Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.8092
Revised: July 20, 2013
Accepted: August 4, 2013
Published online: July 7, 2014
Processing time: 410 Days and 6.8 Hours
Metabolomics is a field of study in systems biology that involves the identification and quantification of metabolites present in a biological system. Analyzing metabolic differences between unperturbed and perturbed networks, such as cancerous and non-cancerous samples, can provide insight into underlying disease pathology, disease prognosis and diagnosis. Despite the large number of review articles concerning metabolomics and its application in cancer research, biomarker and drug discovery, these reviews do not focus on a specific type of cancer. Metabolomics may provide biomarkers useful for identification of early stage gastric cancer, potentially addressing an important clinical need. Here, we present a short review on metabolomics as a tool for biomarker discovery in human gastric cancer, with a primary focus on its use as a predictor of anticancer drug chemosensitivity, diagnosis, prognosis, and metastasis.
Core tip: This article presents a short review on metabolomics as a tool for biomarker discovery in human gastric cancer, with a primary focus on its use as a predictor of anticancer drug chemosensitivity, diagnosis, prognosis, and metastasis.
- Citation: Jayavelu ND, Bar NS. Metabolomic studies of human gastric cancer: Review. World J Gastroenterol 2014; 20(25): 8092-8101
- URL: https://www.wjgnet.com/1007-9327/full/v20/i25/8092.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i25.8092
Gastric cancer is the fourth most common cancer and the second most deadly cancer worldwide[1,2]; it is particularly prevalent in Asian countries[3,4]. According to the American Cancer Society, approximately 738000 people died worldwide from stomach cancer in 2008[5]. At present, no effective treatment is available for this disease, and identification of early stage gastric cancer is difficult because it is often asymptotic or misdiagnosed. Moreover, the prognosis of patients with advanced gastric cancer remains poor due to its high metastatic recurrence[6,7], and the complex molecular mechanisms underlying metastasis are not well characterized[8,9].
Presently, early diagnosis of human gastric cancer or tumor recurrence is primarily based on endoscopy, biopsy and pathological examination. Endoscopy is a widely used method for detecting early stages of gastric cancer[10-12] despite its inconsistent diagnostic efficiency, which stems from variations in the skill and experience of the endoscopist and pathologist. In recent years, several serum biomarkers have been identified as new tools for early screening of gastric cancer in developed countries[11-16]. However, these serum biomarkers are not effective as other screening devices given their low specificity and sensitivity[13]. Recently, epidemiological data have revealed that Helicobacter pylori (H. pylori) infection and dietary factors are the main risk factors associated with gastric cancer[1,2].
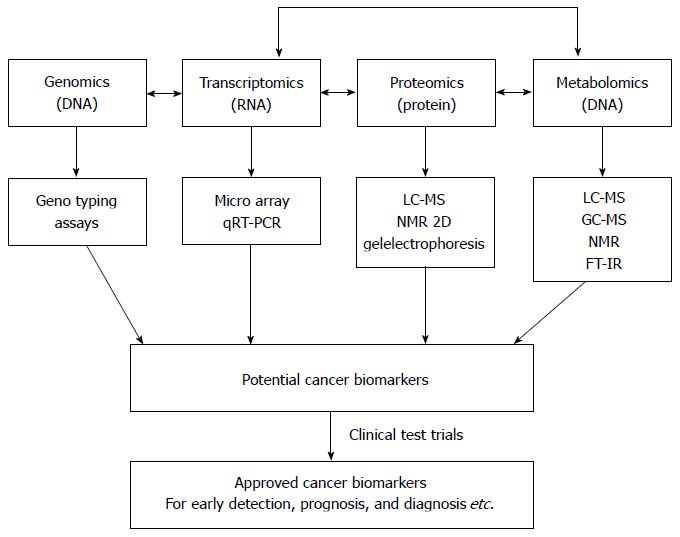
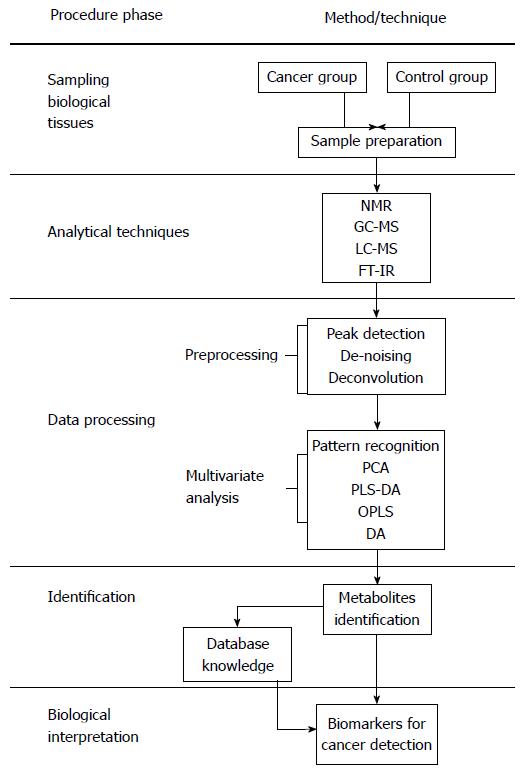
An overview of traditional methods involved in gastric cancer detection, diagnosis and prognosis in comparison with metabolomic methods is presented in Table 1. The field of metabolomics may offer practical solutions to the challenges mentioned above. Metabolomics, the study of the unique metabolite signature in a biological system (cell, tissue, or organism) under a given set of conditions[17], has emerged as a promising technology in the study of human cancers. Metabolites are not merely the end product of gene expression; rather, they are the result of the interaction of the system’s genome with its environment. They are an integral part of any cellular regulatory system[18]. Metabolomics is regarded as one of the new high-throughput, “-omics” technologies. Along with genomics, transcriptomics, and proteomics, metabolomics is a scientific field of study that seeks to achieve the aims of systems biology[18,19]. The biological organization of different “-omes” and the flow of information from the genome to the transcriptome, the proteome and finally the metabolome is presented in Figure 1[20]. Metabolomic studies offer a unique approach for identifying metabolomic pathways that are perturbed under specific conditions[21,22], thereby providing information different from other “-omic” technologies[19]. In recent years, metabolomic studies have been successfully conducted in various cancer systems, including stomach[21], lung[23,24], renal[25,26], breast[27], brain[28] and colorectal[29-32]. Metabolomic studies have also been conducted in human xenograft models[33-38] (transplantation of living cells, tissues or organs from one species to another). These studies can provide valuable information in terms of novel biomarkers that identify cancerous cells. A biomarker[39] often represents a component found in plasma, whose concentration indicates the presence or the severity of disease states. Biomarkers can therefore serve as an indicator of tumor progression and treatment efficacy. Biomarkers can be chemical, physical or biological in nature. Metabolomic studies typically begin with tissue sampling, followed by sample analysis. Nuclear magnetic resonance spectroscopy (NMR) is the most common method of analysis. The large amount of data generated by this analysis is then statistically processed to identify the metabolites that are differentially expressed between the samples, possibly leading to biomarker selection (Figure 2[12,40-42]). The key to identifying potential biomarkers is based on the level of metabolite differences in biological samples taken from cancer patients and normal (control) subjects. Metabolomics also has potential utility in several fields of cancer research, including prognosis[43], diagnosis[44,45] and drug evaluation and development[46-48]. It can also serve as an alternative strategy for personalized cancer therapy[49,50].
| Cancer detection state/stage | Traditional methods | Metabolomics (biomarkers) | Ref. |
| Diagnosis | Endoscopy, biopsy | Lactic acid, butanedioic acid, malic acid, citric acids, pyruvic acid, 3-hydroxypropionic acid, serine, proline | [91,93,100,101] |
| Prognosis | Radiotherapy, chemotherapy surgery | Valine, isoleucine, serine, 3-indoxyl sulfate, hippurate, citrate | [96,99,102] |
| Metastasis | Computed tomography (CT) scanning, endoscopic ultrasonography (EUS), positron emission tomography (PET) | Sarcosine, alanine, proline, serine, myo-inositol, glycerol | [90,91,98,103] |
| Chemosensitivity of drugs | MTT chemosensitivity assay | 1-acyl-lysophosphatidylcholines and polyunsaturated fatty acids | [75,104] |
Several review articles[12,40,51,52] have been published on metabolomic applications in cancer research[20,53-55], biomarker discovery[39,56,57] and natural product drug discovery[18]. However, none of them have focused on a specific type of cancer, particularly gastric cancer. Hence, the aim of this article is to provide a brief overview of the benefits of metabolomic studies to human gastric cancer research, with a special focus on biomarkers. The remainder of the paper is organized as follows. In next section, we briefly discuss different analytical techniques used in metabolomic studies and methods for data analysis. Then, we review several studies of applying metabolomics to gastric cancer research. Finally, future directions and concluding remarks are presented.
A number of analytical techniques are currently used for metabolomic studies depending on the particular metabolite of interest. In general, NMR spectroscopy (in most cases 1H-NMR)[58,59], liquid chromatography (LC)[26,60]/gas chromatography (GC)-mass spectrometry (MS)[31,61,62], Fourier transform spectrometry[63,64] and capillary electrophoresis (CE)-mass spectrometry[65-68] are the major spectroscopic techniques used in metabolomic analysis. Generally, a combination of different methods provides more information than a single method when analyzing the complete metabolome. NMR is one of the most common analytical methods for urine and plasma analysis[69] due to its non-destructive nature, quantitative ability, and safe metabolite identification that provides detailed information on molecular structure. However, NMR suffers from poor sensitivity. GC-/MS and LC-/MS are widely accepted techniques for metabolite separation and analysis. Metabolites must be volatile in nature in order to use the GC-/MS technique efficiently. Fatty acids, organic acids and sugars are the best-suited metabolites for GC-/MS. In contrast, LC-/MS can cover a broad range of metabolites, including both volatile and non-volatile compounds. CE-/MS is best suited for studies involving energy metabolism given its ability to simultaneously quantify charged, low-molecular weight compounds. A short overview of the advantages and limitations of the different metabolomic methods is presented in Table 2. GC-MS, LC-MS and NMR are the most commonly used methods in cancer research, especially gastric cancer.
| Method | Sampling characteristics | Sensitivity | Advantages | Disadvantages | Ref. |
| Nuclear magnetic resonance (NMR) spectroscopy | Non-destructive; minimum sample required | 10-6 | Fully automated with a high degree of reproducibility; relatively easy to identify metabolites from simple one-dimensional spectra | Lower sensitivity than mass spectrometry; co-resonant metabolites can be difficult to quantify; drug metabolites can be co-resonant with metabolites of interest | [20,41,105] |
| Gas chromatography-mass spectrometry (GC-MS) | Requires extraction, sample dried and chemical derivation | 10-12 | A relatively cheap and reproducible method with a high degree of sensitivity | Sample preparation can be time consuming; not all compounds are suitable for gas chromatography | [20,41,106,107] |
| Liquid chromatography-mass spectrometry (LC-MS) | Requires extraction and concentration (vacuum drying), liquid-liquid extraction | 10-15 | This method is increasingly being used in place of GC-MS as sample preparation is not as time consuming; has a sensitivity similar to GC-MS | More costly than GC-MS and depends on the reproducibility of liquid chromatography (more difficult to control than GC); can also suffer from ion suppression | [20,41,108,109] |
| Fourier-transform infrared (FT-IR) spectrometry | Uses vibrational frequencies of metabolites to produce a fingerprint of metabolism | 10-6 | Cheap and good for high-throughput first screening | Very difficult to identify which metabolites are responsible for causing changes; very poor at distinguishing metabolites within a class of compounds | [20,41,110,111] |
| Raman spectroscopy | Non-destructive; minimum sample required, occasionally hydration is needed | 10-6 | Has the advantage over FT-IR in that water has only a weak Raman spectrum; therefore, many functional groups can be observed | Very poor at distinguishing classes of compounds | [20,41,110,111] |
Data integration and analysis is an important component of metabolomic studies because a large amount of data is generated, similar to proteomic and transcriptomic studies. Proper management, pre-processing and analysis of these data pose a significant challenge and require sophisticated multivariate statistical software. A sufficient number of statistical algorithms have been developed for the analysis of metabolomic data, both in a supervised and unsupervised manner. The important unsupervised methods that have been extensively used in metabolomic analysis include principal component analysis (PCA), hierarchical clustering and self-organizing maps. Supervised methods include ANOVA, partial least squares (PLS), hierarchical PLS, k-nearest neighbors (KNN) and discriminant function analysis. The principle details and applications of these methods can be found elsewhere[44,70-72]. A short comparison of these methods including advantages and limitations is provided in review articles[41,52,55].
Chemosensitivity prediction is a challenging task in the treatment of advanced gastric cancer[73]. Chemotherapy with anticancer drugs plays a significant role in the personalized management of gastric cancer[74]. Some patients with gastric cancer do not respond well to these drugs, and in some cases, chemotherapy may cause severe toxicity and functional impairment[75-78]. Hence, it is crucial to select individual patients with high chemosensitivity for the management of cancer by chemotherapy treatment. The two major approaches for predicting the activity of anticancer drugs in gastric cancer are resistance enzyme testing and cell-culture testing (chemosensitivity)[73]. In the past, chemosensitivity predictions have been based on clone formation, cell metabolic activity assays in vitro, proliferation, and tumor growth. Unfortunately, these methods suffer from low specificity, sensitivity and accuracy[75].
In order to overcome these limitations, high-throughput “-omic” methods have been developed as powerful tools for use in different types of cancer treatments[79-82]. Wang et al[75] described a metabolic approach for chemosensitivity prediction in a human xenograft model of gastric cancer treated with cisplatin and 5-fluorouracil. In this approach, mice were divided randomly into control and treatment groups (i.e., resistant, intermediate, and sensitive groups based on relative tumor growth). Blood plasma was collected, and metabolic profiles were obtained by using high performance liquid chromatography coupled with a quadrupole time-of-flight mass spectrometer (HPLC/Q-TOF-MS). From the metabolic data, a predictive model was developed using a KNN algorithm[83] with 90% accuracy, and 18 chemosensitivity metabolites for gastric cancer were proposed in their study. Key metabolites included 1-acyl-lysophosphatidylcholine and polyunsaturated fatty acids, which are hydrolysis products of phosphatidylcholine. The 1-acyl-lysophosphatidylcholine biochemical pathway regulates the activity of enzymes like phospholipases A2 and B1 and lysophosphatidylcholine acetyltransferases[84-88]. Thus, these key metabolites could serve as crucial modulators of gastric cancer chemosensitivity.
Metastasis[22] is the spread of a disease from one organ or part to a non-adjacent organ or part. Most gastric cancer deaths occur as a result of metastasis. It is important to explore the complex mechanisms of gastric cancer metastasis in order to identify the key metabolic markers involved in the process. Several genes involved in gastric cancer metastasis have been reported in the literature[8,9,89]. However, no potential biomarkers were identified as predictors of metastasis and prognosis due to large variations in expression levels. Chen et al[90] have conducted metabolomic studies on human xenograft models to elucidate the underlying mechanisms of gastric cancer metastasis and discover possible biomarkers for diagnosis. Their mice were randomized into control, metastatic, and non-metastatic groups, and tissue samples from each group were collected and analyzed using GC-MS. Their study identified approximately 30 metabolites differentially regulated among the groups. Proline was the most up-regulated tissue metabolite in the metastatic group, with a 2.45-fold increase in expression compared with the non-metastatic group. Glutamine was the most down-regulated metabolite, with a 1.71-fold reduction in expression in the metastatic group compared with the non-metastatic group. All of these metabolites were involved in pathways associated with gastric cancer metastasis, and most of them were found in proline and serine metabolism. Hu et al[91] also conducted similar metabolomic studies, but their metabolic profiles were obtained from urine samples. A PCA model was developed to discriminate the gastric cancer model from control and to differentiate the metastatic and non-metastatic groups. The level of lactic acid was increased in the cancer group compared with the normal group. The noted increase may be attributed to the ‘Warburg effect’, where glucose is converted to lactic acid in cancer cells due to an increased rate of aerobic glycolysis[92]. Chen et al[93] developed a urinary metabolic model based on human xenograft models to distinguish between metastatic and non-metastatic groups. GC-MS studies were also conducted on samples from cancer patients and healthy controls. The metabolites lactic acid, serine, proline, malic acid and fatty acids showed significant metabolic differences between cancerous and non-cancerous groups. From the above discussion, it is clear that proline and serine metabolism plays an important role in metastasis, and metabolic biomarkers derived from those pathways can be used for the treatment of gastric cancer metastasis.
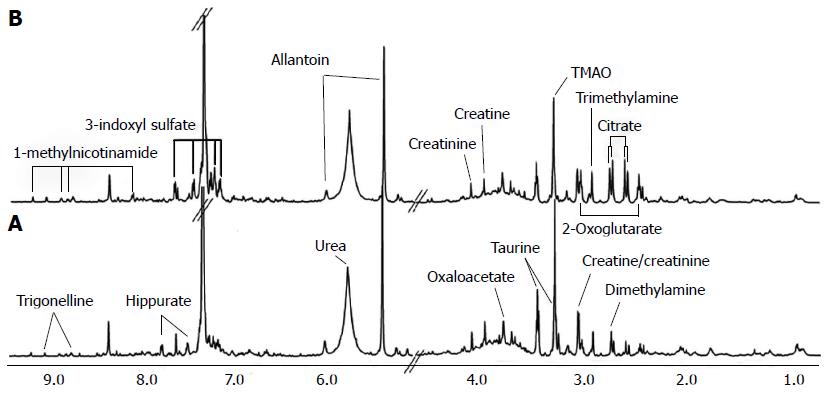
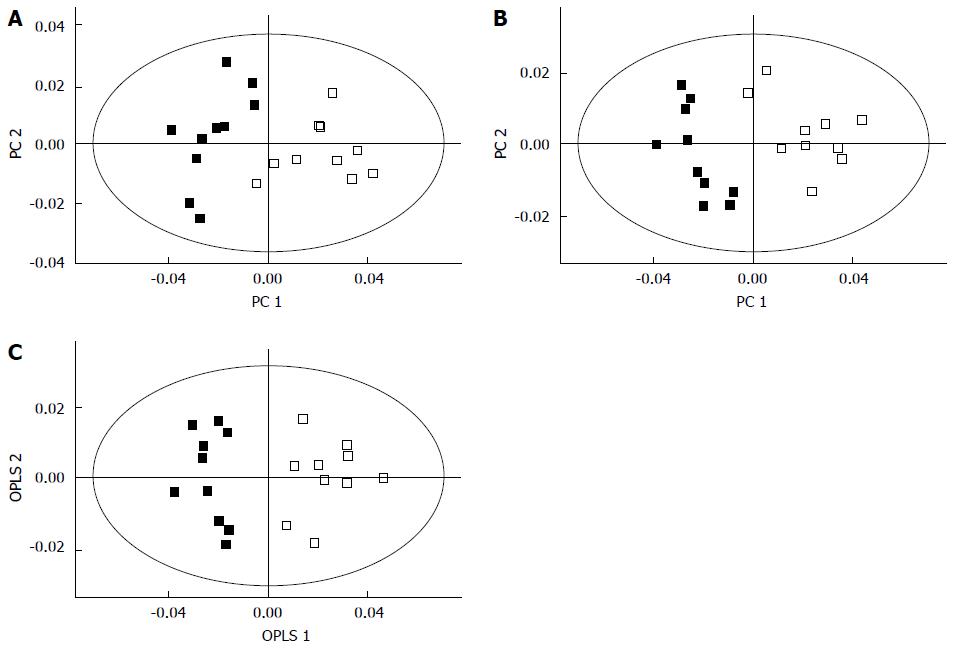
Biomarkers play a vital role in early stage diagnosis, disease prognosis, drug target identification, and patient reaction to a particular treatment. Several biomarkers have been proposed for gastric cancer diagnosis and prognosis. For example, serum amyloid A was proposed as a sensitive diagnostic biomarker[94], and the inhibitor of matrix metalloproteinase-1 was suggested as a potential prognostic biomarker[95]. Kim et al[96] conducted 1H-NMR-based metabolomic studies on mouse models to identify possible urinary biomarkers for human gastric cancer. A comparison of the NMR spectra for the cancer and control groups is shown in Figure 3[96], and the metabolite trimethylamine oxide (TMAO) is significantly reduced in cancer cells compared with the control, and it is clearly visible in the spectra. Pattern recognition methods attempting to discriminate the control from the tumor group indicated (Figure 4[96]) a clear separation between the cancer and control groups, thus implying the presence of significant metabolic differences in certain metabolites between these two groups. TMAO, 3-indoxyl sulfate, hippurate, 2-oxoglutarate, and citrate showed significant changes in concentration between cancer and control groups and were proposed as potential urinary biomarkers for gastric cancer detection. Yu et al[97] established a metabolic model to characterize several different stages of gastric cancer including chronic superficial gastritis (CSG), chronic atrophic gastritis (CAG), intestinal metaplasia (IM), gastric dysplasia (DYS) and GC. CSG showed metabolic patterns distinct from the other groups (i.e., CAG, IM, DYS, and GC, whose plots were closely clustered). IM closely clustered with GC, suggesting that these two stages share similar metabolic patterns. Fifteen metabolites displayed distinct metabolic signatures, facilitating discrimination of CSG and GC and characterization of different stages of GC. These biomarkers can be useful for indicating GC risk. Song et al[98] developed a similar metabolic model based on metabolomic studies of serum samples from cancer and control groups. In this study, the supervised multivariate statistical method orthogonal partial least squares discriminant analysis was applied to discriminate between cancer and non-cancer groups, but this model failed to distinguish the different tumor node metastasis stages of cancer. In addition, approximately 50 metabolites, many involved in amino acid and fatty acid metabolism, displayed significant metabolic differences between cancer and control groups and were proposed as potential markers for the detection of cancer. In an additional metabolomic study on gastric cancer patients, Wu et al[99] identified tissue metabolic markers and confirmed that valine metabolism was involved in the metabolic changes associated with gastric cancer. In another study[100], a metabolic diagnostic model was developed to characterize gastrointestinal cancer (esophageal, gastric, and colorectal cancers) based on serum metabolomics.
Thus, biomarkers discovered from metabolomic studies may play a significant role in gastric cancer with regard to early stage detection, diagnosis, prognosis, drug development and chemosensitivity predictions. The complete details of metabolomics studies on human gastric cancer including study population, sample type and analytical method used are presented in Table 3.
| Patients/xenograft model | Sample | Sample size (cancer + control) | Analytical method | Multivariate method | Major findings | Ref. |
| Both Xenograft model Patients | Urinary sample | 33 | GC-MS | PCA | Lactic acid, serine, proline, malic acid and fatty acids as potential markers for screening and early diagnosis | [93] |
| Patients | Serum | 60 | GC-MS | OPLS-DA | Sarcosine as a potential biomarker for the progression of gastric cancer metastasis | [98] |
| Patients | Plasma | 80 | GC-TOF-MS | PLS-DA | Azelaic acid, glutamate, urate, creatinine, threonate as markers for characterizing the precancerous stages and gastric cancer | [97] |
| Patients | Serum | 50 | GC-MS | PCA | 3-hydroxypropionic acid and pyruvic acids as potential diagnostic markers for gastric cancer | [100] |
| Patients | Tissue | 18 | GC-MS with chemical derivatization | PCA | Valine, isoleucine, serine and phosphoserine for diagnosis and staging of gastric cancers | [99] |
| Xenograft model | Plasma | 80 | HPLC/Q-TOF-MS | PLS and hierarchical PLS | 1-acyl-lysophosphatidylcholines and polyunsaturated fatty acids as potential indicators of chemosensitivity for gastric cancer | [75] |
| Xenograft model | Urinary sample | 24 | GC/MS | PCA | Lactic acid, butanedioic acid, malic acid and citric acids as potential markers for cancer screening. Alanine, proline, myo-inositol and glycerol as key markers for identifying cancer metastasis | [91] |
| Xenograft model | Tissue | 22 | GC/MS | PCA | Serine and proline metabolism pathways were enriched in cancer metastasis and may help elucidate the complex molecular mechanisms governing metastasis | [90] |
The use of metabolomics in human gastric cancer to discover novel biomarkers is an emerging field. The metabolomics field is superior to other “-omic” methods, as it provides accurate quantities of metabolites in a particular biological system. Hence, the biomarkers identified by metabolomics are likely to be reliable. NMR, GC-MS and LC-MS metabolic techniques are widely used in gastric cancer research. Furthermore, a large number of multivariate data analysis methods have been developed to analyze metabolomic data; PCA and PLS are the most prominent examples. However, despite the number of statistical tools available in metabolomics, many of these methods have limitations; thus, room for further development exists.
Metabolomics has also demonstrated promise in the development of diagnostic tools for gastric cancers. These studies are based on small cohorts; therefore, larger studies are needed for validation of biomarker utility and thereafter translation to a clinical setting. The ability to obtain a high quality sample along with sample collection, storage and analysis are all factors that have large consequences on metabolic results. This fact underscores the need for standardized protocols. Metabolomic studies are beneficial for cancer identification, diagnosis and prognosis. Moreover, by combining metabolomics with other ‘‘-omic’’ methods, a more comprehensive understanding of the processes involved in cancer development is likely to be generated.
We would like to thank Arne Kristian Sandvik, Astrid Lægreid and Tone Frost Bathen for their comments on the manuscript.
P- Reviewers: Kita H, Lan K, Nishida T, Tanaka N S- Editor: Gou SX L- Editor: A E- Editor: Wang CH
| 1. | Crew KD, Neugut AI. Epidemiology of upper gastrointestinal malignancies. Semin Oncol. 2004;31:450-464. [PubMed] |
| 2. | Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354-362. [PubMed] |
| 3. | Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009;472:467-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 411] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 4. | Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 578] [Cited by in RCA: 647] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 5. | Society AC. Global Cancer Facts and Figures. 2nd ed. Atlanta: American Cancer Society 2011; . |
| 6. | Macdonald JS. Gastric cancer--new therapeutic options. N Engl J Med. 2006;355:76-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 7. | Cunningham D, Chua YJ. East meets west in the treatment of gastric cancer. N Engl J Med. 2007;357:1863-1865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Rajdev L. Treatment options for surgically resectable gastric cancer. Curr Treat Options Oncol. 2010;11:14-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 9. | Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8:629-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 340] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 10. | Tashiro A, Sano M, Kinameri K, Fujita K, Takeuchi Y. Comparing mass screening techniques for gastric cancer in Japan. World J Gastroenterol. 2006;12:4873-4874. [PubMed] |
| 11. | Sipponen P, Ranta P, Helske T, Kääriäinen I, Mäki T, Linnala A, Suovaniemi O, Alanko A, Härkönen M. Serum levels of amidated gastrin-17 and pepsinogen I in atrophic gastritis: an observational case-control study. Scand J Gastroenterol. 2002;37:785-791. [PubMed] |
| 12. | Lu X, Zhao X, Bai C, Zhao C, Lu G, Xu G. LC-MS-based metabonomics analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;866:64-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 13. | Miki K, Morita M, Sasajima M, Hoshina R, Kanda E, Urita Y. Usefulness of gastric cancer screening using the serum pepsinogen test method. Am J Gastroenterol. 2003;98:735-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 161] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Kitahara F, Kobayashi K, Sato T, Kojima Y, Araki T, Fujino MA. Accuracy of screening for gastric cancer using serum pepsinogen concentrations. Gut. 1999;44:693-697. [PubMed] |
| 15. | Väänänen H, Vauhkonen M, Helske T, Kääriäinen I, Rasmussen M, Tunturi-Hihnala H, Koskenpato J, Sotka M, Turunen M, Sandström R. Non-endoscopic diagnosis of atrophic gastritis with a blood test. Correlation between gastric histology and serum levels of gastrin-17 and pepsinogen I: a multicentre study. Eur J Gastroenterol Hepatol. 2003;15:885-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 16. | Kikuchi S, Kurosawa M, Sakiyama T, Tenjin H, Miki K, Wada O, Inaba Y. Long-term effect of Helicobacter pylori infection on serum pepsinogens. Jpn J Cancer Res. 2000;91:471-476. [PubMed] |
| 17. | Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 924] [Cited by in RCA: 797] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 18. | Rochfort S. Metabolomics reviewed: a new “omics” platform technology for systems biology and implications for natural products research. J Nat Prod. 2005;68:1813-1820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 280] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 19. | Schmidt C. Metabolomics takes its place as latest up-and-coming “omic” science. J Natl Cancer Inst. 2004;96:732-734. [PubMed] |
| 20. | Griffin JL, Shockcor JP. Metabolic profiles of cancer cells. Nat Rev Cancer. 2004;4:551-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 565] [Cited by in RCA: 519] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 21. | Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918-4925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 774] [Article Influence: 48.4] [Reference Citation Analysis (1)] |
| 22. | Klein CA. Cancer. The metastasis cascade. Science. 2008;321:1785-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 204] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 23. | Tan C, Chen H, Xia C. Early prediction of lung cancer based on the combination of trace element analysis in urine and an Adaboost algorithm. J Pharm Biomed Anal. 2009;49:746-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Hori S, Nishiumi S, Kobayashi K, Shinohara M, Hatakeyama Y, Kotani Y, Hatano N, Maniwa Y, Nishio W, Bamba T. A metabolomic approach to lung cancer. Lung Cancer. 2011;74:284-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 158] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 25. | Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, Thompson IM, Weiss RH. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics. 2009;8:558-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 206] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 26. | Kind T, Tolstikov V, Fiehn O, Weiss RH. A comprehensive urinary metabolomic approach for identifying kidney cancerr. Anal Biochem. 2007;363:185-195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 336] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 27. | Gu H, Pan Z, Xi B, Asiago V, Musselman B, Raftery D. Principal component directed partial least squares analysis for combining nuclear magnetic resonance and mass spectrometry data in metabolomics: Application to the detection of breast cancer. Analytica Chimica Acta. 2011;686:57-63. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 28. | Monleón D, Morales JM, Gonzalez-Darder J, Talamantes F, Cortés O, Gil-Benso R, López-Ginés C, Cerdá-Nicolás M, Celda B. Benign and atypical meningioma metabolic signatures by high-resolution magic-angle spinning molecular profiling. J Proteome Res. 2008;7:2882-2888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Monleón D, Morales JM, Barrasa A, López JA, Vázquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 2009;22:342-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 146] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 30. | Mal M, Koh PK, Cheah PY, Chan EC. Development and validation of a gas chromatography/mass spectrometry method for the metabolic profiling of human colon tissue. Rapid Commun Mass Spectrom. 2009;23:487-494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, Ni Y, Zhao A, Cai S, Xu LX. Urinary metabonomic study on colorectal cancer. J Proteome Res. 2010;9:1627-1634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 179] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 32. | Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J Proteome Res. 2009;8:4844-4850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 314] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 33. | Meyer LH, Debatin KM. Diversity of human leukemia xenograft mouse models: implications for disease biology. Cancer Res. 2011;71:7141-7144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 34. | Raheem O, Kulidjian AA, Wu C, Jeong YB, Yamaguchi T, Smith KM, Goff D, Leu H, Morris SR, Cacalano NA. A novel patient-derived intra-femoral xenograft model of bone metastatic prostate cancer that recapitulates mixed osteolytic and osteoblastic lesions. J Transl Med. 2011;9:185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 35. | Jia Y, Liu M, Huang W, Wang Z, He Y, Wu J, Ren S, Ju Y, Geng R, Li Z. Recombinant human endostatin endostar inhibits tumor growth and metastasis in a mouse xenograft model of colon cancer. Pathol Oncol Res. 2012;18:315-323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 36. | Hui X, Chen H, Zhang S, Ma X, Wang X, Huang B. Antitumor activities of recombinant human interferon (IFN)-λ1 in vitro and in xenograft models in vivo for colon cancer. Cancer Lett. 2011;311:141-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 37. | Huynh H, Choo SP, Toh HC, Tai WM, Chung AY, Chow PK, Ong R, Soo KC. Comparing the efficacy of sunitinib with sorafenib in xenograft models of human hepatocellular carcinoma: mechanistic explanation. Curr Cancer Drug Targets. 2011;11:944-953. [PubMed] |
| 38. | Liang F, Wang MY, Huang WB, Li AJ. [Effect of sodium cantharidinate on the angiogenesis of nude mice with human gastric cancer]. Zhong Yao Cai. 2011;34:343-346. [PubMed] |
| 39. | Issaq HJ, Fox SD, Chan KC, Veenstra TD. Global proteomics and metabolomics in cancer biomarker discovery. J Sep Sci. 2011;34:3484-3492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 40. | Issaq HJ, Abbott E, Veenstra TD. Utility of separation science in metabolomic studies. J Sep Sci. 2008;31:1936-1947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Wang QZ, Wu CY, Chen T, Chen X, Zhao XM. Integrating metabolomics into a systems biology framework to exploit metabolic complexity: strategies and applications in microorganisms. Appl Microbiol Biotechnol. 2006;70:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 42. | Dunn WB, Bailey NJ, Johnson HE. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 653] [Cited by in RCA: 561] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 43. | Ippolito JE, Xu J, Jain S, Moulder K, Mennerick S, Crowley JR, Townsend RR, Gordon JI. An integrated functional genomics and metabolomics approach for defining poor prognosis in human neuroendocrine cancers. Proc Natl Acad Sci USA. 2005;102:9901-9906. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 84] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 44. | Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, Geisler JP, Miller G, Sellers T, Cliby W. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer. 2005;113:782-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 241] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 45. | Yan SK, Wei BJ, Lin ZY, Yang Y, Zhou ZT, Zhang WD. A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol. 2008;44:477-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 46. | Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 1375] [Article Influence: 59.8] [Reference Citation Analysis (1)] |
| 47. | Lindon JC, Holmes E, Bollard ME, Stanley EG, Nicholson JK. Metabonomics technologies and their applications in physiological monitoring, drug safety assessment and disease diagnosis. Biomarkers. 2004;9:1-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 319] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 48. | Lindon JC, Holmes E, Nicholson JK. Metabonomics: systems biology in pharmaceutical research and development. Curr Opin Mol Ther. 2004;6:265-272. [PubMed] |
| 49. | Clayton TA, Lindon JC, Cloarec O, Antti H, Charuel C, Hanton G, Provost JP, Le Net JL, Baker D, Walley RJ. Pharmaco-metabonomic phenotyping and personalized drug treatment. Nature. 2006;440:1073-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 646] [Cited by in RCA: 603] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 50. | Di Leo A, Claudino W, Colangiuli D, Bessi S, Pestrin M, Biganzoli L. New strategies to identify molecular markers predicting chemotherapy activity and toxicity in breast cancer. Ann Oncol. 2007;18 Suppl 12:xii8-xi14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 51. | Nicholson JK, Lindon JC. Systems biology: Metabonomics. Nature. 2008;455:1054-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1368] [Cited by in RCA: 1382] [Article Influence: 81.3] [Reference Citation Analysis (0)] |
| 52. | Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics--a review in human disease diagnosis. Anal Chim Acta. 2010;659:23-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 374] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 53. | Griffin JL, Kauppinen RA. Tumour metabolomics in animal models of human cancer. J Proteome Res. 2007;6:498-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 79] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 54. | Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431-440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 582] [Cited by in RCA: 539] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 55. | Blekherman G, Laubenbacher R, Cortes DF, Mendes P, Torti FM, Akman S, Torti SV, Shulaev V. Bioinformatics tools for cancer metabolomics. Metabolomics. 2011;7:329-343. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 56. | Barderas MG, Laborde CM, Posada M, de la Cuesta F, Zubiri I, Vivanco F, Alvarez-Llamas G. Metabolomic profiling for identification of novel potential biomarkers in cardiovascular diseases. J Biomed Biotechnol. 2011;2011:790132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 57. | Jain KK. The Handbook of Biomarkers. 1st ed: Springer Science 2010; . |
| 58. | Serkova NJ, Glunde K. Metabolomics of cancer. Methods Mol Biol. 2009;520:273-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 59. | Jordan KW, Cheng LL. NMR-based metabolomics approach to target biomarkers for human prostate cancer. Expert Rev Proteomics. 2007;4:389-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 60. | Chen Y, Zhang R, Song Y, He J, Sun J, Bai J, An Z, Dong L, Zhan Q, Abliz Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: finding potential biomarkers for breast cancer. Analyst. 2009;134:2003-2011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 61. | Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, Raftery D. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70:8309-8318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 225] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 62. | Yang Q, Shi X, Wang Y, Wang W, He H, Lu X, Xu G. Urinary metabonomic study of lung cancer by a fully automatic hyphenated hydrophilic interaction/RPLC-MS system. J Sep Sci. 2010;33:1495-1503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 63. | Johnson HE, Broadhurst D, Goodacre R, Smith AR. Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry. 2003;62:919-928. [PubMed] |
| 64. | Kim DH, Jarvis RM, Xu Y, Oliver AW, Allwood JW, Hampson L, Hampson IN, Goodacre R. Combining metabolic fingerprinting and footprinting to understand the phenotypic response of HPV16 E6 expressing cervical carcinoma cells exposed to the HIV anti-viral drug lopinavir. Analyst. 2010;135:1235-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 65. | Itoh A, Ohashi Y, Soga T, Mori H, Nishioka T, Tomita M. Application of capillary electrophoresis-mass spectrometry to synthetic in vitro glycolysis studies. Electrophoresis. 2004;25:1996-2002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 66. | Soga T. Capillary electrophoresis-mass spectrometry for metabolomics. Methods Mol Biol. 2007;358:129-137. [PubMed] |
| 67. | Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2:488-494. [PubMed] |
| 68. | Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78-95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 821] [Cited by in RCA: 730] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 69. | Lenz EM, Bright J, Wilson ID, Morgan SR, Nash AF. A 1H NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. J Pharm Biomed Anal. 2003;33:1103-1115. [PubMed] |
| 70. | Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA. 1999;96:2907-2912. [PubMed] |
| 71. | Raamsdonk LM, Teusink B, Broadhurst D, Zhang N, Hayes A, Walsh MC, Berden JA, Brindle KM, Kell DB, Rowland JJ. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 791] [Cited by in RCA: 644] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 72. | Musumarra G, Barresi V, Condorelli DF, Scirè S. A bioinformatic approach to the identification of candidate genes for the development of new cancer diagnostics. Biol Chem. 2003;384:321-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 73. | Kubota T, Weisenthal L. Chemotherapy sensitivity and resistance testing: to be “standard” or to be individualized, that is the question. Gastric Cancer. 2006;9:82-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Wagner AD, Unverzagt S, Grothe W, Kleber G, Grothey A, Haerting J, Fleig WE. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010;(3) CD004064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 350] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 75. | Wang X, Yan SK, Dai WX, Liu XR, Zhang WD, Wang JJ. A metabonomic approach to chemosensitivity prediction of cisplatin plus 5-fluorouracil in a human xenograft model of gastric cancer. Int J Cancer. 2010;127:2841-2850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 76. | Sikora K. Personalized cancer therapy--the key to the future. Pharmacogenomics. 2004;5:225-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 77. | Sikora K. Personalized medicine for cancer: from molecular signature to therapeutic choice. Adv Cancer Res. 2007;96:345-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 78. | Park DJ, Lenz HJ. Determinants of chemosensitivity in gastric cancer. Curr Opin Pharmacol. 2006;6:337-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 79. | McLeod HL. Individualized cancer therapy: molecular approaches to the prediction of tumor response. Expert Rev Anticancer Ther. 2002;2:113-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 80. | Garman KS, Nevins JR, Potti A. Genomic strategies for personalized cancer therapy. Hum Mol Genet. 2007;16 Spec No. 2:R226-R232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 81. | Ma Y, Ding Z, Qian Y, Shi X, Castranova V, Harner EJ, Guo L. Predicting cancer drug response by proteomic profiling. Clin Cancer Res. 2006;12:4583-4589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 82. | Fareed KR, Kaye P, Soomro IN, Ilyas M, Martin S, Parsons SL, Madhusudan S. Biomarkers of response to therapy in oesophago-gastric cancer. Gut. 2009;58:127-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 83. | Brereton RG. Chemometrics: data analysis for the laboratory and chemical plant. USA: John Wiley & Sons 2003; . |
| 84. | Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res. 2009;50 Suppl:S237-S242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 648] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 85. | Burke JE, Dennis EA. Phospholipase A2 biochemistry. Cardiovasc Drugs Ther. 2009;23:49-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 320] [Cited by in RCA: 279] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 86. | Kent C. Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim Biophys Acta. 2005;1733:53-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 87. | Lands WE. Stories about acyl chains. Biochim Biophys Acta. 2000;1483:1-14. [PubMed] |
| 88. | Jackson SK, Abate W, Tonks AJ. Lysophospholipid acyltransferases: novel potential regulators of the inflammatory response and target for new drug discovery. Pharmacol Ther. 2008;119:104-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 89. | Takikawa M, Akiyama Y, Maruyama K, Suzuki A, Liu F, Tai S, Ohshita C, Kawaguchi Y, Bandou E, Yonemura Y. Proteomic analysis of a highly metastatic gastric cancer cell line using two-dimensional differential gel electrophoresis. Oncol Rep. 2006;16:705-711. [PubMed] |
| 90. | Chen JL, Tang HQ, Hu JD, Fan J, Hong J, Gu JZ. Metabolomics of gastric cancer metastasis detected by gas chromatography and mass spectrometry. World J Gastroenterol. 2010;16:5874-5880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 48] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 91. | Hu JD, Tang HQ, Zhang Q, Fan J, Hong J, Gu JZ, Chen JL. Prediction of gastric cancer metastasis through urinary metabolomic investigation using GC/MS. World J Gastroenterol. 2011;17:727-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 44] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 92. | Liu X, Wang X, Zhang J, Lam EK, Shin VY, Cheng AS, Yu J, Chan FK, Sung JJ, Jin HC. Warburg effect revisited: an epigenetic link between glycolysis and gastric carcinogenesis. Oncogene. 2010;29:442-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 93. | Chen JL, Fan J, Tang HQ, Hu JD, Gu JZ. Urinary Metabolomic Analysis of Human Gastric Cancer Mouse Models and Patients Using Gas Chromatography/Mass Spectrometry. J Mol Biomark Diagn. 2011;1-8. |
| 94. | Chan DC, Chen CJ, Chu HC, Chang WK, Yu JC, Chen YJ, Wen LL, Huang SC, Ku CH, Liu YC. Evaluation of serum amyloid A as a biomarker for gastric cancer. Ann Surg Oncol. 2007;14:84-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 95. | Wang CS, Wu TL, Tsao KC, Sun CF. Serum TIMP-1 in gastric cancer patients: a potential prognostic biomarker. Ann Clin Lab Sci. 2006;36:23-30. [PubMed] |
| 96. | Kim KB, Yang JY, Kwack SJ, Park KL, Kim HS, Ryu do H, Kim YJ, Hwang GS, Lee BM. Toxicometabolomics of urinary biomarkers for human gastric cancer in a mouse model. J Toxicol Environ Health A. 2010;73:1420-1430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 97. | Yu L, Aa J, Xu J, Sun M, Qian S, Cheng L, Yang S, Shi R. Metabolomic phenotype of gastric cancer and precancerous stages based on gas chromatography time-of-flight mass spectrometry. J Gastroenterol Hepatol. 2011;26:1290-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 98. | Song H, Peng JS, Dong-Sheng Y, Yang ZL, Liu HL, Zeng YK, Shi XP, Lu BY. Serum metabolic profiling of human gastric cancer based on gas chromatography/mass spectrometry. Braz J Med Biol Res. 2012;45:78-85. [PubMed] |
| 99. | Wu H, Xue R, Tang Z, Deng C, Liu T, Zeng H, Sun Y, Shen X. Metabolomic investigation of gastric cancer tissue using gas chromatography/mass spectrometry. Anal Bioanal Chem. 2010;396:1385-1395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 100. | Ikeda A, Nishiumi S, Shinohara M, Yoshie T, Hatano N, Okuno T, Bamba T, Fukusaki E, Takenawa T, Azuma T. Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomed Chromatogr. 2012;26:548-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 101. | Holdstock G, Bruce S. Endoscopy and gastric cancer. Gut. 1981;22:673-676. [RCA] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 102. | Layke JC, Lopez PP. Gastric cancer: diagnosis and treatment options. Am Fam Physician. 2004;69:1133-1140. [PubMed] |
| 103. | Akagi T, Shiraishi N, Kitano S. Lymph Node Metastasis of Gastric Cancer. Cancers. 2011;3:2141-2159. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 104. | Nakamura R, Saikawa Y, Kubota T, Kumagai A, Kiyota T, Ohashi M, Yoshida M, Otani Y, Kumai K, Kitajima M. Role of the MTT chemosensitivity test in the prognosis of gastric cancer patients after postoperative adjuvant chemotherapy. Anticancer Res. 2006;26:1433-1437. [PubMed] |
| 105. | Grivet JP, Delort AM, Portais JC. NMR and microbiology: from physiology to metabolomics. Biochimie. 2003;85:823-840. [PubMed] |
| 106. | Devantier R, Scheithauer B, Villas-Bôas SG, Pedersen S, Olsson L. Metabolite profiling for analysis of yeast stress response during very high gravity ethanol fermentations. Biotechnol Bioeng. 2005;90:703-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 107. | Schauer N, Steinhauser D, Strelkov S, Schomburg D, Allison G, Moritz T, Lundgren K, Roessner-Tunali U, Forbes MG, Willmitzer L. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005;579:1332-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 423] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 108. | Buchholz A, Takors R, Wandrey C. Quantification of intracellular metabolites in Escherichia coli K12 using liquid chromatographic-electrospray ionization tandem mass spectrometric techniques. Anal Biochem. 2001;295:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 165] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 109. | Smedsgaard J, Nielsen J. Metabolite profiling of fungi and yeast: from phenotype to metabolome by MS and informatics. J Exp Bot. 2005;56:273-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 110. | Ellis DI, Broadhurst D, Kell DB, Rowland JJ, Goodacre R. Rapid and quantitative detection of the microbial spoilage of meat by fourier transform infrared spectroscopy and machine learning. Appl Environ Microbiol. 2002;68:2822-2828. [PubMed] |
| 111. | Kaderbhai NN, Broadhurst DI, Ellis DI, Goodacre R, Kell DB. Functional genomics via metabolic footprinting: monitoring metabolite secretion by Escherichia coli tryptophan metabolism mutants using FT-IR and direct injection electrospray mass spectrometry. Comp Funct Genomics. 2003;4:376-391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |