Published online Dec 14, 2014. doi: 10.3748/wjg.v20.i46.17305
Revised: July 18, 2014
Accepted: September 12, 2014
Published online: December 14, 2014
Processing time: 190 Days and 6.7 Hours
Alternative splicing, which is a common phenomenon in mammalian genomes, is a fundamental process of gene regulation and contributes to great protein diversity. Alternative splicing events not only occur in the normal gene regulation process but are also closely related to certain diseases including cancer. In this review, we briefly demonstrate the concept of alternative splicing and DNA damage and describe the association of alternative splicing and cancer pathogenesis, focusing on the potential relationship of alternative splicing, DNA damage, and gastrointestinal cancers. We will also discuss whether alternative splicing leads to genetic instability, which is considered to be a driving force for tumorigenesis. Better understanding of the role and mechanism of alternative splicing in tumorigenesis may provide new directions for future cancer studies.
Core tip: Alternative splicing is a fundamental process of gene regulation in eukaryotes. Alternative splicing of DNA damage repair proteins is a significant cause of gene mutations, and those mutations in turn affect alternative splicing in cancer. Alternative splicing is associated with tumorigenesis by contributing to genetic instability. Therefore, alternative splicing of DNA damage response-related genes has an important role in tumorigenesis, survival, and growth of gastrointestinal cancers. In summary, the alternative splicing variants of these genes could be potential targets for both diagnosis and treatment of gastrointestinal cancers.
- Citation: Rahmutulla B, Matsushita K, Nomura F. Alternative splicing of DNA damage response genes and gastrointestinal cancers. World J Gastroenterol 2014; 20(46): 17305-17313
- URL: https://www.wjgnet.com/1007-9327/full/v20/i46/17305.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i46.17305
Alternative splicing is a fundamental process of gene regulation, which results in a single gene that codes for multiple proteins by excluding and/or including particular exons from pre-mRNA produced from that gene[1]. The process is performed by the spliceosome composed of five small nuclear ribonucleoproteins (snRNPs; U1, U2, U4, U5, and U6) and more than 100 different polypeptides[2]. In this process, many different types of proteins are translated from mRNA of the same gene origin and contribute to protein diversity. For example, at least 60% of human gene products undergo alternative splicing[3], approximately 100000 alternative splicing events have been identified in the human genome, and up to 95% of human multi-exonic genes have been alternatively spliced[4]. There are approximately 20000-35000 protein-coding genes in a mammalian genome[5], but the number of proteins generated by alternative splicing is much higher[6] because many of these genes have multiple splicing patterns compensate up to thousands[7]. Thus, alternative splicing is a common phenomenon in the process of mammalian gene regulation and generation of protein diversity.
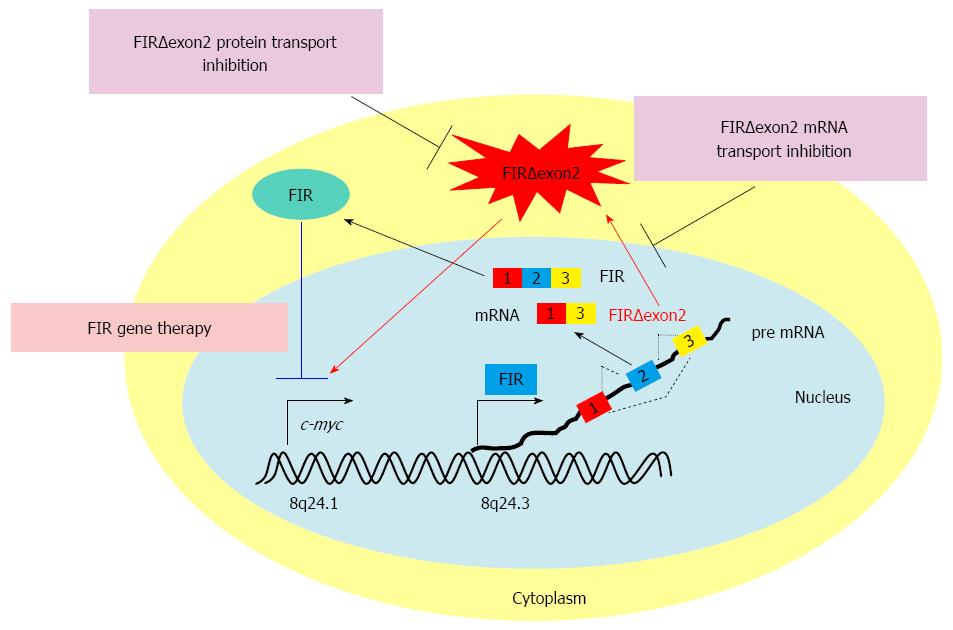
Alternative splicing events may occur in both normal and disease-related gene regulation processes. The frequency of alternative splicing is higher in cancerous tissues than in normal tissues[8]. Occasionally, alternative splicing variants are expressed in cancer cells but not in normal cells. For example, far upstream element-binding protein (FBP)-interacting repressor (FIR) splice variants lacking or containing exon 2 and/or exon 5 are expressed in the majority of hepatocellular carcinomas (HCCs) but not in normal hepatocytes[9]. A well-known tumor suppressor gene p53 is alternatively spliced to produce at least twelve protein isoforms, which have important roles in cancer formation and progression[10]. It has been suggested that missense or silent mutations affect splicing[11-15]. According to the human gene mutation database, approximately 84% of hereditary diseases are associated with point mutations[16]. Teraoka et al[17] suggested that 48% of these mutations result in defective splicing in the ATM gene in patients with ataxia-telangiectasia, and ATM has also been reported to be alternatively spliced in several types of cancer[18-20]. López-Bigas et al[11] estimated that more than 60% of all human disease-related mutations affect splicing. Lim et al[21] suggested that 22% of disease alleles that were originally classified as missense mutations may also affect splicing and approximately one third of all disease-causing mutations alter pre-mRNA splicing. Alternative splicing variants of many genes and some well-known splicing factors have been reported to be associated with numerous cancers. For example, Ikaros family genes include Ikaros, Helios, and Aiolos. The Ikaros gene (ZNFN1A1) is a member of the Kruppel transcription factor family characterized by the presence of zinc-finger domains located at their N- and C-termini and is alternatively spliced to give a number of variants[22]. Ikaros itself acts as a tumor suppressor in the lymphoid lineage[23], but alternative splicing variants, such as Ik11, are aberrantly expressed in B-cell lymphoproliferative disorders and involved in tumor pathogenesis[24]. Helios was found to be abnormally spliced in adult T-cell leukemia, and deregulation of Helios expression promotes T-cell growth[25]. The splicing factor SRSF6 is an oncoprotein reported to be over-expressed in lung and colon cancers[26]. Another splicing factor hnRNP has been suggested to be an oncogenic driver in glioblastoma[27,28]. Recurrent somatic mutations of splicing machinery genes, such as SF3B1, U2AF1, ZRSR2, and SRSF2, have been reported in numerous malignancies, including myelodysplastic syndromes, leukemias, and ovarian and gastric cancers[29-33]. Pre-mRNA processing factor 6 (PRPF6), a member of the tri-snRNP spliceosome complex, is required for alternative splicing of a number of genes, including ZAK kinase, and splicing activity of PRPF6 is important for colon cancer cell growth[34,35]. In addition to the associations shown in the above examples, many studies have suggested that alternative splicing is indeed closely related to certain diseases such as gastrointestinal cancers[18,36-48].
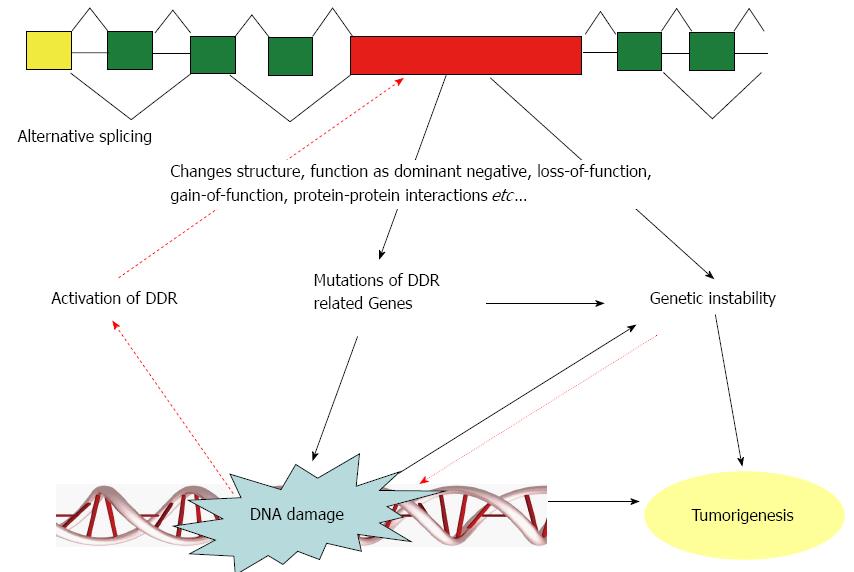
Impaired DNA damage responses induce genetic instability. DNA double-stranded breaks represent one of the most severe types of DNA damage and promote genetic instability that is lethal to cells if left unrepaired[49,50]. Genetic instability includes two major categories: one is microsatellite instability, which involves subtle changes in DNA sequences (faulty DNA repair), and the other is chromosomal instability (CIN), which is characterized by gains and losses of whole or parts of chromosomes, and CIN is considered to be a driving force for tumorigenesis[51,52]. Single-stranded or double-stranded DNA breaks increase the susceptibility of chromosomal gross structural alterations that lead to CIN[51]. CIN is closely associated with the intrinsic multidrug resistance of cancer[51,53]. The possible association of DNA damage, alternative splicing, and genetic instability is schematically shown in Figure 1.
Chromosomal alterations are found in nearly all human cancers[54]. As mentioned above, severe types of DNA damage promote genetic instability and are an integral component of human neoplasia[55]. Alternative splicing affects the stability of transcripts by introducing premature STOP codons and directing mRNA degradation through the nonsense-mediated mRNA decay pathway[56]. Alternative splicing of DNA damage response genes promotes genetic instability. Therefore, alternative splicing is closely associated with DNA damage and tumorigenesis. Previous studies have shown that gastrointestinal cancers are closely associated with alternative splicing of DNA damage-related genes that cause genetic instability. For example, ATM is involved in the homologous recombination (HR) pathway of DNA repair, and MRE11 is a component of the DNA damage sensor MRN; these genes are found to be alternatively spliced in colon cancer cells[18,36]. Germline mutations in the DNA mismatch repair genes, MSH2, MLH1, MSH6 and PMS2, are the cause of colon cancer, called Lynch syndrome[44,45] and they are reported to be spliced in a number of gastrointestinal cancers[37-39,57]. Splicing factor 3b (SF3b) is a subcomplex of the U2 snRNP in the spliceosome[58]. SAP155 (a subunit of SF3b) is required for proper FIR expression and vice versa, and SAP155 knockdown or SF3b inhibition disrupts alternative splicing of FIR pre-mRNA and generates FIRΔexon2[59]. FIR also acts as a molecular sensor for bleomycin-induced DNA damage by potentially interacting with DNA-PKcs and Ku-86/XRCC5[60] and has been reported to be alternatively spliced in colorectal cancer[40] as well as in HCCs[9]. Multifunctional splicing factor U2AF65, which has biotinylated triplex DNA affinity, has been reported to be associated with colorectal cancers[61]. Poly (ADP-ribose) polymerase (PARP)-1 is involved in single- stranded DNA damage repair and has a control role in the HR pathway[62]. PARP-1 is activated by Helicobacter pylori in the development and proliferation of gastric cancer[63]. The tumor suppressor genes, BRCA1 and BRCA2, are involved in DNA damage repair through their association with the HR mediator, RAD51, and their mutations are usually known to contribute to the tumorigenesis of hereditary breast and ovarian cancers[64]. Recent studies have further suggested that BRCA1 mutations in females below the age of 50 years increase the risk of colorectal cancer[65], and BRCA2 mutations are closely associated with pancreatic carcinogenesis[66,67]. RING finger protein 43, which is an E3-type ubiquitin ligase, has been reported to be mutated in pancreatic cancer[46] and gastric cancer[47] and was recently reported to act as a regulator of ATM-ATR DNA damage response; its mutation is associated with a high risk of developing sessile-serrated adenomas[48], which are believed to lead to colorectal cancer. The genes reported to have alternative splicing mutations in gastrointestinal cancers are summarized in Table 1. From the above examples, we can conclude that alternative splicing mutations in DNA damage response genes are closely associated with gastrointestinal carcinogenesis.
| Genes | Role in DDR | Gastrointestinal Cancers | Reference papers |
| DDR-related genes in gastrointestinal cancers | |||
| ATM | DNA damage response kinase involved in HR pathway of DNA repair | Colon cancer cells | [18] |
| MSH2, MLH1, MHS6, PMS2 | Involved in DNA mismatch repair | Colorectal cancer and gastric cancers | [37-39,44,45,57] |
| MRE11 | Component of DNA damage sensor complex MRN | Colorectal cancer | [36] |
| PARP-1 | Involved in single stranded DNA damage repair | Gastric cancer | [63] |
| Plays role in controlling HR pathway | |||
| RNF43 | Function as a regulator of ATM/ATR/DNA damage response | Pancreatic cancer | [46] |
| Gastric cancer | [47] | ||
| Sessile serrated adenomas | [48] | ||
| AP4 | Activation by cellular stresses result in DNA damage inducer of EMT | Mediates EMT in colorectal cancer lines cancer | [84,85] |
| BRCA1 | Involved in HR | Colorectal cancer | [65] |
| BRCA2 | Involved in HR | Pancreatic cancer | [66,67] |
| U2AF65 | With biotinylated triplex DNA affinity | Colorectal cancer | [61] |
| FIR (PUF60) | Originally a transcriptional facor, also reported as a molecular | Colorectal cancer | [40,96,97] |
| sensor for bleomycin-induced DNA damage pathway | Hepatocellular carcinoma | [9] | |
| Other genes in gastrointestinal cancers | |||
| P53 | Tumor suppressor, guardian of the genome | Colon cancer, head and neck cancer | [10] |
| CD44 | Class I transmembrane glycoprotein involved in cell adhesion, cell-cell | Intestinal tumorigenesis | [41] |
| interactions, migration and important player in stem cells and cancer | |||
| OPN-b | Osteopontin splice variant, contributed to gastric cancer cell survival | Gastric cancer | [71] |
| by regulation of Bcl-2 family proteins and CD44v expressions | |||
| p27 (CDKN1B) | Cell cycle regulatory gene | Small intestine neuroendocrine tumors | [72] |
| c-KIT | Stem cell growth factor receptor, also known as CD117 | Gastrointestinal stromal tumors | [75] |
| Prrx1 | Paired related homoeobox 1, a newly reported EMT inducer | Pancreatic cancer | [94] |
| Colorectal cancer | [95] | ||
| HDM2 | Human double minute 2, negative regulator of p53 | Colorectal cancer | [79] |
| PKM2 | Pyruvate kinase M2 gene, inactive state is associated with tumor cell proliferation, could switch between PKM2 to PKM1 | Impaired colorectal cancer growth | [82] |
| BRAF | Raf kinase family member BRAF is a proto-oncogen replays a role in | Malignant melanomas | [77] |
| regulating the MAP kinase/ERKs signaling pathway | Colorectal cancer | ||
| BMP | Bone morphogenetic proteins, are a group of growth factors, function in the formation of bone and cartilage,constitute morphogenetic signals etc. | Gastric cancer | [42] |
| PRPF6 | Pre-mRNA processing factor 6, a member of the tri-snRNP spliceosome complex | Colon cancer | [34,35] |
| Dystrophin | Cause of Duchenne muscular dystrophy | Metastatic GIST | [76] |
| FGFR2 | The fibroblast growth factor receptor 2, encodes for a fibroblast | Pancreatic ductal adenocarcinoma | [91] |
| growth factor-activated transmembrane receptor tyrosine kinase | Hepatic cancer metastasis | ||
| Splicing factors in other cancers | |||
| SRSF6 | Splicing facor | Lung and colon cancers | [26] |
| hnRNP | Splicing facor | Glioblastoma | [27,28] |
| SF3B1, U2AF1 | Splicing factors | Associated with numerous malignancies | [29-33] |
| ZRSR2, SRSF2 | |||
| Ik11 (Ikaros) | Alternative splicing variant of Ikaros, a member of Ikaros family genes | B-cell lympho-proliferative disorders | [24] |
| Helios | A member of Ikaros family genes | T-cell leukemia | [25] |
| PUF60 (FIR) | FIR lacks exon5 of PUF60. FIR/PUF60 interacts with SF3B1 | Colon cancer, leukemia | [40,80,97] |
| hnRNPM | RNA-binding protein heterogeneous nuclear ribonucleoprotein M | Breast cancer metastasis | [92] |
As mentioned above, alternative splicing is closely associated with gastrointestinal cancers and has an important role in their tumorigenesis. Gastrointestinal cancers are malignancies of the gastrointestinal tract and accessory organs of digestion, including the esophagus, stomach, biliary system, pancreas, small intestine, large intestine, rectum, and anus. They account for a large proportion of human malignancies and are a major cause of morbidity and mortality worldwide[68]. Among the gastrointestinal cancers, colorectal cancer is the third most frequently diagnosed cancer worldwide after lung and breast cancers, with 1.23 million diagnosed cases (9.7% of cancer diagnoses) in 2008[69]. There are many genetic and epigenetic changes that occur during colorectal carcinogenesis, including mutations of oncogenes, tumor suppressor genes, and mismatch repair genes; genetic instability; allelic losses in specific chromosomal arms; and methylation changes in gene promoters[70]. In addition, alternative splicing mutations have an important role in gastrointestinal carcinogenesis. In particular, alternatively spliced CD44 variants promote intestinal tumorigenesis induced by the activation of Wnt signaling[41]. Osteopontin splice variant (OPN-b) is found to be dominantly elevated in gastric cancer cell lines, and OPN-b has been shown to promote gastric cancer cell survival by regulation of Bcl-2 family proteins and CD44v expressions[71]. The cyclin-dependent kinase inhibitor gene, which encodes P27, has been reported to have recurrent somatic mutations in small intestinal neuroendocrine tumors[72]. P27 was shown to be associated with proliferative activity of gastric cancer[73,74]. Approximately 85%-95% of gastrointestinal stromal tumors (GIST) have mutations in the c-KIT gene[75]. Dystrophin is expressed in the nonneoplastic and benign counterparts of GIST, but inactivation of dystrophin was observed in 96% of metastatic GIST. Deletion of the dystrophin-encoding and muscular dystrophy-associated DMD gene through alternative splicing led to inactivation of larger dystrophin isoforms and contributed to tumor formation and metastasis[76]. Mutations in the bone morphogenetic protein signaling pathway led to the development of juvenile polyposis syndrome, which increases the risk of gastric cancer development[42]. The Raf kinase family member, BRAF, is a proto-oncogene that has been reported to be frequently mutated in numerous human cancers, such as somatic missense mutations, in 66% of malignant melanomas and at lower frequency in colorectal cancers[77]. Murine double minute 2, which is a negative regulator of the tumor suppressor gene p53, was shown to be alternatively spliced under DNA damage and contributed to numerous tumorigenesis, and its alternative splicing is mediated by FBP1 (FUBP1)[78]. The human counterpart is the negative regulator of p53, human double minute 2, which is frequently mutated by alternative splicing in colorectal cancer[79]. FUBP1 is a c-myc transcriptional activator[80]. Coupling of splicing and transcription should be considered and analyzed for better understanding of carcinogenesis. The pyruvate kinase muscle (PKM) gene is alternatively spliced to either M1 (PKM1) or M2 (PKM2) isoforms. PKM2 mostly promotes cancer cell growth, and PKM1 is usually expressed in normal differentiated tissues[81,82]. PKM2 itself is not necessary for tumor cell proliferation, and the inactive state of PKM2 has been shown to be associated with tumor cell proliferation, whereas nonproliferating tumor cells require activation of PKM[83]. MicroRNAs, such as miR-124, miR-137, and miR-340, have been shown to regulate alternative splicing of the PKM gene to switch PKM expression from PKM2 to PKM1 and contribute to impaired colorectal cancer growth[82]. Studies have suggested many alternative splicing isoforms of genes, such as VEGFA, UGT1A, PXR, cyclin D1, BIRC5 (survivin), DPD, K-RAS, SOX9, and SLC39A14, are potential therapeutic targets of colorectal cancers[43]. In brief, alternative splicing variants are potential targets for both diagnosis and treatment of gastrointestinal cancers.
Alternative splicing variants of certain genes not only have important roles in tumorigenesis but also significantly contribute to cancer metastasis. For example, the transcription factor, AP4, is encoded by the p53 tumor-suppressor gene and activated by numerous cellular stresses, which generally result in DNA damage[84]. AP4 is an inducer of epithelial-mesenchymal transition (EMT) and mediates c-MYC-induced EMT in colorectal cancer cell lines[85]. EMT of tumor cells contributes to metastasis[86,87]. Mesenchymal-epithelial transition (MET), which presumably contributes to tumor suppression[88], has been shown to be induced by p53 activation. Most recently, Peng et al[89] summarized the role of EMT in gastric cancer and suggested that loss of E-cadherin via its transcriptional repressors, such as Snail, ZEB, and Twist, is a key step in EMT activation, which significantly contributes to gastric carcinogenesis. Fibroblast growth factor receptor 2 (FGFR2) encodes for a fibroblast growth factor-activated transmembrane receptor tyrosine kinase and has been shown to be associated with EMT-related alternative splicing[90]; its alternative splicing generates the IIIb and IIIc isoforms. FGFR-2 IIIb expression correlates with venous invasion of pancreatic ductal adenocarcinoma, whereas FGFR-2 IIIc expression correlates with faster development of liver metastasis[91]. RNA-binding protein heterogeneous nuclear ribonucleoprotein M promotes breast cancer metastasis by activating the switch of alternative splicing that occurs during EMT[92]. Recently, splicing of paired related homoeobox 1 (Prrx1) has been reported to be a novel EMT-MET switch. Alternative splicing of Prrx1 results in two variants, Prrx1a and Prrx1b, and the ratio of Prrx1a (with inhibition domain)/Prrx1b (lack of inhibition domain)[93] switches EMT-MET of cells and controls migration and invasion of pancreatic cancer[94]. Notably, Prrx1 is involved in metastasis and poor prognosis in colorectal cancer[95].
Alternative splicing variants can be potential targets for the diagnosis and treatment of many cancers, including gastrointestinal cancers (Figure 2)[43,96]. Novel splicing variants of FIR were generated by SAP155 siRNA, and these variants were also found to be activated in human colorectal cancer tissues[97]. Circulating FIR and FIRΔexon2 mRNAs are potential novel screening markers for colorectal cancer testing with conventional carcino-embryonic antigen and carbohydrate antigen 19-9. Given the central role of c-Myc in the development of many cancers, one direction toward the development of cancer gene therapies directed against c-Myc may go through FIR and its variants. The Sendai virus vector of FIR has shown strong tumor growth suppression with no significant side effects in an animal xenograft model and is potentially applicable to future clinical cancer treatment[98].
Alternative splicing is a fundamental process of gene regulation in eukaryotes. It is a common phenomenon in mammalian genomes because most human genes undergo this process[4]. Alternative splicing leads to genetic instability, such as CIN, which drives tumorigenesis. DNA damage is one of the major reasons for genetic instabilities, and major components of the DNA damage repair pathway are alternatively spliced in certain cancers. Therefore, alternative splicing is closely associated with tumorigenesis by contributing to genetic instability. Alternative splicing of DNA damage repair proteins is a significant cause of gene mutations, which reciprocally affects alternative splicing in cancer. DNA damage promotes genetic instability, and genetic instability further promotes tumorigenesis (Figure 1). Genetic instability caused by certain types of DNA damage may be critical for the development of all colorectal cancers[55]. Many genes involved in the DNA damage repair pathway are alternatively spliced in gastrointestinal cancers (Table 1). Thus, the alternative splicing in DNA damage response-related genes has an important role in the tumorigenesis, survival, and growth of gastrointestinal cancers. Establishing a well-organized database of alternative splicing would be helpful for facilitation of the process of considering a set of splice isoforms or their common regulatory network as targets of diagnostic or therapeutic strategies. Better understanding of the role and mechanism of alternative splicing in tumorigenesis may lead to novel directions for future cancer studies.
P- Reviewer: Fan YM, Kamocki Z S- Editor: Gou SX L- Editor: A E- Editor: Ma S
| 1. | Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291-336. [PubMed] |
| 2. | Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 4. | Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413-1415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2521] [Cited by in RCA: 2825] [Article Influence: 166.2] [Reference Citation Analysis (0)] |
| 5. | Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet. 2000;25:232-234. [PubMed] |
| 6. | Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956-960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 958] [Cited by in RCA: 940] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 7. | Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367-370. [PubMed] |
| 8. | Kim E, Goren A, Ast G. Insights into the connection between cancer and alternative splicing. Trends Genet. 2008;24:7-10. [PubMed] |
| 9. | Malz M, Bovet M, Samarin J, Rabenhorst U, Sticht C, Bissinger M, Roessler S, Bermejo JL, Renner M, Calvisi DF. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014;60:1241-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Surget S, Khoury MP, Bourdon JC. Uncovering the role of p53 splice variants in human malignancy: a clinical perspective. Onco Targets Ther. 2013;7:57-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 177] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 11. | López-Bigas N, Audit B, Ouzounis C, Parra G, Guigó R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005;579:1900-1903. [PubMed] |
| 12. | Mazoyer S, Puget N, Perrin-Vidoz L, Lynch HT, Serova-Sinilnikova OM, Lenoir GM. A BRCA1 nonsense mutation causes exon skipping. Am J Hum Genet. 1998;62:713-715. [PubMed] |
| 13. | Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285-298. [PubMed] |
| 14. | Sanz DJ, Acedo A, Infante M, Durán M, Pérez-Cabornero L, Esteban-Cardeñosa E, Lastra E, Pagani F, Miner C, Velasco EA. A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin Cancer Res. 2010;16:1957-1967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 15. | Rouleau E, Lefol C, Moncoutier V, Castera L, Houdayer C, Caputo S, Bièche I, Buisson M, Mazoyer S, Stoppa-Lyonnet D. A missense variant within BRCA1 exon 23 causing exon skipping. Cancer Genet Cytogenet. 2010;202:144-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 16. | Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577-581. [PubMed] |
| 17. | Teraoka SN, Telatar M, Becker-Catania S, Liang T, Onengüt S, Tolun A, Chessa L, Sanal O, Bernatowska E, Gatti RA. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet. 1999;64:1617-1631. [PubMed] |
| 18. | Ejima Y, Yang L, Sasaki MS. Aberrant splicing of the ATM gene associated with shortening of the intronic mononucleotide tract in human colon tumor cell lines: a novel mutation target of microsatellite instability. Int J Cancer. 2000;86:262-268. [PubMed] |
| 19. | Ham MF, Takakuwa T, Luo WJ, Liu A, Horii A, Aozasa K. Impairment of double-strand breaks repair and aberrant splicing of ATM and MRE11 in leukemia-lymphoma cell lines with microsatellite instability. Cancer Sci. 2006;97:226-234. [PubMed] |
| 20. | Thorstenson YR, Roxas A, Kroiss R, Jenkins MA, Yu KM, Bachrich T, Muhr D, Wayne TL, Chu G, Davis RW. Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003;63:3325-3333. [PubMed] |
| 21. | Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci USA. 2011;108:11093-11098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Bellavia D, Mecarozzi M, Campese AF, Grazioli P, Gulino A, Screpanti I. Notch and Ikaros: not only converging players in T cell leukemia. Cell Cycle. 2007;6:2730-2734. [PubMed] |
| 23. | Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289-299. [PubMed] |
| 24. | Capece D, Zazzeroni F, Mancarelli MM, Verzella D, Fischietti M, Di Tommaso A, Maccarone R, Plebani S, Di Ianni M, Gulino A. A novel, non-canonical splice variant of the Ikaros gene is aberrantly expressed in B-cell lymphoproliferative disorders. PLoS One. 2013;8:e68080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | Asanuma S, Yamagishi M, Kawanami K, Nakano K, Sato-Otsubo A, Muto S, Sanada M, Yamochi T, Kobayashi S, Utsunomiya A. Adult T-cell leukemia cells are characterized by abnormalities of Helios expression that promote T cell growth. Cancer Sci. 2013;104:1097-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Ørntoft TF, Mu D, Karni R. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J Pathol. 2013;229:630-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 27. | Golan-Gerstl R, Cohen M, Shilo A, Suh SS, Bakàcs A, Coppola L, Karni R. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71:4464-4472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 28. | Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, Pan YX, Cartegni L. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011;30:4084-4097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 130] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 29. | Je EM, Yoo NJ, Kim YJ, Kim MS, Lee SH. Mutational analysis of splicing machinery genes SF3B1, U2AF1 and SRSF2 in myelodysplasia and other common tumors. Int J Cancer. 2013;133:260-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 30. | Visconte V, Makishima H, Maciejewski JP, Tiu RV. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia. 2012;26:2447-2454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 31. | Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1445] [Cited by in RCA: 1629] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 32. | Wan Y, Wu CJ. SF3B1 mutations in chronic lymphocytic leukemia. Blood. 2013;121:4627-4634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 33. | Yoshida K, Ogawa S. Splicing factor mutations and cancer. Wiley Interdiscip Rev RNA. 2014;5:445-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 128] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 34. | Lokody I. Alternative splicing: aberrant splicing promotes colon tumour growth. Nat Rev Cancer. 2014;14:382-383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 35. | Adler AS, McCleland ML, Yee S, Yaylaoglu M, Hussain S, Cosino E, Quinones G, Modrusan Z, Seshagiri S, Torres E. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes Dev. 2014;28:1068-1084. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 36. | Giannini G, Ristori E, Cerignoli F, Rinaldi C, Zani M, Viel A, Ottini L, Crescenzi M, Martinotti S, Bignami M. Human MRE11 is inactivated in mismatch repair-deficient cancers. EMBO Rep. 2002;3:248-254. [PubMed] |
| 37. | Betz B, Theiss S, Aktas M, Konermann C, Goecke TO, Möslein G, Schaal H, Royer-Pokora B. Comparative in silico analyses and experimental validation of novel splice site and missense mutations in the genes MLH1 and MSH2. J Cancer Res Clin Oncol. 2010;136:123-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 38. | Pagenstecher C, Wehner M, Friedl W, Rahner N, Aretz S, Friedrichs N, Sengteller M, Henn W, Buettner R, Propping P. Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants. Hum Genet. 2006;119:9-22. [PubMed] |
| 39. | Renkonen E, Lohi H, Järvinen HJ, Mecklin JP, Peltomäki P. Novel splicing associations of hereditary colon cancer related DNA mismatch repair gene mutations. J Med Genet. 2004;41:e95. [PubMed] |
| 40. | Matsushita K, Tomonaga T, Shimada H, Shioya A, Higashi M, Matsubara H, Harigaya K, Nomura F, Libutti D, Levens D. An essential role of alternative splicing of c-myc suppressor FUSE-binding protein-interacting repressor in carcinogenesis. Cancer Res. 2006;66:1409-1417. [PubMed] |
| 41. | Guo W, Frenette PS. Alternative CD44 splicing in intestinal stem cells and tumorigenesis. Oncogene. 2014;33:537-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Oshima H, Oguma K, Du YC, Oshima M. Prostaglandin E2, Wnt, and BMP in gastric tumor mouse models. Cancer Sci. 2009;100:1779-1785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Miura K, Fujibuchi W, Unno M. Splice isoforms as therapeutic targets for colorectal cancer. Carcinogenesis. 2012;33:2311-2319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 44. | Burt RW, DiSario JA, Cannon-Albright L. Genetics of colon cancer: impact of inheritance on colon cancer risk. Annu Rev Med. 1995;46:371-379. [PubMed] |
| 45. | Lawes DA, Pearson T, Sengupta S, Boulos PB. The role of MLH1, MSH2 and MSH6 in the development of multiple colorectal cancers. Br J Cancer. 2005;93:472-477. [PubMed] |
| 46. | Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, Eshleman JR, Goggins MG, Wolfgang CL, Canto MI. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc Natl Acad Sci USA. 2011;108:21188-21193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 482] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 47. | Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 694] [Cited by in RCA: 836] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 48. | Gala MK, Mizukami Y, Le LP, Moriichi K, Austin T, Yamamoto M, Lauwers GY, Bardeesy N, Chung DC. Germline mutations in oncogene-induced senescence pathways are associated with multiple sessile serrated adenomas. Gastroenterology. 2014;146:520-529. [PubMed] |
| 49. | Collis SJ, DeWeese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2005;24:949-961. [PubMed] |
| 50. | Metcalf JL, Bradshaw PS, Komosa M, Greer SN, Stephen Meyn M, Ohh M. K63-ubiquitylation of VHL by SOCS1 mediates DNA double-strand break repair. Oncogene. 2014;33:1055-1065. [PubMed] |
| 51. | Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IP, Howell M. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858-1870. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 378] [Cited by in RCA: 367] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 52. | Lentini L, Amato A, Schillaci T, Di Leonardo A. Simultaneous Aurora-A/STK15 overexpression and centrosome amplification induce chromosomal instability in tumour cells with a MIN phenotype. BMC Cancer. 2007;7:212. [PubMed] |
| 53. | Woodford-Richens KL, Rowan AJ, Gorman P, Halford S, Bicknell DC, Wasan HS, Roylance RR, Bodmer WF, Tomlinson IP. SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway. Proc Natl Acad Sci USA. 2001;98:9719-9723. [PubMed] |
| 54. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649. [PubMed] |
| 55. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623-627. [PubMed] |
| 57. | Zhu M, Chen HM, Wang YP. Missense mutations of MLH1 and MSH2 genes detected in patients with gastrointestinal cancer are associated with exonic splicing enhancers and silencers. Oncol Lett. 2013;5:1710-1718. [PubMed] |
| 58. | Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Lührmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978-4988. [PubMed] |
| 59. | Matsushita K, Kajiwara T, Tamura M, Satoh M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Yoshimoto R, Ito A. SAP155-mediated splicing of FUSE-binding protein-interacting repressor serves as a molecular switch for c-myc gene expression. Mol Cancer Res. 2012;10:787-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 60. | Rahmutulla B, Matsushita K, Satoh M, Seimiya M, Tsuchida S, Kubo S, Shimada H, Ohtsuka M, Miyazaki M, Nomura F. Alternative splicing of FBP-interacting repressor coordinates c-Myc, P27Kip1/cyclinE and Ku86/XRCC5 expression as a molecular sensor for bleomycin-induced DNA damage pathway. Oncotarget. 2014;5:2404-2417. [PubMed] |
| 61. | Nelson LD, Bender C, Mannsperger H, Buergy D, Kambakamba P, Mudduluru G, Korf U, Hughes D, Van Dyke MW, Allgayer H. Triplex DNA-binding proteins are associated with clinical outcomes revealed by proteomic measurements in patients with colorectal cancer. Mol Cancer. 2012;11:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 62. | Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31:4959-4964. [PubMed] |
| 63. | Nossa CW, Jain P, Tamilselvam B, Gupta VR, Chen LF, Schreiber V, Desnoyers S, Blanke SR. Activation of the abundant nuclear factor poly(ADP-ribose) polymerase-1 by Helicobacter pylori. Proc Natl Acad Sci USA. 2009;106:19998-20003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 64. | Lord CJ, Ashworth A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med. 2013;19:1381-1388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 349] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 65. | Phelan CM, Iqbal J, Lynch HT, Lubinski J, Gronwald J, Moller P, Ghadirian P, Foulkes WD, Armel S, Eisen A. Incidence of colorectal cancer in BRCA1 and BRCA2 mutation carriers: results from a follow-up study. Br J Cancer. 2014;110:530-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 66. | Kaur G, Saif MW. Translational research in pancreatic adenocarcinoma. JOP. 2014;15:121-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 67. | Koorstra JB, Hustinx SR, Offerhaus GJ, Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8:110-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 138] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 68. | Mactier KE, Glaire MA, Basavaraju U, El-Omar EM, Hold GL. MicroRNAs in gastrointestinal malignancy: a tool in cancer prevention? Eur J Cancer Prev. 2014;23:540-549. [PubMed] |
| 69. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11128] [Cited by in RCA: 11837] [Article Influence: 845.5] [Reference Citation Analysis (4)] |
| 70. | Jass JR. Colorectal cancer: a multipathway disease. Crit Rev Oncog. 2006;12:273-287. [PubMed] |
| 71. | Tang X, Li J, Yu B, Su L, Yu Y, Yan M, Liu B, Zhu Z. Osteopontin splice variants differentially exert clinicopathological features and biological functions in gastric cancer. Int J Biol Sci. 2013;9:55-66. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 72. | Francis JM, Kiezun A, Ramos AH, Serra S, Pedamallu CS, Qian ZR, Banck MS, Kanwar R, Kulkarni AA, Karpathakis A. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet. 2013;45:1483-1486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 247] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 73. | Aoyagi K, Kouhuji K, Miyagi M, Imaizumi T, Kizaki J, Isobe T, Shirouzu K. Expression of p27Kip1 protein in gastric carcinoma. Hepatogastroenterology. 2013;60:390-394. [PubMed] |
| 74. | Jee H, Lee SH, Park JW, Lee BR, Nam KT, Kim DY. Connexin32 inhibits gastric carcinogenesis through cell cycle arrest and altered expression of p21Cip1 and p27Kip1. BMB Rep. 2013;46:25-30. [PubMed] |
| 75. | De Vogelaere K, Aerts M, Haentjens P, De Grève J, Delvaux G. Gastrointestinal stromal tumor of the stomach: progresses in diagnosis and treatment. Acta Gastroenterol Belg. 2013;76:403-406. [PubMed] |
| 76. | Wang Y, Marino-Enriquez A, Bennett RR, Zhu M, Shen Y, Eilers G, Lee JC, Henze J, Fletcher BS, Gu Z. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet. 2014;46:601-606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 77. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [PubMed] |
| 78. | Jacob AG, Singh RK, Mohammad F, Bebee TW, Chandler DS. The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre-mRNA. J Biol Chem. 2014;289:17350-17364. [PubMed] |
| 79. | Yu Z, Zhang B, Cui B, Wang Y, Han P, Wang X. Identification of spliced variants of the proto-oncogene HDM2 in colorectal cancer. Cancer. 2012;118:1110-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 80. | Avigan MI, Strober B, Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538-18545. [PubMed] |
| 81. | Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 792] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 82. | Sun Y, Zhao X, Zhou Y, Hu Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol Rep. 2012;28:1346-1352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 139] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 83. | Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 420] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 85. | Jackstadt R, Röh S, Neumann J, Jung P, Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J Exp Med. 2013;210:1331-1350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 86. | Samatov TR, Tonevitsky AG, Schumacher U. Epithelial-mesenchymal transition: focus on metastatic cascade, alternative splicing, non-coding RNAs and modulating compounds. Mol Cancer. 2013;12:107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 87. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 47165] [Article Influence: 3368.9] [Reference Citation Analysis (5)] |
| 88. | Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 391] [Cited by in RCA: 433] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 89. | Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol. 2014;20:5403-5410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 139] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (1)] |
| 90. | Savagner P, Vallés AM, Jouanneau J, Yamada KM, Thiery JP. Alternative splicing in fibroblast growth factor receptor 2 is associated with induced epithelial-mesenchymal transition in rat bladder carcinoma cells. Mol Biol Cell. 1994;5:851-862. [PubMed] |
| 91. | Matsuda Y, Yoshimura H, Suzuki T, Uchida E, Naito Z, Ishiwata T. Inhibition of fibroblast growth factor receptor 2 attenuates proliferation and invasion of pancreatic cancer. Cancer Sci. 2014;105:1212-1219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 92. | Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014;28:1191-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 93. | Norris RA, Kern MJ. The identification of Prx1 transcription regulatory domains provides a mechanism for unequal compensation by the Prx1 and Prx2 loci. J Biol Chem. 2001;276:26829-26837. [PubMed] |
| 94. | Reichert M, Takano S, von Burstin J, Kim SB, Lee JS, Ihida-Stansbury K, Hahn C, Heeg S, Schneider G, Rhim AD. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013;27:288-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 95. | Takahashi Y, Sawada G, Kurashige J, Uchi R, Matsumura T, Ueo H, Takano Y, Akiyoshi S, Eguchi H, Sudo T. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br J Cancer. 2013;109:307-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 96. | Matsushita K, Tomonaga T, Kajiwara T, Shimada H, Itoga S, Hiwasa T, Kubo S, Ochiai T, Matsubara H, Nomura F. c-myc suppressor FBP-interacting repressor for cancer diagnosis and therapy. Front Biosci (Landmark Ed). 2009;14:3401-3408. [PubMed] |
| 97. | Kajiwara T, Matsushita K, Itoga S, Tamura M, Tanaka N, Tomonaga T, Matsubara H, Shimada H, Habara Y, Matsuo M. SAP155-mediated c-myc suppressor far-upstream element-binding protein-interacting repressor splicing variants are activated in colon cancer tissues. Cancer Sci. 2013;104:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 98. | Matsushita K, Shimada H, Ueda Y, Inoue M, Hasegawa M, Tomonaga T, Matsubara H, Nomura F. Non-transmissible Sendai virus vector encoding c-myc suppressor FBP-interacting repressor for cancer therapy. World J Gastroenterol. 2014;20:4316-4328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |