronic HBV patient cohorts.
For the first time, the authors proved that the HBV genotype C preS1 deletion might significantly contribute to disease progression in chronic HBV subjects by extending the duration of Hepatitis B virus e antigen (HBeAg) seropositive status and increasing HBV replications. This study provides novel insight into the understanding of the higher infectivity and virulence of genotype C compared with other genotypes.
The FRET-based RT-PCR method for detection of the preS1 deletion developed in this study might hold potential for the early prediction of the risk of liver disease progression in chronic HBV subjects.
The FRET-based RT-PCR could be used for molecular epidemiologic purposes. It can permit not only the simultaneous identification of coexisting quasispecies but also the direct identification of target mutations from primary specimens, such as serum through melting curve analyses of the amplification product.
For the first time, the authors discovered the clinical implications of the preS1 start codon deletion via a molecular epidemiologic approach using FRET-based RT-PCR, which might significantly contribute to disease progression in chronic HBV subjects infected with genotype C by extending the duration of HBeAg seropositive status and increasing HBV replications. The results are interesting and may explain the higher infectivity and virulence observed in genotype C compared with other genotypes.
P- Reviewer: Alam S, Bolhassani A, Chung YH, Gao C, Nakajima H S- Editor: Yu J L- Editor: A E- Editor: Wang CH
| 1. | Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34 Suppl 1:S1-S3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 319] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 2. | Chen DS. From hepatitis to hepatoma: lessons from type B viral hepatitis. Science. 1993;262:369-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 269] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 3. | Korea Centers for Disease Control. Data Resource Profile: The Korea National Health and Nutrition Examination Survey. Korea: KCDC 2007; . |
| 4. | Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology. 2007;50:52-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 107] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 5. | Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 315] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 6. | Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012;7:e47372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 7. | Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013;8:e54486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 8. | Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, Yang S, Lee M, Yoon JH, Kim YJ. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80:1337-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 9. | Lee SA, Cho YK, Lee KH, Hwang ES, Kook YH, Kim BJ. Gender disparity in distribution of the major hydrophilic region variants of hepatitis B virus genotype C according to hepatitis B e antigen serostatus. J Med Virol. 2011;83:405-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Lee SA, Kim K, Kim H, Kim BJ. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J Hepatol. 2012;56:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 11. | Lee SA, Kim KJ, Kim DW, Kim BJ. Male-Specific W4P/R Mutation in the Pre-S1 Region of Hepatitis B Virus, Increasing the Risk of Progression of Liver Diseases in Chronic Patients. J Clin Microbiol. 2013;51:3928-3936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Lee SA, Mun HS, Kim H, Lee HK, Kim BJ, Hwang ES, Kook YH, Kim BJ. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J Med Virol. 2011;83:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Mun HS, Lee SA, Kim H, Hwang ES, Kook YH, Kim BJ. Novel F141L pre-S2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J Virol. 2011;85:123-132. [PubMed] |
| 14. | Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, Yoon JH, Kim YJ, Lee HS, Hyun JW. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008;80:1189-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 15. | Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, Yoon JH, Lee HS, Cha CY, Kook YH. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76:194-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Kim BJ. Hepatitis B virus mutations related to liver disease progression of Korean patients. World J Gastroenterol. 2014;20:460-467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 31] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Ganem D. Assembly of hepadnaviral virions and subviral particles. Curr Top Microbiol Immunol. 1991;168:61-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 63] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 18. | Itoh Y, Takai E, Ohnuma H, Kitajima K, Tsuda F, Machida A, Mishiro S, Nakamura T, Miyakawa Y, Mayumi M. A synthetic peptide vaccine involving the product of the pre-S(2) region of hepatitis B virus DNA: protective efficacy in chimpanzees. Proc Natl Acad Sci USA. 1986;83:9174-9178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 148] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Wang HC, Huang W, Lai MD, Su IJ. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 224] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 20. | Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023-2032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 256] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 21. | Caselmann WH, Meyer M, Kekulé AS, Lauer U, Hofschneider PH, Koshy R. A trans-activator function is generated by integration of hepatitis B virus preS/S sequences in human hepatocellular carcinoma DNA. Proc Natl Acad Sci USA. 1990;87:2970-2974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 99] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Lay MJ, Wittwer CT. Real-time fluorescence genotyping of factor V Leiden during rapid-cycle PCR. Clin Chem. 1997;43:2262-2267. [PubMed] |
| 23. | Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat Struct Biol. 2000;7:730-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 672] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 24. | Nie H, Evans AA, London WT, Block TM, Ren XD. Quantification of complex precore mutations of hepatitis B virus by SimpleProbe real time PCR and dual melting analysis. J Clin Virol. 2011;51:234-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 25. | Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, Sung JJ. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494-1498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 369] [Cited by in RCA: 387] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 26. | Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, Shih WL, Kao JH, Chen DS, Chen CJ. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97:265-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 422] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 27. | Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:1134-1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 483] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 28. | Chu CJ, Hussain M, Lok AS. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology. 2002;122:1756-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 359] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 29. | Funk ML, Rosenberg DM, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J Viral Hepat. 2002;9:52-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 265] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 30. | Zhang D, Ma S, Zhang X, Zhao H, Ding H, Zeng C. Prevalent HBV point mutations and mutation combinations at BCP/preC region and their association with liver disease progression. BMC Infect Dis. 2010;10:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |