Published online Oct 28, 2014. doi: 10.3748/wjg.v20.i40.14958
Revised: April 14, 2014
Accepted: June 13, 2014
Published online: October 28, 2014
Processing time: 298 Days and 7.3 Hours
AIM: To develop a method to differentiate pancreatic cancer patients from healthy or benign individuals when carbohydrate antigen (CA) 19-9 is normal.
METHODS: Forty-one serum samples from patients with pancreatic lesions and blood samples from 20 healthy individuals were collected at the first stage of the experiment according to the enrolment criteria. General characteristics and some clinical features were carefully compared to ensure that the results were reasonable. All the blood samples were analyzed by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) combined with CM10 chips and a related bioinformatics analysis program to generate diagnostic models with different proteins. Forty-seven consecutive samples were tested at the next stage to verify the veracity and efficiency of the models.
RESULTS: The sex, age, and serum CA19-9 levels among the three groups (malignant, benign, and healthy) were statistically matched (P values were 0.957, 0.145, and 0.382, respectively). Two patterns were generated. Pattern 1 with four proteins theoretically had a specificity and sensitivity of 100% in distinguishing pancreatic cancer from healthy individuals, while it was 86.7% and 86.4%, respectively, in the subsequent practical verification. The positive predictive value (PPV) of the model was 86.4%. One of the four proteins was expressed highly in pancreatic cancer while the other three were expressed weakly. Pattern 2 consisted of six proteins that showed a specificity of 70.0% and sensitivity of 77.3% for differentiating malignancy from benign tumors. Its PPV reached 85.0%. Only one of these six proteins showed high expression in the malignant group.
CONCLUSION: SELDI-TOF-MS may facilitate diagnosis or differential diagnosis of pancreatic cancer when CA19-9 is normal. Pattern 1 may serve as a useful screening tool.
Core tip: Our study focused on patients with malignant disease and normal carbohydrate antigen 19-9 levels who could be easily misdiagnosed clinically. With careful design, our results could provide early diagnostic models to discriminate cancer patients from healthy volunteers or people with benign disease, as well as some potentially important proteins that could serve as tumor/cancer-specific or -associated antigens in future clinical studies.
- Citation: Jin XL, Xu B, Wu YL. Detection of pancreatic cancer with normal carbohydrate antigen 19-9 using protein chip technology. World J Gastroenterol 2014; 20(40): 14958-14964
- URL: https://www.wjgnet.com/1007-9327/full/v20/i40/14958.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i40.14958
Pancreatic cancer is one of the most life-threatening malignancies due to its difficulty in early detection[1]. As a result, a substantial proportion of patients with pancreatic cancer lose their chance of radical cure for advanced local disease at the time of diagnosis. Because it lacks specific symptoms, clinical diagnosis usually relies on biomarkers and techniques such as dynamic computed tomography and endoscopic ultrasonography[1-5]. Compared with the latter, accurate serological tests are more acceptable, noninvasive detection techniques for routine screening and early diagnosis[1-3]. Nowadays, carbohydrate antigen (CA)19-9 serves as the most widely used biomarker for pancreatic cancer[1,6,7]. However, it is not sufficiently reliable[2,6-8]; many patients with pancreatic cancer are CA19-9 negative, which makes it more difficult to distinguish them from healthy individuals or patients with benign disease through routine serum testing.
In recent years, proteomics has become an efficient method for screening for early diagnostic markers and potentially therapeutic molecular targets. Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) combined with Protein Chip technology provides a fast, sensitive, and high-throughput method to analyze a large number of unknown complicated protein samples in a short period of time[9,10]. SELDI-TOF-MS has been successfully used in the diagnosis, discrimination, and surveillance of some common malignant diseases[11-13].
In this study, we aim to provide a noninvasive and efficient method for screening for pancreatic cancer in a healthy population or in patients with benign diseases, by analyzing serum samples with SELDI-TOF-MS combined with Protein Chip technology, when CA19-9 is not applicable.
We enrolled all patients with pancreatic lesions (except pancreatitis) and normal levels of CA19-9 who were admitted to the Department of Surgery, Second Affiliated Hospital, Zhejiang University School of Medicine from January 2008 to December 2010. A serum CA19-9 level < 37 U/mL was defined as normal, as recommended in the literature[6,7,14]. With informed consent, we collected 41 serum samples from patients with pancreatic diseases (23 with cancer and 18 with benign disease), as well as blood samples from 20 healthy volunteers. Diagnosis was based on operative biopsies, endoscopic biopsies, or surgical specimens, and was confirmed pathologically by two pathologists. All serum samples were collected preoperatively in the morning before breakfast. The sera were kept at room temperature for 1-2 h, centrifuged at 2800 rpm for 10 min, and stored at -80 °C for further study.
We then consecutively collected 32 serum samples from patients who had suspicion of occupying pancreatic lesions but with negative serum CA19-9, as well as from 15 healthy volunteers. These specimens were tested with diagnostic methods derived from the first stage on the second day immediately after collection, usually before any medical intervention. Data were collected and compared with those from later pathological diagnosis.
Sinapinic acid (SPA) was purchased from Fluka (St Louis, MO, United States). ProteinChip Biosystems (Ciphergen PBS II plus SELDI-TOF-MS) and weak cationic exchange (CM10) chips were purchased from Ciphergen Biosystems (Fremont, CA, United States). All other reagents were acquired from Sigma (St Louis, MO, United States).
All serum samples were centrifuged at 10000 g for 4 min on ice to obtain the supernatants. Five microliters of each serum sample and 10 μL U9 buffer (9 mol/L urea, 2% CHAPS, 1% dithiothreitol) were intensively mixed in a 96-well cell culture plate. The plate was agitated on a platform shaker at 4 °C for 30 min before 185 μL sodium acetate (100 mmol/L, pH 4) was added to the U9/serum mixture, and agitation was performed as before. One hundred microliters of each diluted sample was applied to each spot of a bioprocessor containing the CM10 arrays, which were already activated according the instructions. The bioprocessor was sealed and agitated for 1 h at 4 °C. Before the chips were removed from the bioprocessor, they were washed five times (three times with sodium acetate and twice with deionized water) and air dried. One microliter of a saturated solution (5 mL/L trifluoroacetic acid plus SPA in 0.5 L/L acetonitrile) was added to each chip twice.
Air-dried chips were analyzed by the Protein Biological System II plus mass spectrometry reader. Data were collected by averaging 180 laser shots with intensity of 200, detector sensitivity of 8, highest mass of 100 kDa, and optimized range of 2-20 kDa. Calibration of the instrument was performed using the All-In-One Protein Standard.
The protein peaks were detected with an automatic peak detection pass using noise of spectra infiltration and signal-to-noise ratio. Peak clusters were completed to cluster the peaks in different samples that had similar masses. All these were performed using ProteinChip Software 3.1 (Ciphergen Biosystems). The peak intensities were preprocessed by scaling all the data to the range: -1-1.
Pattern recognition techniques were applied to diverse areas including gene microarray and mass spectrometry. We utilized a Multi-Layer Perception (MLP) artificial neural network (ANN) with a scaled conjugate gradient (SCG) optimized back propagation algorithm for discriminating pancreatic cancers.
We estimated the power of each peak in discriminating different group samples by an integrated approach that included t test, receiver operating characteristic (ROC) curves, and mean square error (MSE) of the ANN.
The ANN to calculate the MSE had four layers. In addition to the output layer and input layer, the ANN also had two hidden layers each with 100 nodes. The ANN was used on 1000 epochs and MSE was obtained. The MSE was calculated as the difference between the target output and the ANN prediction value. The lower MSE value of the peak showed the higher relative importance value for the ability to discriminate the different groups accurately. Leave-one-out cross-validation (LOOCV) approach was applied to estimate the accuracy of the classification. The process continued until each sample was reserved one time as a test sample.
The capability of each peak in distinguishing multiple groups was estimated by the P value of the Wilcoxon t test. The top 10 peaks with the lowest P values were selected for further analysis. All the combinations of 10 peaks were estimated by the LOOCV ANN. The combination of peaks with the highest accuracy was selected as the set of potential biomarkers. The ANN model with the highest Youden’s index was selected as the final model for detecting pancreatic cancer.
In the cancer group during the first stage, pathological diagnosis proved that all 23 patients had pancreatic ductal adenocarcinoma, with five stage I, 14 stage II, two stage III, and two stage IV, according to TNM staging; while there were five different types of tumor among the benign disease group (Table 1). In more detail, seven had mucinous cystadenoma, four solid pseudopapillary tumors, three insulinomas, two serous cystadenomas, and two intraductal papillary mucinous tumors (Table 1). The malignant group comprised 13 men and 10 women with a median age of 66 years (range, 30-80 years), while the benign group comprised 10 men and 8 women with a median age of 60 years (range, 49-79 years) (Table 1). Twelve healthy men and eight healthy women formed the third group with a median age of 53 years (range, 36-75 years) (Table 1). Although the serum levels of CA19-9 among the three groups seemed clearly different, the difference was not significant (P = 0.255) (Table 1).
| Training set | Blind test set | P-value1 | |||||||
| Malignant | Benign | Healthy | P-value | Malignant | Benign | Healthy | P-value | ||
| Sex | |||||||||
| Male | 13 (56.52) | 10 (55.56) | 12 (60.00) | 0.957 | 10 (45.45) | 5 (50.00) | 5 (33.33) | 0.662 | 0.665 |
| Female | 10 (43.48) | 8 (44.44) | 8 (40.00) | 12 (54.55) | 5 (50.00) | 10 (66.67) | |||
| Age (yr, mean ± SD) | 62.61 ± 12.71 | 60.61 ± 8.51 | 55.50 ± 13.13 | 0.145 | 65.14 ± 13.02 | 59.10 ± 14.15 | 63.07 ± 12.60 | 0.402 | 0.18 |
| Pathology | |||||||||
| Ductal adenocarcinoma | 23 (100.00) | 22 (100.00) | |||||||
| Mucinous cystadenoma | 7 (38.89) | 2 (20.00) | |||||||
| Solid pseudopapillary tumor | 4 (22.22) | ||||||||
| Insulinoma | 3 (16.67) | 3 (30.00) | |||||||
| Serous cystadenoma | 2 (11.11) | 3 (30.00) | |||||||
| Intraductal papillary mucinous tumor | 2 (11.11) | 2 (20.00) | |||||||
| CA19-9 (U/mL, mean ± SD) | 10.66 ± 8.92 | 11.94 ± 6.93 | 9.67 ± 6.94 | 0.382 | 12.45 ± 9.69 | 15.51 ± 11.60 | 10.00 ± 8.27 | 0.557 | 0.667 |
Among the individuals enrolled in the next stage, 22 patients suspected of having pancreatic diseases were proved to have malignant disease, and had a median age of 68 years (range, 40-91 years). Ten men and twelve women were included in this group (Table 1). The other two groups had 10 and 15 samples, respectively, and were sex- and age-matched on the basis of statistical analysis (Table 1). The respective distribution of serum CA19-9 levels among these three independent populations was nearly the same, as was sex and age (P = 0.557) (Table 1). Compared with the samples in the first stage, none of these three characteristics showed a significant difference (Table 1).
Among all the patients with malignancies and healthy volunteers, the first 23 cancer carriers and 20 healthy individuals comprised the training set, while the remaining 22 and 15 individuals in the respective groups collected at the second stage formed the blind test set (in statistical design).
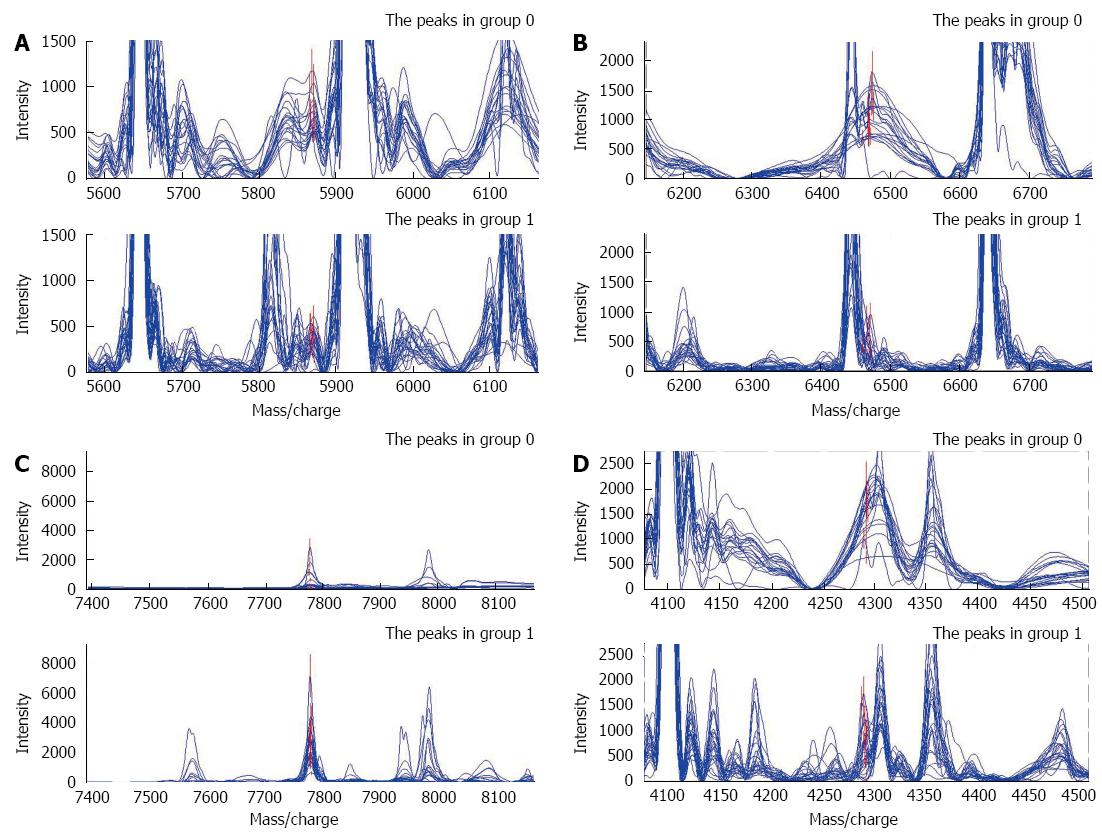
After filtrating noise and identifying peak clusters, we selected qualified peaks generated from 43 training set samples and ranked them in accordance with the P values of the Wilcoxon rank sum test. The top 10 peaks with the smallest P values were further gathered and imported to the support vector machine (SVM) with a random combination. The accuracy of each model in discriminating pancreatic cancer from healthy controls was calculated, and the combination with the highest Youden’s index was finally selected as the diagnostic model. A model consisting of four potential biomarkers with m/z of 5870.0549, 6468.2454, 7777.958, and 4291.7922 Da was generated. Compared with healthy individuals, pancreatic cancer patients presented with a higher level of the protein of 7777.958 Da; while the remaining three peaks were expressed weakly in pancreatic cancer but highly in healthy individuals (Figure 1). The descriptive statistics of these four peaks was shown in Table 2. Evaluated by leave-one-out cross-validation, Pattern 1 comprised four peaks and had a specificity and sensitivity of 100%.
| Intensity in malignant disease (n= 23) | Intensity in healthy individuals (n= 20) | ||
| m/z | (mean ± SD) | (mean ± SD) | P-value |
| 5870.055 | 357.47 ± 137.81 | 688.97 ± 191.35 | 2.61E-07 |
| 6468.245 | 363.30 ± 265.95 | 1107.33 ± 356.79 | 3.84E-07 |
| 7777.958 | 2582.30 ± 1399.85 | 453.67 ± 726.81 | 5.62E-07 |
| 4291.792 | 669.14 ± 497.32 | 1562.50 ± 413.97 | 3.95E-06 |
All 32 suspicious serum samples (22 of which exhibited cancer according to later pathological results) were analyzed with Pattern 1. Compared with pathological diagnosis, test results were correctly made in 13 of 15 healthy individuals and 19 of 22 pancreatic cancer patients. The specificity and sensitivity of the blind test was 86.7% and 86.4%, respectively. One false positive was seen in the benign disease group, which slightly reduced its PPV to 86.4% in practical application (Table 3).
| n | Training set | Blind test set | ||||
| Specificity | Sensitivity | Specificity | Sensitivity | PPV | ||
| Pattern 1 | 4 | 100% | 100% | 86.7% | 86.4% | 86.4% |
| Pattern 2 | 6 | 83.3% | 91.3% | 70.0% | 77.3% | 85.0% |
As in Pattern 1, 23 malignant serum samples and 18 benign ones were analyzed as the training set in Pattern 2, while 22 and 10 samples collected in the next stage comprised the blind test set.
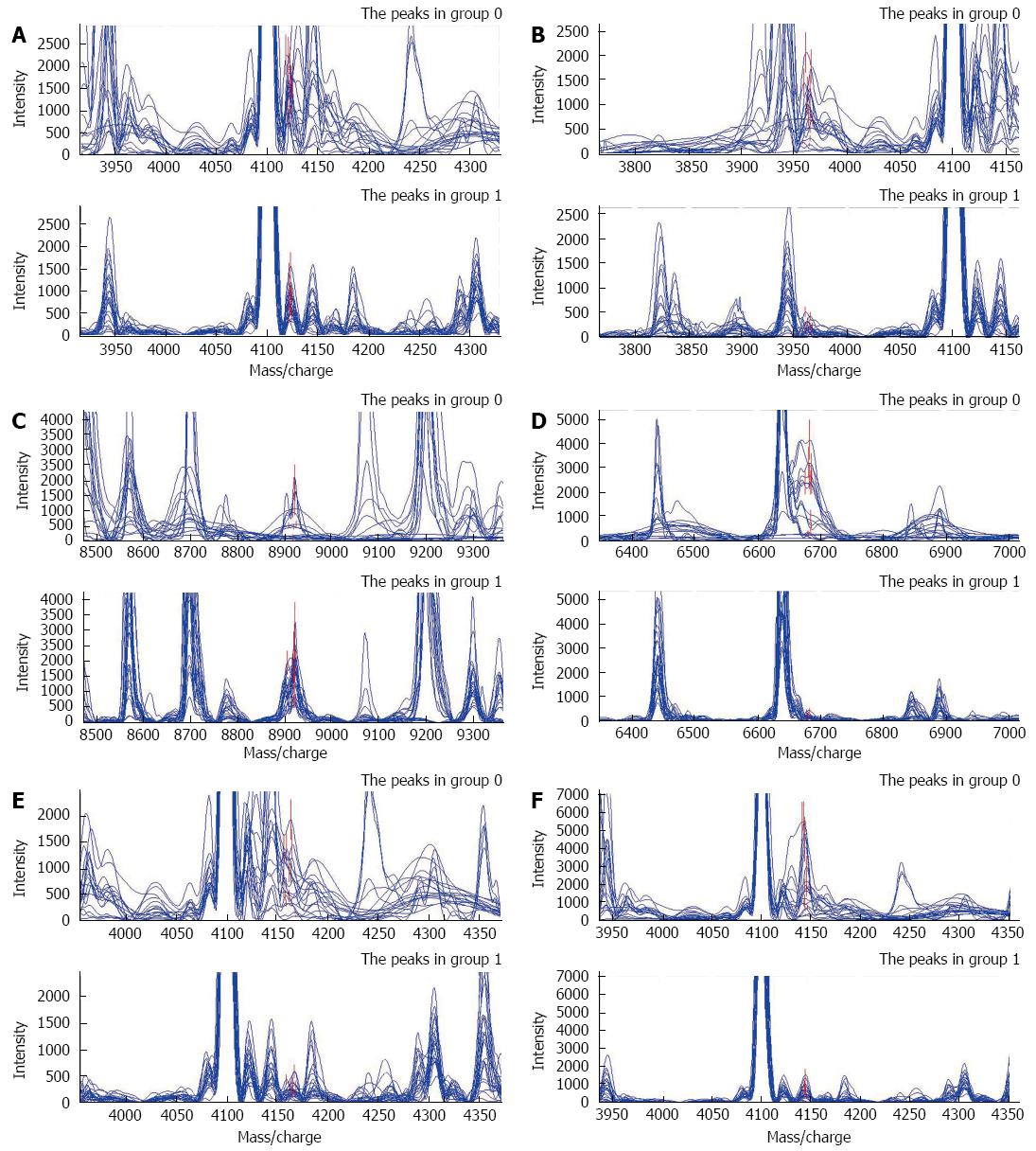
Pattern 2 was constructed on the basis of the analysis of the spectra between pancreatic cancer and benign pancreatic tumors by the ANN. Six potential biomarkers with m/z of 4122.6162, 3964.2531, 8920.0244, 6683.7967, 4163.9423, and 4144.4237 Da were finally selected. Coincidently, among the six proteins, only one protein was highly expressed in the cancer group, while the remaining five proteins were expressed weakly (Figure 2). The descriptive statistics of these six peaks are shown in Table 4. This six-peak model possessed a specificity of 83.33% and a sensitivity of 91.30% by means of LOOCV.
| Intensity in malignant disease (n= 23) | Intensity in benign disease (n= 18) | ||
| m/z | (mean ± SD) | (mean ± SD) | P-value |
| 4122.616 | 762.41 ± 314.68 | 1541.63 ± 453.43 | 5.17E-06 |
| 3964.253 | 211.84 ± 118.31 | 875.93 ± 547.64 | 3.51E-05 |
| 8920.024 | 1574.91 ± 686.01 | 540.98 ± 635.90 | 3.93E-05 |
| 6683.797 | 226.35 ± 94.75 | 1389.22 ± 1308.83 | 1.32E-04 |
| 4163.942 | 272.72 ± 142.38 | 791.27 ± 509.13 | 2.23E-04 |
| 4144.424 | 631.43 ± 392.13 | 2292.02 ± 1801.45 | 3.71E-04 |
Thirty-two samples in the blind test set were analyzed on the second day after collection, as for Pattern 1. Compared with their pathological diagnosis, the model correctly diagnosed seven benign diseases and 17 pancreatic cancers. The specificity and sensitivity in the blind test dropped to 70.0% and 77.3%, respectively (Table 3).
Although the clinical utility of CA19-9 in the diagnosis of pancreatic cancer is still unclear and controversial[1,8,15-17], it is frequently used by clinicians as an indicator. Nevertheless, normal CA19-9 level is not uncommon among pancreatic cancer patients, which may cause a diagnostic dilemma and even result in neglecting the possibility of malignancy, thus leading to delayed medical intervention. As far as we are aware, this situation has not been researched previously. Therefore, we designed the present study to focus on those patients with normal serum CA19-9 levels through SELDI-TOF-MS combined with ANN analysis, which has already proved useful in the detection of other digestive solid tumors such as colorectal cancer[18] and hepatocellular carcinoma[19].
In our study, Pattern 1 demonstrated four potential biomarkers to distinguish pancreatic cancer patients from healthy individuals when it was not possible to use CA19-9. To avoid false detection of protein peaks and make the results reasonable and repeatable were the main challenges among mass-spectrometry-generated data analysis. ANN analysis and leave-one-out cross-validation were applied to filter the weak or fake peaks and generate the most powerful pattern for discrimination. An adequate number of samples were accumulated until the difference among the distribution of sex, age and serum CA19-9 level became statistically insignificant (Table 1), to ensure the reasonability of the pattern. Ideal sensitivity and specificity (both 100%) were obtained in the training set. Sensitivity of 86.4%, specificity of 86.7%, and PPV of 86.4% were late observations in the following blind test. Although they were not as good as the training test (theoretical values), the practical identification capability of Pattern 1 was still acceptable. Hence, it could be helpful as a fast and noninvasive diagnostic modality to discriminate pancreatic cancer when serum CA19-9 level is normal.
CA19-9 was once the most widely-investigated screening biomarker for pancreatic cancer, but was unable to detect pancreatic cancer reliably. Poor results were observed in a large study by Kim et al[8] with 70940 asymptomatic individuals enrolled and followed. The PPV of pancreatic cancer with CA19-9 was only 0.9%, and 98.5% of the population were CA19-9 negative (< 37 U/mL). It has been reported that serum CA19-9 level of pancreatic adenocarcinoma is associated with tumor size and stage: small cancers or those in early stage were more likely to present with normal CA19-9 level[2,8]. Therefore, the markers we identified with SELDI-TOF-MS in Pattern 1 could probably serve as a potentially powerful screening tool. This needs to be confirmed in a large prospective study.
Patients with benign pancreatic diseases are more likely to have a normal CA19-9 level[3,20], therefore we compared the proteomic spectra between pancreatic cancer and benign tumors, which resulted in another diagnostic pattern model. Six candidate biomarkers provided a specificity of 70.0% and a sensitivity of 77.3% to differentiate pancreatic cancer from benign tumors. Some of the proteins identified by SELDI-TOF-MS are associated with inflammation induced by tumor-induced nonspecific tissue injury[21], and bioactive molecules from different parts of the pancreas or different tumors differ in their constitution and quantity[22], thus it is not surprising that compared with different combinations (e.g., cancer and healthy controls, or cancer and benign tumors), SELDI-TOF-MS profiling generated a different pattern. The inclusion of benign tumors helped us to distinguish these patients from cancer carriers, and to differentiate tumor-induced markers from cancer-specific molecules. Despite the reduction in sensitivity and specificity that might have been related to the different proportions of each benign tumor, the model was still helpful for its acceptable PPV.
Although we could make an accurate diagnosis without clarifying the identity of each protein peak by SELDI-TOF-MS, it is practical to purify and identify these proteins, which may promote the development of antibody-based clinical tests[22] and target-molecule-based clinical treatment. Subsequent research is needed to validate the findings with large samples, ideally in a multicenter setting.
In conclusion, our study probably provides clinicians with a helpful alternative for differentiation of pancreatic cancer patients from healthy controls or patients with benign disease. Pattern 1 may also serve as a useful screening tool for pancreatic cancer. SELDI-TOF-MS profiling is effective for analysis of serum proteins due to its high specificity and sensitivity. The proteins that we discovered may play an important role as tumor/cancer-specific or -associated antigens in future clinical practice. A large clinical trial is needed to confirm the diagnostic utility of our models.
Pancreatic cancer is one of the most lethal malignancies because of lack of efficient screening and diagnostic modalities. The most widely-used biomarker carbohydrate antigen (CA)19-9, is not reliable, especially at the beginning or early stage of the disease. Additionally, quite a few patients with advanced disease are CA19-9 negative. Surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) recently became an efficient technique in screening for early diagnostic markers and the potentially therapeutic molecular targets of tumors. The authors provided a non-invasive and efficient method for screening pancreatic cancer in a healthy population or in patients with benign diseases by SELDI-TOF-MS combined with protein chip technology.
Finding tumor-specific biomarkers which could facilitate the diagnosis or differential diagnosis of pancreatic cancer or to investigate those proteins that may serve as potential screening tools remains a research hotspot.
This study focused on pancreatic cancer patients with normal CA19-9 levels that would be easily misdiagnosed in clinical practice. The results may help clinicians to establish the diagnosis for cancer patients when CA19-9 is not applicable, and to discover some potentially important proteins which may serve as tumor/cancer-specific or tumor/cancer-associate antigen in clinical practice.
The two diagnostic models generated in this study provide clinicians with a helpful alternative in differentiating pancreatic cancer patients from healthy individuals or benign patients. Pattern 1 may also serves as a useful screening tool for pancreatic cancer. The discovered proteins may play an important role as tumor/cancer-specific or tumor/cancer-associate antigen in cancer diagnosis and treatment.
CA19-9: A tumor/cancer-associate antigen which is mainly associated with hepatopancreatobiliary diseases. Patients with pancreatic cancer often present with extremely high levels of CA19-9.
A study based on a reasonable hypothesis of finding a biomarker signature using gene chip technology to distinguish between benign pancreatic conditions and malignant pancreatic carcinoma. The paper is interesting.
P- Reviewer: Dobrucali AM, Yip D S- Editor: Ma YJ L- Editor: Rutherford A E- Editor: Wang CH
| 1. | Duffy MJ, Sturgeon C, Lamerz R, Haglund C, Holubec VL, Klapdor R, Nicolini A, Topolcan O, Heinemann V. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann Oncol. 2010;21:441-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 266] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 2. | Ballehaninna UK, Chamberlain RS. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J Gastrointest Oncol. 2012;3:105-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 352] [Reference Citation Analysis (3)] |
| 3. | Zubarik R, Gordon SR, Lidofsky SD, Anderson SR, Pipas JM, Badger G, Ganguly E, Vecchio J. Screening for pancreatic cancer in a high-risk population with serum CA 19-9 and targeted EUS: a feasibility study. Gastrointest Endosc. 2011;74:87-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 4. | Joergensen MT, Brünner N, De Muckadell OB. Comparison of circulating MMP-9, TIMP-1 and CA19-9 in the detection of pancreatic cancer. Anticancer Res. 2010;30:587-592. [PubMed] |
| 5. | Sandblom G, Granroth S, Rasmussen IC. TPS, CA 19-9, VEGF-A, and CEA as diagnostic and prognostic factors in patients with mass lesions in the pancreatic head. Ups J Med Sci. 2008;113:57-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 6. | Morris-Stiff G, Taylor MA. Ca19-9 and pancreatic cancer: Is it really that good? J Gastrointest Oncol. 2012;3:88-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 7. | Ballehaninna UK, Chamberlain RS. Serum CA 19-9 as a Biomarker for Pancreatic Cancer-A Comprehensive Review. Indian J Surg Oncol. 2011;2:88-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 8. | Kim JE, Lee KT, Lee JK, Paik SW, Rhee JC, Choi KW. Clinical usefulness of carbohydrate antigen 19-9 as a screening test for pancreatic cancer in an asymptomatic population. J Gastroenterol Hepatol. 2004;19:182-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 265] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Wright GL. SELDI proteinchip MS: a platform for biomarker discovery and cancer diagnosis. Expert Rev Mol Diagn. 2002;2:549-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Weinberger SR, Boschetti E, Santambien P, Brenac V. Surface-enhanced laser desorption-ionization retentate chromatography mass spectrometry (SELDI-RC-MS): a new method for rapid development of process chromatography conditions. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:307-316. [PubMed] |
| 11. | Yang S, Nan Y, Tian Y, Zhang W, Zhou B, Bu L, Huo S, Chen G, Yu J, Zheng S. Study of distinct protein profiles for early diagnosis of NSCLC using LCM and SELDI-TOF-MS. Med Oncol. 2008;25:380-386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Xu WH, Chen YD, Hu Y, Yu JK, Wu XG, Jiang TJ, Zheng S, Zhang SZ. Preoperatively molecular staging with CM10 ProteinChip and SELDI-TOF-MS for colorectal cancer patients. J Zhejiang Univ Sci B. 2006;7:235-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | Yu JK, Zheng S, Tang Y, Li L. An integrated approach utilizing proteomics and bioinformatics to detect ovarian cancer. J Zhejiang Univ Sci B. 2005;6:227-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Kim HJ, Kim MH, Myung SJ, Lim BC, Park ET, Yoo KS, Seo DW, Lee SK, Min YI. A new strategy for the application of CA19-9 in the differentiation of pancreaticobiliary cancer: analysis using a receiver operating characteristic curve. Am J Gastroenterol. 1999;94:1941-1946. [PubMed] |
| 15. | Singh S, Tang SJ, Sreenarasimhaiah J, Lara LF, Siddiqui A. The clinical utility and limitations of serum carbohydrate antigen (CA19-9) as a diagnostic tool for pancreatic cancer and cholangiocarcinoma. Dig Dis Sci. 2011;56:2491-2496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 16. | Molina V, Visa L, Conill C, Navarro S, Escudero JM, Auge JM, Filella X, Lopez-Boado MA, Ferrer J, Fernandez-Cruz L. CA 19-9 in pancreatic cancer: retrospective evaluation of patients with suspicion of pancreatic cancer. Tumour Biol. 2012;33:799-807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 17. | Liao Q, Zhao YP, Yang YC, Li LJ, Long X, Han SM. Combined detection of serum tumor markers for differential diagnosis of solid lesions located at the pancreatic head. Hepatobiliary Pancreat Dis Int. 2007;6:641-645. [PubMed] |
| 18. | Chen YD, Zheng S, Yu JK, Hu X. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin Cancer Res. 2004;10:8380-8385. [PubMed] |
| 19. | Wang JX, Zhang B, Yu JK, Liu J, Yang MQ, Zheng S. Application of serum protein fingerprinting coupled with artificial neural network model in diagnosis of hepatocellular carcinoma. Chin Med J (Engl). 2005;118:1278-1284. [PubMed] |
| 20. | Duraker N, Hot S, Polat Y, Höbek A, Gençler N, Urhan N. CEA, CA 19-9, and CA 125 in the differential diagnosis of benign and malignant pancreatic diseases with or without jaundice. J Surg Oncol. 2007;95:142-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 21. | Rai AJ, Zhang Z, Rosenzweig J, Shih IeM, Pham T, Fung ET, Sokoll LJ, Chan DW. Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med. 2002;126:1518-1526. [PubMed] |
| 22. | Koopmann J, Zhang Z, White N, Rosenzweig J, Fedarko N, Jagannath S, Canto MI, Yeo CJ, Chan DW, Goggins M. Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin Cancer Res. 2004;10:860-868. [PubMed] |